Moderate Phosphorus Addition to Field-Grown Bananas Enhanced Soil Microbial Enzyme Activities but Had Negligible Impacts on Bacterial, Fungal, and Nematode Diversity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Design of Field Experiment
2.2. Plant Performance
2.3. Sample Collection
2.4. Soil Chemical and Biochemical Measurements
2.5. Characterisation of Nematode Communities
2.6. DNA Extraction
2.7. PCR Amplification and Sequencing of 16S and ITS rRNA Genes
2.8. Sequence Data Processing
2.9. Matching Sequences to the Common Core of Banana
2.10. Statistical Analyses
3. Results
3.1. Soil Physicochemical Properties and Plant Performance
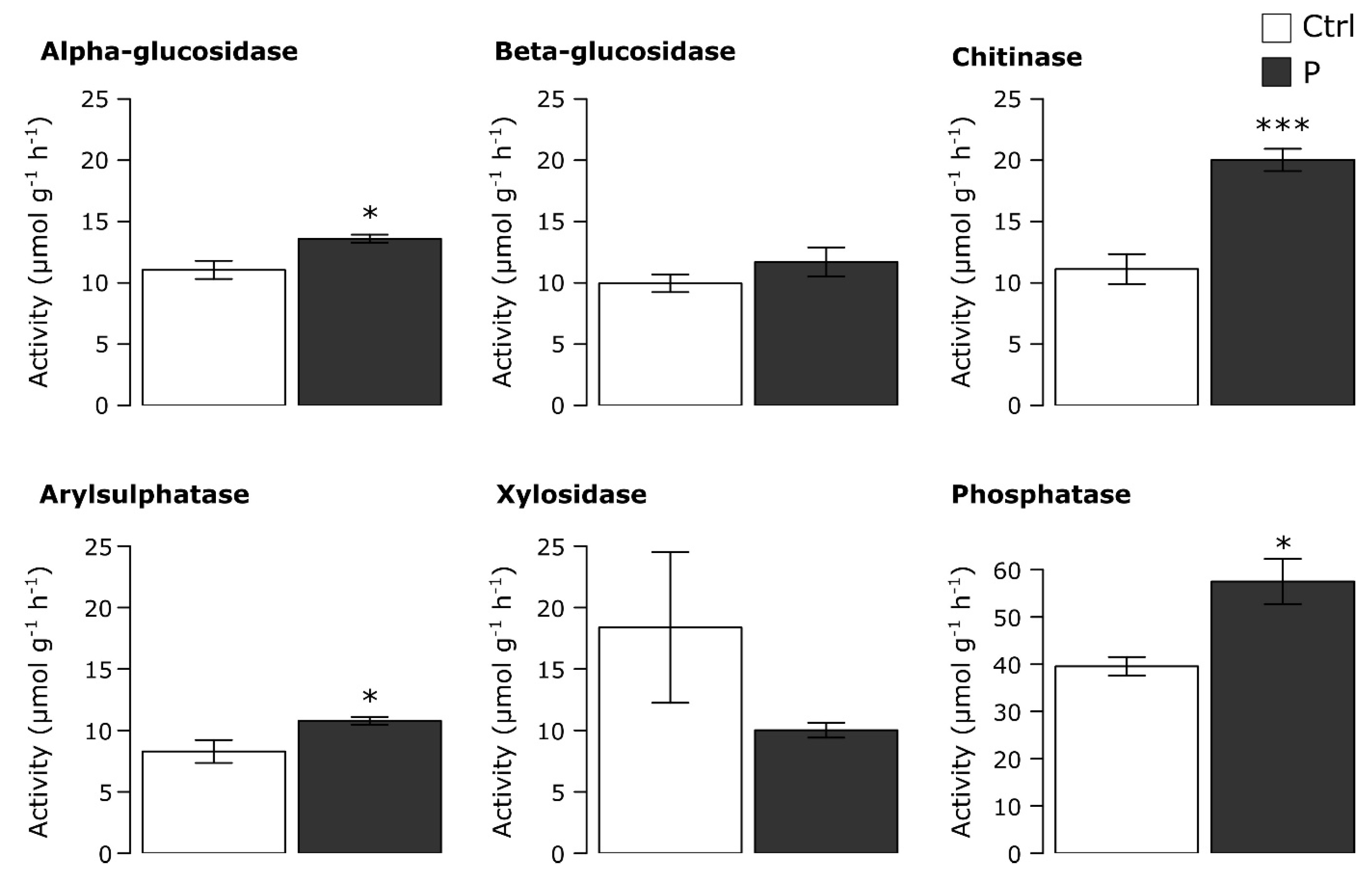
3.2. Potential Soil Microbial Enzyme Activities
3.3. Effects of P Addition on the Diversity of Bacterial and Fungal Communities
3.4. Effects of P Addition on the Relative Abundance of Core Bacteria and Fungi
3.5. Nematode Numbers, Diversity, and Guild Representation
4. Discussion
4.1. P Addition Accelerated Crop Cycling but Did Not Impact Yield or Plant Height
4.2. Impacts of P on the Potential Activities of Soil Microbial Enzymes
4.3. Bacterial and Fungal Diversity Was Not Influenced by P Addition
4.4. The Core Microbiome Is Not Effectively Manipulated by P
4.5. Nematodes Are Not Affected by the Addition of P
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAO. Bananas: Commodity in Focus. Available online: https://www.fao.org/markets-and-trade/commodities/bananas/en/ (accessed on 25 July 2023).
- Aurore, G.; Parfait, B.; Fahrasmane, L. Bananas, raw materials for making processed food products. Trends Food Sci. Tech. 2009, 20, 78–91. [Google Scholar] [CrossRef]
- Lahav, E. Banana nutrition. In Bananas and Plantains; Gowen, S., Ed.; Springer: Dordrecht, The Netherlands, 1995; pp. 258–316. ISBN 978-94-011-0737-2. [Google Scholar]
- Department of Environment and Science. Prescribed Methodology for Banana Cultivation; Reef Water Quality Program; Queensland Government: Brisbane, QLD, Australia, 2022.
- Queensland Government. Requirements for Banana Growers. Reef Protection Regulations. Available online: https://www.qld.gov.au/environment/agriculture/sustainable-farming/reef/reef-regulations/bananas (accessed on 7 March 2024).
- Dai, Z.; Liu, G.; Chen, H.; Chen, C.; Wang, J.; Ai, S.; Wei, D.; Li, D.; Ma, B.; Tang, C.; et al. Long-term nutrient inputs shift soil microbial functional profiles of phosphorus cycling in diverse agroecosystems. ISME J. 2020, 14, 757–770. [Google Scholar] [CrossRef]
- McKnight, M.M.; Qu, Z.; Copeland, J.K.; Guttman, D.S.; Walker, V.K. A practical assessment of nano-phosphate on soybean (Glycine max) growth and microbiome establishment. Sci. Rep. 2020, 10, 9151. [Google Scholar] [CrossRef] [PubMed]
- Pantigoso, H.A.; Manter, D.K.; Vivanco, J.M. Phosphorus addition shifts the microbial community in the rhizosphere of blueberry (Vaccinium corymbosum L.). Rhizosphere 2018, 7, 1–7. [Google Scholar] [CrossRef]
- Xiao, D.; Che, R.; Liu, X.; Tan, Y.; Yang, R.; Zhang, W.; He, X.; Xu, Z.; Wang, K. Arbuscular mycorrhizal fungi abundance was sensitive to nitrogen addition but diversity was sensitive to phosphorus addition in karst ecosystems. Biol. Fertil. Soils 2019, 55, 457–469. [Google Scholar] [CrossRef]
- Declerck, S.; Plenchette, C.; Strullu, D.G. Mycorrhizal dependency of banana (Musa acuminata, AAA group) cultivar. Plant Soil 1995, 176, 183–187. [Google Scholar] [CrossRef]
- Olatunji, O.A.; Gong, S.; Tariq, A.; Pan, K.; Sun, X.; Chen, W.; Zhang, L.; Dakhil, M.A.; Huang, D.; Tan, X. The effect of phosphorus addition, soil moisture, and plant type on soil nematode abundance and community composition. J. Soils Sediments 2019, 19, 1139–1150. [Google Scholar] [CrossRef]
- Jiang, N.; Wei, K.; Pu, J.; Zhang, Y.; Xie, H.; Bao, H.; Chen, L. Effects of the reduction in chemical fertilizers on soil phosphatases encoding genes (phoD and phoX) under crop residue mulching. Appl. Soil Ecol. 2022, 175, 104428. [Google Scholar] [CrossRef]
- Neal, A.; McLaren, T.; Lourenço Campolino, M.; Hughes, D.; Marcos Coelho, A.; Gomes de Paula Lana, U.; Aparecida Gomes, E.; Morais de Sousa, S. Crop type exerts greater influence upon rhizosphere phosphohydrolase gene abundance and phylogenetic diversity than phosphorus fertilization. FEMS Microbiol. Ecol. 2021, 97, fiab033. [Google Scholar] [CrossRef]
- Wang, L.; Wen, Y.; Tong, R.; Zhang, H.; Chen, H.; Hu, T.; Liu, G.; Wang, J.; Zhu, L.; Wu, T. Understanding Responses of Soil Microbiome to the Nitrogen and Phosphorus Addition in Metasequoia glyptostroboides Plantations of Different Ages. Microb. Ecol. 2022, 84, 565–579. [Google Scholar] [CrossRef]
- Fan, P.; Lai, C.; Yang, J.; Hong, S.; Yang, Y.; Wang, Q.; Wang, B.; Zhang, R.; Jia, Z.; Zhao, Y.; et al. Crop rotation suppresses soil-borne Fusarium wilt of banana and alters microbial communities. Arch. Agron. Soil Sci. 2022, 68, 447–459. [Google Scholar] [CrossRef]
- Shen, Z.; Ruan, Y.; Xue, C.; Zhong, S.; Li, R.; Shen, Q. Soils naturally suppressive to banana Fusarium wilt disease harbor unique bacterial communities. Plant Soil 2015, 393, 21–33. [Google Scholar] [CrossRef]
- Birt, H.W.G.; Pattison, A.B.; Skarshewski, A.; Daniells, J.; Raghavendra, A.; Dennis, P.G. The core bacterial microbiome of banana (Musa spp.). Environ. Microbiome 2022, 17, 46. [Google Scholar] [CrossRef] [PubMed]
- Birt, H.W.G.; Pattison, A.B.; Skarshewski, A.; Daniells, J.; Raghavendra, A.; Dennis, P.G. The core fungal microbiome of banana (Musa spp.). Front. Microbiol. 2023, 14, 1127779. [Google Scholar] [CrossRef] [PubMed]
- Cannon, M.G.; Smith, C.D.; Murtha, G.G. Soils of the Cardwell-Tully Area, North Queensland; Division of Soils Divisional Report; CSIRO: Canberra, ACT, Australia, 1992; ISBN 0-643-05081-7. [Google Scholar]
- Armour, J.D.; Nelson, P.N.; Daniells, J.W.; Rasiah, V.; Inman-Bamber, N.G. Nitrogen leaching from the root zone of sugarcane and bananas in the humid tropics of Australia. Agric. Ecosyst. Environ. 2013, 180, 68–78. [Google Scholar] [CrossRef]
- Kernot, I.; Lindsay, S. Tropical Banana Information Kit; Agrilink: Your growing guide to better farming series; Queensland Department of Primary Industries: Brisbane, QLD, Australia, 1998; ISBN 978-0-7242-6725-5.
- Armour, J. Nutrient Management Plan for the Banana Industry (of North Queensland); Australian Banana Growers’ Council: Rocklea, QLD, Australia, 2018. [Google Scholar]
- Rayment, G.E.; Lyons, D.J. Soil Chemical Methods—Australasia; CSIRO Publishing: Melbourne, VIC, Australia, 2011; ISBN 0-643-06768-X. [Google Scholar]
- Moody, P.; Cong, P. Soil Constraints and Management Package (SCAMP): Guidelines for Management of Tropical Upland Soils; Australian Centre for International Agricultural Research: Canberra, ACT, Australia, 2008.
- Edwards, T. What is Soil Organic Carbon? Available online: https://www.agric.wa.gov.au/measuring-and-assessing-soils/what-soil-organic-carbon (accessed on 23 February 2024).
- Lapis-Gaza, H.R.; Pattison, A.B. Functional Soil Biological Measurements to Support Healthy Soils. In The Plant Microbiome: Methods and Protocols; Carvalhais, L.C., Dennis, P.G., Eds.; Springer: New York, NY, USA, 2021; pp. 265–281. ISBN 978-1-07-161040-4. [Google Scholar]
- Zarcinas, B.A.; Cartwright, B.; Spouncer, L.R. Nitric acid digestion and multi-element analysis of plant material by inductively coupled plasma spectrometry. Commun. Soil Sci. Plant Anal. 1987, 18, 131–146. [Google Scholar] [CrossRef]
- Whiteheads, A.G.; Hemming, J.R. A comparison of some quantitative methods of extracting small vermiform nematodes from soil. Ann. Appl. Biol. 1965, 55, 25–38. [Google Scholar] [CrossRef]
- Goodey, T.; Goodey, J.B. Soil and Freshwater Nematodes, 2nd ed.; Butler & Tanner Ltd.: London, UK, 1963. [Google Scholar]
- Yeates, G.W.; Bongers, T.; de Goede, R.G.M.; Freckman, D.W.; Georgieva, S.S. Feeding habits in soil nematode families and genera—An outline for soil ecologists. J. Nematol. 1993, 25, 315–331. [Google Scholar] [PubMed]
- Chan, C.M.; Lyons, R.; Dennis, P.G.; Lant, P.; Pratt, S.; Laycock, B. Effect of Toxic Phthalate-Based Plasticizer on the Biodegradability of Polyhydroxyalkanoate. Environ. Sci. Technol. 2022, 56, 17732–17742. [Google Scholar] [CrossRef]
- Deng, H.; Zhang, B.; Yin, R.; Wang, H.; Mitchell, S.M.; Griffiths, B.S.; Daniell, T.J. Long-term effect of re-vegetation on the microbial community of a severely eroded soil in sub-tropical China. Plant Soil 2010, 328, 447–458. [Google Scholar] [CrossRef]
- Pennanen, T.; Caul, S.; Daniell, T.J.; Griffiths, B.S.; Ritz, K.; Wheatley, R.E. Community-level responses of metabolically-active soil microorganisms to the quantity and quality of substrate inputs. Soil Biol. Biochem. 2004, 36, 841–848. [Google Scholar] [CrossRef]
- Redford, A.J.; Bowers, R.M.; Knight, R.; Linhart, Y.; Fierer, N. ecology of the phyllosphere: Geographic and phylogenetic variability in the distribution of bacteria on tree leaves. Environ. Microbiol. 2010, 12, 2885–2893. [Google Scholar] [CrossRef]
- Toju, H.; Tanabe, A.S.; Yamamoto, S.; Sato, H. High-coverage ITS primers for the DNA-based identification of ascomycetes and basidiomycetes in environmental samples. PLoS ONE 2012, 7, e40863. [Google Scholar] [CrossRef] [PubMed]
- White, T.; Bruns, T.; Lee, S.; Taylor, J.; Innis, M.; Gelfand, D.; Sninsky, J. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In Pcr Protocols: A Guide to Methods and Applications; Academic Press: Cambridge, MA, USA, 1990; Volume 31, pp. 315–322. [Google Scholar]
- Rohland, N.; Reich, D. Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture. Genome Res. 2012, 22, 939–946. [Google Scholar] [CrossRef] [PubMed]
- Ihrmark, K.; Bödeker, I.T.M.; Cruz-Martinez, K.; Friberg, H.; Kubartova, A.; Schenck, J.; Strid, Y.; Stenlid, J.; Brandström-Durling, M.; Clemmensen, K.E.; et al. New primers to amplify the fungal ITS2 region—Evaluation by 454-sequencing of artificial and natural communities. FEMS Microbiol. Ecol. 2012, 82, 666–677. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Forstner, C.; Orton, T.G.; Skarshewski, A.; Wang, P.; Kopittke, P.M.; Dennis, P.G. Effects of graphene oxide and graphite on soil bacterial and fungal diversity. Sci. Total Environ. 2019, 671, 140–148. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed]
- Bengtsson-Palme, J.; Ryberg, M.; Hartmann, M.; Branco, S.; Wang, Z.; Godhe, A.; De Wit, P.; Sánchez-García, M.; Ebersberger, I.; de Sousa, F.; et al. Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods Ecol. Evol. 2013, 4, 914–919. [Google Scholar] [CrossRef]
- Abarenkov, K.; Nilsson, R.H.; Larsson, K.-H.; Taylor, A.F.S.; May, T.W.; Frøslev, T.G.; Pawlowska, J.; Lindahl, B.; Põldmaa, K.; Truong, C.; et al. The UNITE database for molecular identification and taxonomic communication of fungi and other eukaryotes: Sequences, taxa and classifications reconsidered. Nucleic Acids Res. 2024, 52, D791–D797. [Google Scholar] [CrossRef]
- Zhang, Z.; Schwartz, S.; Wagner, L.; Miller, W. A Greedy Algorithm for Aligning DNA Sequences. J. Comput. Biol. 2000, 7, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- McDonald, D.; Clemente, J.C.; Kuczynski, J.; Rideout, J.R.; Stombaugh, J.; Wendel, D.; Wilke, A.; Huse, S.; Hufnagle, J.; Meyer, F. The Biological Observation Matrix (BIOM) format or: How I learned to stop worrying and love the ome-ome. Gigascience 2012, 1, 1–6. [Google Scholar] [CrossRef] [PubMed]
- McDonald, D.; Jiang, Y.; Balaban, M.; Cantrell, K.; Zhu, Q.; Gonzalez, A.; Morton, J.T.; Nicolaou, G.; Parks, D.H.; Karst, S.M.; et al. Greengenes2 unifies microbial data in a single reference tree. Nat. Biotechnol. 2023, 42, 715–718. [Google Scholar] [CrossRef]
- Chaumeil, P.-A.; Mussig, A.J.; Hugenholtz, P.; Parks, D.H. GTDB-Tk: A toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 2020, 36, 1925–1927. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.; Knight, R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- Shannon, C.E. A Mathematical Theory of Communication. Bell Syst. Tech. J. 1948, 27, 379–423. [Google Scholar] [CrossRef]
- Chao, A. Nonparametric Estimation of the Number of Classes in a Population. Scand. J. Stat. 1984, 11, 265–270. [Google Scholar]
- Faith, D.P. Conservation evaluation and phylogenetic diversity. Biol. Conserv. 1992, 61, 1–10. [Google Scholar] [CrossRef]
- Anderson, M.J. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001, 26, 32–46. [Google Scholar] [CrossRef]
- Oksanen, J.; Simpson, G.L.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Solymos, P.; Stevens, M.H.H.; Szoecs, E.; et al. Vegan: Community Ecology Package. 2022. Available online: https://cran.r-project.org/web/packages/vegan/index.html (accessed on 28 August 2024).
- Legendre, P.; Gallagher, E.D. Ecologically meaningful transformations for ordination of species data. Oecologia 2001, 129, 271–280. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Stastistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Wang, C.; Mori, T.; Mao, Q.; Zhou, K.; Wang, Z.; Zhang, Y.; Mo, H.; Lu, X.; Mo, J. Long-term phosphorus addition downregulates microbial investments on enzyme productions in a mature tropical forest. J. Soils Sediments 2020, 20, 921–930. [Google Scholar] [CrossRef]
- Zheng, M.; Huang, J.; Chen, H.; Wang, H.; Mo, J. Responses of soil acid phosphatase and beta-glucosidase to nitrogen and phosphorus addition in two subtropical forests in southern China. Eur. J. Soil Biol. 2015, 68, 77–84. [Google Scholar] [CrossRef]
- Luo, R.; Fan, J.; Wang, W.; Luo, J.; Kuzyakov, Y.; He, J.-S.; Chu, H.; Ding, W. Nitrogen and phosphorus enrichment accelerates soil organic carbon loss in alpine grassland on the Qinghai-Tibetan Plateau. Sci. Total. Environ. 2019, 650, 303–312. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Gan, B.; Li, Q.; Xiao, W.; Song, X. Effects of Nitrogen and Phosphorus Addition on Soil Extracellular Enzyme Activity and Stoichiometry in Chinese Fir (Cunninghamia lanceolata) Forests. Front. Plant Sci. 2022, 13, 834184. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhou, K.; Mori, T.; Mo, J.; Zhang, W. Effects of phosphorus and nitrogen fertilization on soil arylsulfatase activity and sulfur availability of two tropical plantations in southern China. Forest Ecol. Manag. 2019, 453, 117613. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, C.; Li, Y. Contrasting effects of nitrogen and phosphorus additions on soil nitrous oxide fluxes and enzyme activities in an alpine wetland of the Tibetan Plateau. PLoS ONE 2019, 14, e0216244. [Google Scholar] [CrossRef]
- Burke, C.; Steinberg, P.; Rusch, D.; Kjelleberg, S.; Thomas, T. Bacterial community assembly based on functional genes rather than species. Proc. Natl. Acad. Sci. USA 2011, 108, 14288–14293. [Google Scholar] [CrossRef] [PubMed]
- Frossard, A.; Gerull, L.; Mutz, M.; Gessner, M.O. Disconnect of microbial structure and function: Enzyme activities and bacterial communities in nascent stream corridors. ISME J. 2012, 6, 680–691. [Google Scholar] [CrossRef] [PubMed]
- LeBrun, E.S.; King, R.S.; Back, J.A.; Kang, S. Microbial Community Structure and Function Decoupling Across a Phosphorus Gradient in Streams. Microb. Ecol. 2018, 75, 64–73. [Google Scholar] [CrossRef] [PubMed]
- Szejgis, J.; Carrillo, Y.; Jeffries, T.C.; Dijkstra, F.A.; Chieppa, J.; Horn, S.; Bristol, D.; Maisnam, P.; Eldridge, D.; Nielsen, U.N. Altered rainfall greatly affects enzyme activity but has limited effect on microbial biomass in Australian dryland soils. Soil Biol. Biochem. 2024, 189, 109277. [Google Scholar] [CrossRef]
- Cabanás, C.G.-L.; Fernández-González, A.J.; Cardoni, M.; Valverde-Corredor, A.; López-Cepero, J.; Fernández-López, M.; Mercado-Blanco, J. The banana root endophytome: Differences between mother plants and suckers and evaluation of selected bacteria to control fusarium oxysporum f.sp. cubense. J. Fungi 2021, 7, 194. [Google Scholar] [CrossRef]
- Liu, Y.; Shi, G.; Mao, L.; Cheng, G.; Jiang, S.; Ma, X.; An, L.; Du, G.; Johnson, N.C.; Feng, H. Direct and indirect influences of 8 yr of nitrogen and phosphorus fertilization on Glomeromycota in an alpine meadow ecosystem. New Phytol. 2012, 194, 523–535. [Google Scholar] [CrossRef] [PubMed]
- Krüger, M.; Stockinger, H.; Krüger, C.; Schüßler, A. DNA-based species level detection of Glomeromycota: One PCR primer set for all arbuscular mycorrhizal fungi. New Phytol. 2009, 183, 212–223. [Google Scholar] [CrossRef] [PubMed]
- Nel, B.; Steinberg, C.; Labuschagne, N.; Viljoen, A. Isolation and characterization of nonpathogenic Fusarium oxysporum isolates from the rhizosphere of healthy banana plants. Plant Pathol. 2006, 55, 207–216. [Google Scholar] [CrossRef]
- Forsyth, L.M.; Smith, L.J.; Aitken, E.A.B. Identification and characterization of non-pathogenic Fusarium oxysporum capable of increasing and decreasing Fusarium wilt severity. Mycol. Res. 2006, 110, 929–935. [Google Scholar] [CrossRef]
- Nel, B.; Steinberg, C.; Labuschagne, N.; Viljoen, A. The potential of nonpathogenic Fusarium oxysporum and other biological control organisms for suppressing fusarium wilt of banana. Plant Pathol. 2006, 55, 217–223. [Google Scholar] [CrossRef]
- Belgrove, A.; Steinberg, C.; Viljoen, A. Evaluation of Nonpathogenic Fusarium oxysporum and Pseudomonas fluorescens for Panama Disease Control. Plant Dis. 2011, 95, 951–959. [Google Scholar] [CrossRef]
- Sun, F.; Tariq, A.; Chen, H.; He, Q.; Guan, Y.; Pan, K.; Chen, S.; Li, J.; Zhao, C.; Wang, H.; et al. Effect of nitrogen and phosphorus application on agricultural soil food webs. Arch. Agron. Soil. Sci. 2017, 63, 1176–1186. [Google Scholar] [CrossRef]
- Qi, Y.; Sun, X.; Peng, S.; Tan, X.; Zhou, S. Effects of fertilization on soil nematode communities in an alpine meadow of Qinghai-Tibet plateau. Front. Ecol. Evol. 2023, 11, 1122505. [Google Scholar] [CrossRef]
- Todd, T.C. Effects of management practices on nematode community structure in tallgrass prairie. Appl. Soil. Ecol. 1996, 3, 235–246. [Google Scholar] [CrossRef]
- Zhong, S.; He, Y.; Zeng, H.; Mo, Y.; Jin, Z. Effects of banana wilt disease on soil nematode community structure and diversity. Afr. J. Biotechnol. 2011, 10, 12759–12767. [Google Scholar] [CrossRef]
- Nielsen, U.N.; Prior, S.; Delroy, B.; Walker, J.K.M.; Ellsworth, D.S.; Powell, J.R. Response of belowground communities to short-term phosphorus addition in a phosphorus-limited woodland. Plant Soil 2015, 391, 321–331. [Google Scholar] [CrossRef]
- Ferris, H. Nematodes and the soil food web: Understanding healthy soils. In Proceedings of the California Conference on Biological Control IV, Berkeley, CA, USA, 13–15 July 2004. [Google Scholar]
- Orr, R.; Northfield, T.D.; Pattison, A.; Nelson, P.N. Soil physicochemical characteristics and leaf nutrient contents on banana farms of North Queensland, Australia. Crop Pasture Sci. 2023, 74, 483–493. [Google Scholar] [CrossRef]





| Soil Property | Ctrl | P | |
|---|---|---|---|
| pH | 5.54 ± 0.24 | 5.5 ± 0.31 | |
| Colwell P (mg/kg) | 19.2 ± 1.92 | 56.4 ± 22.28 | ** |
| Phosphorus buffering index | 218 ± 8.37 | 202 ± 14.83 | |
| Total C (%) | 1.70 ± 0.12 | 1.68 ± 0.19 | |
| Labile C (mg/kg) | 0.3 ± 0.07 | 0.3 ± 0.06 | |
| Nitrate N (mg/kg) | 3.08 ± 1.30 | 3.80 ± 1.61 | |
| Organic matter (%) | 2.16 ± 0.18 | 2.4 ± 0.21 | |
| Organic carbon (%) | 1.25 ± 0.10 | 1.40 ± 0.10 | |
| Electrical conductivity (ds/m) | 0.14 ± 0.01 | 0.14 ± 0.02 |
| Plant Crop | Ratoon 1 | |||||||
|---|---|---|---|---|---|---|---|---|
| Agronomic Variable | Ctrl | P | p Value | Ctrl | P | p Value | ||
| Vegetative | ||||||||
| Bunch emergence time (d) | ||||||||
| 50% | 261 | 240 | <0.001 | *** | 524 | 484 | <0.001 | *** |
| 85% | 302 | 276 | <0.001 | *** | 580 | 548 | <0.001 | *** |
| Plant height (cm) | 245 ± 5.7 | 231 ± 3.5 | 0.129 | 329 ± 3.2 | 326 ± 3.3 | 0.634 | ||
| Foliar P (mg g−1) | 0.24 ± 0.035 | 0.25 ± 0.054 | 0.675 | 0.28 ± 0.078 | 0.24 ± 0.72 | 0.251 | ||
| Fruit | ||||||||
| Harvested bunches (d) | ||||||||
| 50% | 339 | 326 | <0.001 | *** | 641 | 610 | <0.001 | *** |
| 85% | 366 | 354 | <0.001 | *** | 718 | 680 | <0.001 | *** |
| Bunch weight (kg) | 23.2 ± 0.71 | 20.1 ± 0.63 | 0.041 | * | 32.0 ± 0.88 | 30.8 ± 0.78 | 0.391 | |
| Average Yield (kg plant−1 yr−1) | 21.5 ± 0.47 | 19.9 ± 0.47 | 0.078 | 28.8 ± 0.58 | 27.8 ± 0.62 | 0.372 | ||
| Target | Response Variable | Predictor Variable | F Value | p Value | |
|---|---|---|---|---|---|
| Bacteria | Shannon’s Diversity Index | Compartment | 11.63 | 0.004 | ** |
| Treatment | 0.02 | 0.900 | |||
| Compartment: Treatment | 0.10 | 0.751 | |||
| Fungi | Shannon’s Diversity Index | Compartment | 22.43 | <0.001 | *** |
| Treatment | 0.10 | 0.761 | |||
| Compartment: Treatment | 0.08 | 0.785 | |||
| Nematodes | Shannon’s Diversity Index | Treatment | 2.33 | 0.165 |
| Target | Response Variable | Predictor Variable | F Value | R2 Value | p Value | |
|---|---|---|---|---|---|---|
| Bacteria | OTU relative abundances | Compartment | 14.90 | 46.0 | <0.001 | *** |
| (Hellinger transformed) | Treatment | 0.76 | 2.3 | 0.520 | ||
| Compartment: Treatment | 0.71 | 2.2 | 0.559 | |||
| Weighted UniFrac distances | Compartment | 25.13 | 58.8 | <0.001 | *** | |
| Treatment | 0.47 | 1.1 | 0.655 | |||
| Compartment: Treatment | 1.11 | 2.6 | 0.284 | |||
| Fungi | OTU relative abundances | Compartment | 4.65 | 20.9 | <0.001 | *** |
| (Hellinger transformed) | Treatment | 0.99 | 4.4 | 0.387 | ||
| Compartment: Treatment | 0.66 | 2.9 | 0.941 | |||
| Nematodes | Taxon frequencies | Treatment | 1.67 | 17.3 | 0.233 | |
| Response Variable | F Value | p Value | |
|---|---|---|---|
| Total nematodes | 1.51 | 0.254 | |
| Feeding groups | |||
| Plant parasitic nematodes | 1.7 | 0.231 | |
| Fungivorous nematodes | 2.29 | 0.169 | |
| Bactivorous nematodes | 0.01 | 0.925 | |
| Predatory nematodes | 1.48 | 0.259 | |
| Omnivorous nematodes | 3.34 | 0.105 | |
| Guild | |||
| Fu2 | 2.29 | 0.169 | |
| Ba1 | 0.00 | 0.955 | |
| Ba2 | 0.03 | 0.870 | |
| Ba3 | 1.98 | 0.197 | |
| Ca3 | 0.00 | 1.000 | |
| Ca4 | 15.36 | 0.004 | ** |
| Om4 | 3.34 | 0.105 | |
| pp2 ectoparasites | 1.70 | 0.228 | |
| pp3 endoparasites | 1.00 | 0.347 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Clarke, A.-B.C.; Lapis-Gaza, H.R.; Irvine-Brown, S.; Lyons, R.; Sun, J.; Pattison, A.B.; Dennis, P.G. Moderate Phosphorus Addition to Field-Grown Bananas Enhanced Soil Microbial Enzyme Activities but Had Negligible Impacts on Bacterial, Fungal, and Nematode Diversity. Appl. Microbiol. 2024, 4, 1582-1599. https://doi.org/10.3390/applmicrobiol4040108
Clarke A-BC, Lapis-Gaza HR, Irvine-Brown S, Lyons R, Sun J, Pattison AB, Dennis PG. Moderate Phosphorus Addition to Field-Grown Bananas Enhanced Soil Microbial Enzyme Activities but Had Negligible Impacts on Bacterial, Fungal, and Nematode Diversity. Applied Microbiology. 2024; 4(4):1582-1599. https://doi.org/10.3390/applmicrobiol4040108
Chicago/Turabian StyleClarke, Anna-Belle C., Hazel R. Lapis-Gaza, Stuart Irvine-Brown, Rebecca Lyons, Jiarui Sun, Anthony B. Pattison, and Paul G. Dennis. 2024. "Moderate Phosphorus Addition to Field-Grown Bananas Enhanced Soil Microbial Enzyme Activities but Had Negligible Impacts on Bacterial, Fungal, and Nematode Diversity" Applied Microbiology 4, no. 4: 1582-1599. https://doi.org/10.3390/applmicrobiol4040108
APA StyleClarke, A.-B. C., Lapis-Gaza, H. R., Irvine-Brown, S., Lyons, R., Sun, J., Pattison, A. B., & Dennis, P. G. (2024). Moderate Phosphorus Addition to Field-Grown Bananas Enhanced Soil Microbial Enzyme Activities but Had Negligible Impacts on Bacterial, Fungal, and Nematode Diversity. Applied Microbiology, 4(4), 1582-1599. https://doi.org/10.3390/applmicrobiol4040108






