Phenotypic and Genotypic Characterization of Antimicrobial Resistance in Salmonella enterica Serovars from Colombian Pig Farms
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Selection of Pig Farms
2.2. Sample Types
2.3. Isolation and Identification of Salmonella spp.
2.3.1. Water
2.3.2. Rectal Swabs
2.3.3. Serotype Determination
2.4. Antimicrobial Susceptibility Testing
2.5. Analysis of Whole Genome Sequencing (WGS) of Salmonella enterica Isolates
2.6. Statistical Analyses
3. Results
3.1. Prevalence of Salmonella spp.
3.2. Serotypes Identified
3.2.1. Serotyping According to the Kauffmann–White–Le Minor Scheme
3.2.2. Serotyping According to Whole Genome Sequencing
3.2.3. Comparison of Salmonella Serotypes Found in Rectal Swabs and Water Samples at the Farm Level
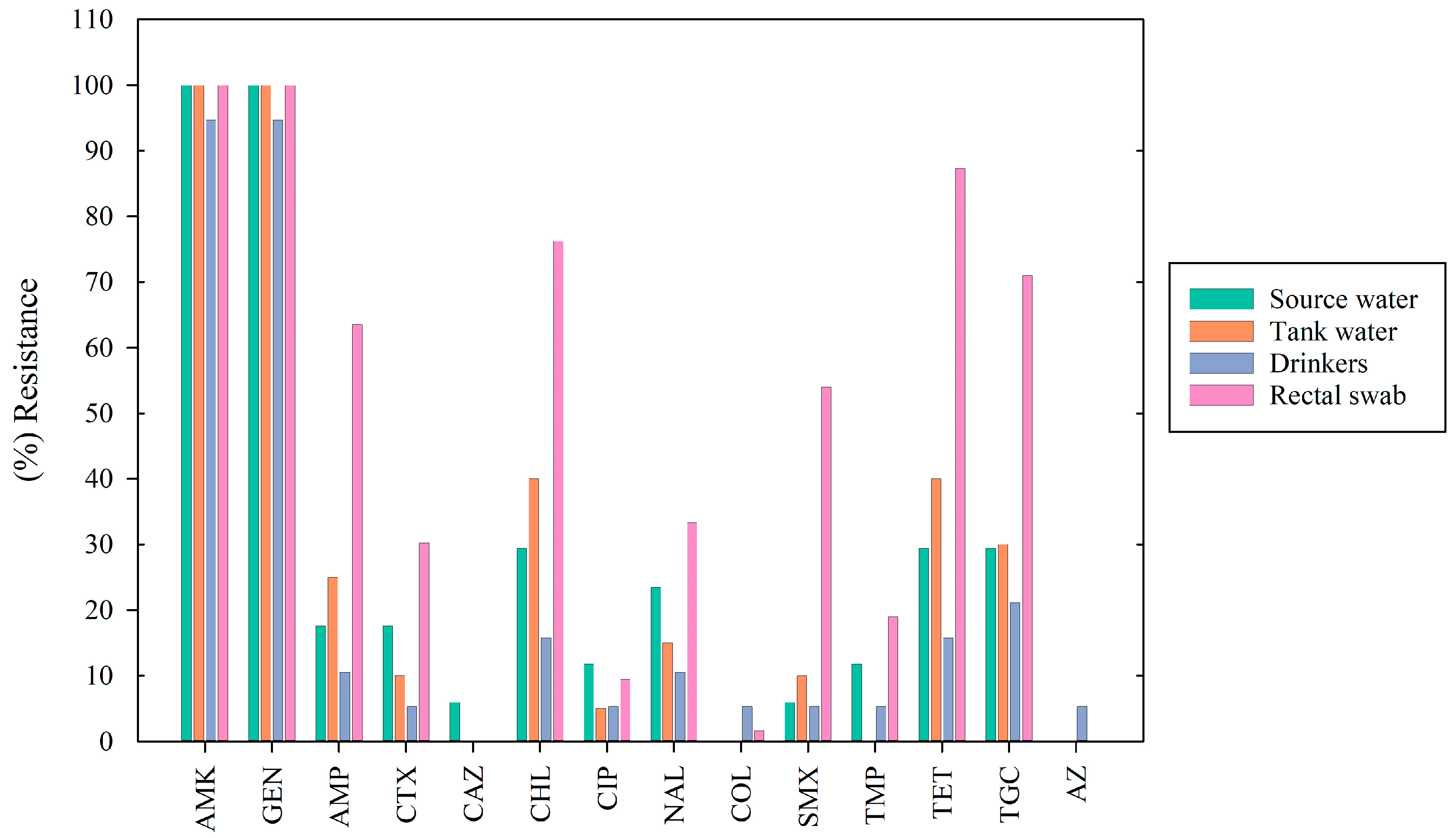
3.3. Antimicrobial Susceptibility Testing
3.4. Resistance Genes Identified from WGS
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, C.-M.; Xu, L.-M.; Mou, X.; Xu, H.; Liu, J.; Miao, Y.-H.; Wang, X.C.; Li, X. Characterization and Evolution of Antibiotic Resistance of Salmonella in Municipal Wastewater Treatment Plants. J. Environ. Manag. 2019, 251, 109547. [Google Scholar] [CrossRef] [PubMed]
- Vico, J.P.; Lorenzutti, A.M.; Zogbi, A.P.; Aleu, G.; Sánchez, I.C.; Caffer, M.I.; Rosmini, M.R.; Mainar-Jaime, R.C. Prevalence, Associated Risk Factors, and Antimicrobial Resistance Profiles of Non-Typhoidal Salmonella in Large Scale Swine Production in Córdoba, Argentina. Res. Vet. Sci. 2020, 130, 161–169. [Google Scholar] [CrossRef]
- Stevens, M.P.; Humphrey, T.J.; Maskell, D.J. Molecular Insights into Farm Animal and Zoonotic Salmonella Infections. Phil. Trans. R. Soc. B 2009, 364, 2709–2723. [Google Scholar] [CrossRef]
- Wiedemann, A.; Virlogeux-Payant, I.; Chaussé, A.-M.; Schikora, A.; Velge, P. Interactions of Salmonella with Animals and Plants. Front. Microbiol. 2015, 5, 791. [Google Scholar] [CrossRef]
- OMSA. Capítulo 3.10.3. Salmonelosis. OMSA—Organización Mundial de Sanidad Animal. Available online: https://www.woah.org/fileadmin/Home/esp/Health_standards/tahm/3.10.03_SALMONELLOSIS.pdf (accessed on 2 August 2024).
- Bonardi, S. Salmonella in the Pork Production Chain and Its Impact on Human Health in the European Union. Epidemiol. Infect. 2017, 145, 1513–1526. [Google Scholar] [CrossRef] [PubMed]
- Evangelopoulou, G.; Kritas, S.; Christodoulopoulos, G.; Burriel, A.R. The Commercial Impact of Pig Salmonella spp. Infections in Border-Free Markets during an Economic Recession. Vet. World 2015, 8, 257–272. [Google Scholar] [CrossRef] [PubMed]
- Andres, V.M.; Davies, R.H. Biosecurity Measures to Control Salmonella and Other Infectious Agents in Pig Farms: A Review. Compr. Rev. Food Sci. Food Saf. 2015, 14, 317–335. [Google Scholar] [CrossRef]
- Baer, A.A.; Miller, M.J.; Dilger, A.C. Pathogens of Interest to the Pork Industry: A Review of Research on Interventions to Assure Food Safety. Compr. Rev. Food Sci. Food Saf. 2013, 12, 183–217. [Google Scholar] [CrossRef]
- Dickson, J.S. Salmonella in the Pork Production Chain—Hogs, Pigs, and Pork. Available online: https://swine.extension.org/salmonella-in-the-pork-production-chain/ (accessed on 2 August 2024).
- Giraldo-Cardona, J.P.; Gualdrón-Ramírez, D.; Chamorro-Tobar, I.; Pulido-Villamarín, A.; Santamaría-Durán, N.; Castañeda-Salazar, R.; Zambrano-Moreno, C.; Carrascal-Camacho, A.K. Salmonella spp. Prevalence, Antimicrobial Resistance and Risk Factor Determination in Colombian Swine Farms. Pesq. Vet. Bras. 2019, 39, 816–822. [Google Scholar] [CrossRef]
- De Busser, E.V.; De Zutter, L.; Dewulf, J.; Houf, K.; Maes, D. Salmonella Control in Live Pigs and at Slaughter. Vet. J. 2013, 196, 20–27. [Google Scholar] [CrossRef]
- Casanova-Higes, A.; Marín-Alcalá, C.M.; Andrés-Barranco, S.; Cebollada-Solanas, A.; Alvarez, J.; Mainar-Jaime, R.C. Weaned Piglets: Another Factor to Be Considered for the Control of Salmonella Infection in Breeding Pig Farms. Vet. Res. 2019, 50, 45. [Google Scholar] [CrossRef]
- Zhang, S.; Li, S.; Gu, W.; den Bakker, H.; Boxrud, D.; Taylor, A.; Roe, C.; Driebe, E.; Engelthaler, D.M.; Allard, M.; et al. Zoonotic Source Attribution of Salmonella enterica Serotype Typhimurium Using Genomic Surveillance Data, United States. Emerg. Infect. Dis. 2019, 25, 82. [Google Scholar] [CrossRef]
- Barreto, M.; Castillo-Ruiz, M.; Retamal, P. Salmonella enterica: A Review or the Trilogy Agent, Host and Environment and Its Importance in Chile. Rev. Chil. Infectol. 2016, 33, 547–557. [Google Scholar] [CrossRef]
- Campos, J.; Mourão, J.; Peixe, L.; Antunes, P. Non-Typhoidal Salmonella in the Pig Production Chain: A Comprehensive Analysis of Its Impact on Human Health. Pathogens 2019, 8, 19. [Google Scholar] [CrossRef]
- Cuenca-Arias, P.; Montaño, L.A.; Villarreal, J.M.; Wiesner, M. Caracterización molecular y fenotípica de aislamientos clínicos de Salmonella Typhimurium variante monofásica (1,4,[5],12:i:-) recuperados en Colombia. Biomédica 2020, 40, 722–733. [Google Scholar] [CrossRef] [PubMed]
- CDC. National Notifiable Diseases Surveillance System, 2019 Annual Tables of Infectious Disease. Data, 2021. Available online: https://wonder.cdc.gov/nndss/static/2019/annual/2019-table2o-H.pdf (accessed on 2 August 2024).
- Allard, M.W.; Bell, R.; Ferreira, C.M.; Gonzalez-Escalona, N.; Hoffmann, M.; Muruvanda, T.; Ottesen, A.; Ramachandran, P.; Reed, E.; Sharma, S.; et al. Genomics of Foodborne Pathogens for Microbial Food Safety. Curr. Opin. Biotechnol. 2018, 49, 224–229. [Google Scholar] [CrossRef] [PubMed]
- Nair, D.V.T.; Venkitanarayanan, K.; Kollanoor Johny, A. Antibiotic-Resistant Salmonella in the Food Supply and the Potential Role of Antibiotic Alternatives for Control. Foods 2018, 7, 167. [Google Scholar] [CrossRef]
- Roasto, M.; Bonardi, S.; Mäesaar, M.; Alban, L.; Gomes-Neves, E.; Vieira-Pinto, M.; Vågsholm, I.; Elias, T.; Lindegaard, L.L.; Blagojevic, B. Salmonella enterica Prevalence, Serotype Diversity, Antimicrobial Resistance and Control in the European Pork Production Chain. Trends Food Sci. Technol. 2023, 131, 210–219. [Google Scholar] [CrossRef]
- Resistencia a los Antimicrobianos. Available online: https://www.who.int/es/news-room/fact-sheets/detail/antimicrobial-resistance (accessed on 2 August 2024).
- Vidal, J.; Clavijo, V.; Castellanos, L.; Kathiresan, J.; Kumar, A.; Mehta, K.; Chaparro-Gutiérrez, J. Multidrug-Resistant Salmonella spp. in Fecal Samples of Pigs with Suspected Salmonellosis in Antioquia, Colombia, 2019–2021. Rev. Panam. Salud Publica 2023, 47, e46. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, R.G.; Rosario, D.K.A.; Cunha-Neto, A.; Mano, S.B.; Figueiredo, E.E.S.; Conte-Junior, C.A. Worldwide Epidemiology of Salmonella Serovars in Animal-Based Foods: A Meta-Analysis. Appl. Environ. Microbiol. 2019, 85, e00591-19. [Google Scholar] [CrossRef]
- Instituto Colombiano Agropecuario—ICA. Available online: https://www.ica.gov.co/areas/pecuaria/servicios/epidemiologia-veterinaria/censos-2016/censo-2018 (accessed on 2 August 2024).
- Wilkins, W.; Rajić, A.; Waldner, C.; McFall, M.; Chow, E.; Muckle, A.; Rosengren, L. Distribution of Salmonella Serovars in Breeding, Nursery, and Grow-to-Finish Pigs, and Risk Factors for Shedding in Ten Farrow-to-Finish Swine Farms in Alberta and Saskatchewan. Can. J. Vet. Res. 2010, 74, 81–90. [Google Scholar] [PubMed]
- 9260 Introduction to Detecting Pathogenic Bacteria. In Standard Methods For the Examination of Water and Wastewater; American Public Health Association: Washington, DC, USA, 2017; Available online: https://www.standardmethods.org/doi/abs/10.2105/SMWW.2882.201 (accessed on 21 November 2024).
- 3MTM-Molecular Detection Assay 2, Salmonella. Available online: https://www.neogen.com/categories/pathogens/molecular-detection-assay-2-salmonella/?utm_medium=SocialShare (accessed on 2 August 2024).
- ISO 6579-1:2017; Microbiology of the Food Chain—Horizontal Method for the Detection, Enumeration and Serotyping of Salmonella Part 1: Detection of Salmonella spp. ISO: Geneva, Switzerland, 2017.
- Grimont, P.A.D.; Weill, F.X. Antigenic Formulae of the Salmonella Serovars, 9th ed.; WHO Collaborating Centre for Reference and Research on Salmonella: Paris, France, 2007. [Google Scholar]
- Hernández Toro, I.Y.; Carrascal-Camacho, A.K.; Pulido-Villamarín, A.d.P. Manual Para la Serotipificación de Cepas de Salmonella spp. Obtenidas en la Cadena Porcícola en Colombia; Editorial Pontificia Universidad Javeriana: Bogotá, Colombia, 2023. [Google Scholar]
- Alessiani, A.; Goffredo, E.; Mancini, M.; Occhiochiuso, G.; Faleo, S.; Didonna, A.; Fischetto, R.; Suglia, F.; De Vito, D.; Stallone, A.; et al. Evaluation of Antimicrobial Resistance in Salmonella Strains Isolated from Food, Animal and Human Samples between 2017 and 2021 in Southern Italy. Microorganisms 2022, 10, 812. [Google Scholar] [CrossRef] [PubMed]
- Willis, C.; Jørgensen, F.; Cawthraw, S.; Aird, H.; Lai, S.; Kesby, M.; Chattaway, M.; Lock, I.; Quill, E.; Raykova, G. A Survey of Salmonella, Escherichia coli, and Antimicrobial Resistance in Frozen, Part-Cooked, Breaded, or Battered Chicken Products on Retail Sale in the UK. J. Appl. Microbiol. 2023, 134, lxad093. [Google Scholar] [CrossRef]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-Resistant, Extensively Drug-Resistant and Pandrug-Resistant Bacteria: An International Expert Proposal for Interim Standard Definitions for Acquired Resistance. Clin. Microbiol. Inf. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Jibril, A.H.; Okeke, I.N.; Dalsgaard, A.; Kudirkiene, E.; Akinlabi, O.C.; Bello, M.B.; Olsen, J.E. Prevalence and Risk Factors of Salmonella in Commercial Poultry Farms in Nigeria. PLoS ONE 2020, 15, e0238190. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality Assessment Tool for Genome Assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, C.E.; Kruczkiewicz, P.; Laing, C.R.; Lingohr, E.J.; Gannon, V.P.J.; Nash, J.H.E.; Taboada, E.N. The Salmonella In Silico Typing Resource (SISTR): An Open Web-Accessible Tool for Rapidly Typing and Subtyping Draft Salmonella Genome Assemblies. PLoS ONE 2016, 11, e0147101. [Google Scholar] [CrossRef]
- Zankari, E.; Allesøe, R.; Joensen, K.G.; Cavaco, L.M.; Lund, O.; Aarestrup, F.M. PointFinder: A Novel Web Tool for WGS-Based Detection of Antimicrobial Resistance Associated with Chromosomal Point Mutations in Bacterial Pathogens. J. Antimicrob. Chemother. 2017, 72, 2764–2768. [Google Scholar] [CrossRef]
- Ramirez-Hernandez, A.; Carrascal-Camacho, A.K.; Varón-García, A.; Brashears, M.M.; Sanchez-Plata, M.X. Genotypic Characterization of Antimicrobial Resistant Salmonella spp. Strains from Three Poultry Processing Plants in Colombia. Foods 2021, 10, 491. [Google Scholar] [CrossRef]
- Wiser, B.; Niebuhr, S.E.; Dickson, J.S. Impact of Interventions on the Survival of Salmonella enterica I 4,[5],1 2:I:- in Pork. J. Food Prot. 2022, 85, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Roldan-Henao, M.; Dalsgaard, A.; Cardona-Castro, N.; Restrepo-Rivera, L.; Veloza-Angulo, L.C.; Alban, L. Pilot Study of the Productivity and Salmonella seroprevalence in Pigs Administered Organic Acids. Front. Vet. Sci. 2023, 10, 1123137. [Google Scholar] [CrossRef] [PubMed]
- Ostanello, F.; Lucia, A.D.D. On-Farm Risk Factors Associated with Salmonella in Pig Herds. Large Anim. Rev. 2020, 26, 133–140. [Google Scholar]
- Diep, B.; Barretto, C.; Portmann, A.-C.; Fournier, C.; Karczmarek, A.; Voets, G.; Li, S.; Deng, X.; Klijn, A. Salmonella Serotyping; Comparison of the Traditional Method to a Microarray-Based Method and an in Silico Platform Using Whole Genome Sequencing Data. Front. Microbiol. 2019, 10, 2554. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Yin, Y.; Jones, M.B.; Zhang, Z.; Deatherage Kaiser, B.L.; Dinsmore, B.A.; Fitzgerald, C.; Fields, P.I.; Deng, X. Salmonella Serotype Determination Utilizing High-Throughput Genome Sequencing Data. J. Clin. Microbiol. 2015, 53, 1685–1692. [Google Scholar] [CrossRef]
- Arnold, K.; Lim, S.; Rakler, T.; Rovira, A.; Satuchne, C.; Yechezkel, E.; Wiseman, A.; Pima, Y.; Yakunin, E.; Rokney, A.; et al. Using Genetic Markers for Detection and Subtyping of the Emerging Salmonella enterica Subspecies Enterica Serotype Muenchen. Poultry Sci. 2022, 101, 102181. [Google Scholar] [CrossRef] [PubMed]
- Gymoese, P.; Sørensen, G.; Litrup, E.; Olsen, J.E.; Nielsen, E.M.; Torpdahl, M. Investigation of Outbreaks of Salmonella enterica Serovar Typhimurium and Its Monophasic Variants Using Whole-Genome Sequencing, Denmark. Emerg. Infect. Dis. 2017, 23, 1631–1639. [Google Scholar] [CrossRef] [PubMed]
- Greening, B.; Whitham, H.K.; Aldous, W.K.; Hall, N.; Garvey, A.; Mandernach, S.; Kahn, E.B.; Nonnenmacher, P.; Snow, J.; Meltzer, M.I.; et al. Public Health Response to Multistate Salmonella Typhimurium Outbreak Associated with Prepackaged Chicken Salad, United States, 2018. Emerg. Infect. Dis. 2022, 28, 1254–1256. [Google Scholar] [CrossRef] [PubMed]
- Petrovska, L.; Mather, A.E.; AbuOun, M.; Branchu, P.; Harris, S.R.; Connor, T.; Hopkins, K.L.; Underwood, A.; Lettini, A.A.; Page, A.; et al. Microevolution of Monophasic Salmonella Typhimurium during Epidemic, United Kingdom, 2005–2010. Emerg. Infect. Dis. 2016, 22, 617–624. [Google Scholar] [CrossRef]
- Chattaway, M.A.; Langridge, G.C.; Wain, J. Salmonella Nomenclature in the Genomic Era: A Time for Change. Sci. Rep. 2021, 11, 7494. [Google Scholar] [CrossRef]
- Micallef, S.A.; Rosenberg Goldstein, R.E.; George, A.; Kleinfelter, L.; Boyer, M.S.; McLaughlin, C.R.; Estrin, A.; Ewing, L.; Jean-Gilles Beaubrun, J.; Hanes, D.E.; et al. Occurrence and Antibiotic Resistance of Multiple Salmonella Serotypes Recovered from Water, Sediment and Soil on Mid-Atlantic Tomato Farms. Environ. Res. 2012, 114, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wu, Q.; Zhang, J.; Huang, J.; Chen, L.; Wu, S.; Zeng, H.; Wang, J.; Chen, M.; Wu, H.; et al. Prevalence, Bacterial Load, and Antimicrobial Resistance of Salmonella Serovars Isolated From Retail Meat and Meat Products in China. Front. Microbiol. 2019, 10, 2121. [Google Scholar] [CrossRef]
- Marin, C.; Chinillac, M.C.; Cerdà-Cuéllar, M.; Montoro-Dasi, L.; Sevilla-Navarro, S.; Ayats, T.; Marco-Jimenez, F.; Vega, S. Contamination of Pig Carcass with Salmonella enterica Serovar Typhimurium Monophasic Variant 1,4[5],12:I:- Originates Mainly in Live Animals. Sci. Total Environ. 2020, 703, 134609. [Google Scholar] [CrossRef] [PubMed]
- European Food Safety Authority; European Centre for Disease Prevention and Control. The European Union One Health 2021 Zoonoses Report. EFSA J. 2022, 20, e07666. [Google Scholar] [CrossRef]
- Luo, Q.; Wu, Y.; Bao, D.; Xu, L.; Chen, H.; Yue, M.; Draz, M.S.; Kong, Y.; Ruan, Z. Genomic Epidemiology of Mcr Carrying Multidrug-Resistant ST34 Salmonella enterica Serovar Typhimurium in a One Health Context: The Evolution of a Global Menace. Sci. Total Environ. 2023, 896, 165203. [Google Scholar] [CrossRef] [PubMed]
- Bawn, M.; Alikhan, N.-F.; Thilliez, G.; Kirkwood, M.; Wheeler, N.E.; Petrovska, L.; Dallman, T.J.; Adriaenssens, E.M.; Hall, N.; Kingsley, R.A. Evolution of Salmonella enterica Serotype Typhimurium Driven by Anthropogenic Selection and Niche Adaptation. PLOS Genetics 2020, 16, e1008850. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.; Elbediwi, M.; Nambiar, R.B.; Yang, H.; Lin, J.; Yue, M. Genomic Characterization of Antimicrobial-Resistant Salmonella enterica in Duck, Chicken, and Pig Farms and Retail Markets in Eastern China. Microbiol. Spectr. 2022, 10, e01257-22. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Yang, M.; Cai, H.; Liu, Y.; Gorris, L.; Aslam, M.Z.; Jia, K.; Sun, T.; Wang, X.; Dong, Q. Antibiotic Resistance of Salmonella Typhimurium Monophasic Variant 1,4,[5],12:I:-In China: A Systematic Review and Meta-Analysis. Antibiotics 2022, 11, 532. [Google Scholar] [CrossRef] [PubMed]
- 2019 Salmonella Infections Linked to Butterball Brand Ground Turkey|Outbreak of Salmonella Infections Linked to Butterball Ground Turkey|March 2019|Salmonella|CDC. Available online: https://archive.cdc.gov/#/details?url=https://www.cdc.gov/salmonella/schwarzengrund-03-19/index.html (accessed on 2 August 2024).
- Duc, V.M.; Shin, J.; Nagamatsu, Y.; Fuhiwara, A.; Toyofuku, H.; Obi, T.; Chuma, T. Increased Salmonella Schwarzengrund Prevalence and Antimicrobial Susceptibility of Salmonella enterica Isolated from Broiler Chickens in Kagoshima Prefecture in Japan between 2013 and 2016. J. Vet. Med. Sci. 2020, 82, 585–589. [Google Scholar] [CrossRef]
- Li, I.-C.; Wu, H.-H.; Chen, Z.-W.; Chou, C.-H. Prevalence of IncFIB Plasmids Found among Salmonella enterica Serovar Schwarzengrund Isolates from Animal Sources in Taiwan Using Whole-Genome Sequencing. Pathogens 2021, 10, 1024. [Google Scholar] [CrossRef] [PubMed]
- Gomes, V.T.M.; Moreno, L.Z.; Silva, A.P.S.; Thakur, S.; La Ragione, R.M.; Mather, A.E.; Moreno, A.M. Characterization of Salmonella enterica Contamination in Pork and Poultry Meat from São Paulo/Brazil: Serotypes, Genotypes and Antimicrobial Resistance Profiles. Pathogens 2022, 11, 358. [Google Scholar] [CrossRef] [PubMed]
- Pires, A.F.A.; Funk, J.A.; Bolin, C. Risk Factors Associated with Persistence of Salmonella Shedding in Finishing Pigs. Prev. Vet. Med. 2014, 116, 120–128. [Google Scholar] [CrossRef]
- Traoré, O.; Nyholm, O.; Siitonen, A.; Bonkoungou, I.J.O.; Traoré, A.S.; Barro, N.; Haukka, K. Prevalence and Diversity of Salmonella enterica in Water, Fish and Lettuce in Ouagadougou, Burkina Faso. BMC Microbiol. 2015, 15, 151. [Google Scholar] [CrossRef] [PubMed]
- Hormaeche, E.; Peluffo, C.A.; De Pereyra, V.R. A New Salmonella Type, Salmonella carrau, with Special Reference to the 1,7… Phases of the Kauffmann-White Classification. J. Bacteriol. 1944, 47, 323–326. [Google Scholar] [CrossRef]
- Karczmarczyk, M.; Martins, M.; McCusker, M.; Mattar, S.; Amaral, L.; Leonard, N.; Aarestrup, F.M.; Fanning, S. Characterization of Antimicrobial Resistance in Salmonella enterica Food and Animal Isolates from Colombia: Identification of a qnrB19-Mediated Quinolone Resistance Marker in Two Novel Serovars. FEMS Microbiol. Lett. 2010, 313, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Somda, N.S.; Bonkoungou, I.J.O.; Sambe-Ba, B.; Drabo, M.S.; Wane, A.A.; Sawadogo-Lingani, H.; Savadogo, A. Diversity and Antimicrobial Drug Resistance of Non-Typhoid Salmonella Serotypes Isolated in Lettuce, Irrigation Water and Clinical Samples in Burkina Faso. J. Agric. Food Res. 2021, 5, 100167. [Google Scholar] [CrossRef]
- Lopes, G.V.; Pissetti, C.; da Cruz Payão Pellegrini, D.; da Silva, L.E.; Cardoso, M. Resistance Phenotypes and Genotypes of Salmonella enterica subsp. enterica Isolates from Feed, Pigs, and Carcasses in Brazil. J. Food Prot. 2015, 78, 407–413. [Google Scholar] [CrossRef]
- Possebon, F.S.; Tiba Casas, M.R.; Nero, L.A.; Yamatogi, R.S.; Araújo, J.P., Jr.; Pinto, J.P.D.A.N. Prevalence, Antibiotic Resistance, PFGE and MLST Characterization of Salmonella in Swine Mesenteric Lymph Nodes. Prev. Vet. Med. 2020, 179, 105024. [Google Scholar] [CrossRef] [PubMed]
- Mrema, N.; Mpuchane, S.; Gashe, B.A. Prevalence of Salmonella in Raw Minced Meat, Raw Fresh Sausages and Raw Burger Patties from Retail Outlets in Gaborone, Botswana. Food Control 2006, 17, 207–212. [Google Scholar] [CrossRef]
- Li, Y.-C.; Pan, Z.-M.; Kang, X.-L.; Geng, S.-Z.; Liu, Z.-Y.; Cai, Y.-Q.; Jiao, X.-A. Prevalence, Characteristics, and Antimicrobial Resistance Patterns of Salmonella in Retail Pork in Jiangsu Province, Eastern China. J. Food Prot. 2014, 77, 236–245. [Google Scholar] [CrossRef]
| Number of Positive Samples Detected | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Serotype | Industrial Commercial Farms (n = 75) | Technified Farms (n = 28) | ||||||||
| Source Water | Tank Water | Drinkers | Rectal Swab | % Positive | Source Water | Tank Water | Drinkers | Rectal Swab | % Positive | |
| S. Typhimurium monophasic variant | 1 | 1 | 0 | 25 | 32.5 | 0 | 1 | 2 | 19 | 6.1 |
| S. Typhimurium | 2 | 1 | 1 | 1 | 6.0 | 0 | 0 | 0 | 2 | 5.6 |
| S. Braenderup | 1 | 1 | 1 | 0 | 3.6 | 1 | 1 | 0 | 0 | 5.6 |
| S. Carrau | 1 | 0 | 3 | 0 | 4.8 | 0 | 1 | 0 | 0 | 2.8 |
| S. Saintpaul | 1 | 1 | 1 | 1 | 4.8 | 0 | 0 | 0 | 1 | 2.8 |
| S. Schwarzengrund | 1 | 1 | 2 | 0 | 4.8 | 1 | 0 | 0 | 0 | 2.8 |
| S. Soahanina | 2 | 1 | 1 | 0 | 4.8 | 0 | 0 | 0 | 0 | 0.0 |
| S. Derby | 0 | 0 | 0 | 3 | 3.6 | 0 | 0 | 0 | 0 | 0.0 |
| S. Isangi | 0 | 0 | 0 | 0 | 0.0 | 1 | 1 | 0 | 1 | 8.3 |
| S. Newport | 0 | 1 | 1 | 0 | 2.4 | 0 | 0 | 1 | 0 | 2.8 |
| S. Sandiego | 0 | 1 | 2 | 0 | 3.6 | 0 | 0 | 0 | 0 | 0.0 |
| S. Anatum | 0 | 1 | 0 | 2 | 3.6 | 0 | 0 | 0 | 0 | 0.0 |
| S. Amager | 0 | 0 | 0 | 0 | 0.0 | 0 | 0 | 0 | 2 | 5.6 |
| S. Give | 0 | 0 | 0 | 2 | 2.4 | 0 | 0 | 0 | 0 | 0.0 |
| S. Kedougou | 0 | 1 | 1 | 0 | 2.4 | 0 | 0 | 0 | 0 | 0.0 |
| S. Rubislaw | 1 | 0 | 1 | 0 | 2.4 | 0 | 0 | 0 | 0 | 0.0 |
| S. Glostrup | 1 | 0 | 0 | 0 | 1.2 | 0 | 0 | 0 | 0 | 0.0 |
| S. Meleagridis | 0 | 0 | 0 | 1 | 1.2 | 0 | 0 | 0 | 0 | 0.0 |
| S. Miami | 1 | 0 | 0 | 0 | 1.2 | 0 | 0 | 0 | 0 | 0.0 |
| S. Montevideo | 1 | 0 | 0 | 0 | 1.2 | 0 | 0 | 0 | 0 | 0.0 |
| S. Muenster | 0 | 0 | 0 | 1 | 1.2 | 0 | 0 | 0 | 0 | 0.0 |
| S. Nottingham | 0 | 0 | 0 | 1 | 1.2 | 0 | 0 | 0 | 0 | 0.0 |
| S. Oranienburg | 0 | 1 | 0 | 0 | 1.2 | 0 | 0 | 0 | 0 | 0.0 |
| S. Panama | 1 | 0 | 0 | 0 | 1.2 | 0 | 0 | 0 | 0 | 0.0 |
| S. Poona | 0 | 1 | 0 | 0 | 1.2 | 0 | 0 | 0 | 0 | 0.0 |
| S. Uganda | 0 | 0 | 0 | 1 | 1.2 | 0 | 0 | 0 | 0 | 0.0 |
| S. diarizonae IIIb 60:r:z35 | 0 | 1 | 0 | 0 | 1.2 | 0 | 0 | 0 | 0 | 0.0 |
| S. enterica I H:y:- | 0 | 1 | 1 | 0 | 2.4 | 0 | 0 | 0 | 0 | 0.0 |
| S. houtenae IV 50:z4,z32:- | 0 | 0 | 0 | 0 | 0.0 | 0 | 1 | 0 | 0 | 2.8 |
| S. houtenae IV 11:z4,z23:- | 0 | 1 | 1 | 0 | 2.4 | 0 | 0 | 0 | 0 | 0.0 |
| Total | 14 | 15 | 16 | 38 | 83 | 3 | 5 | 3 | 25 | 36 |
| No. | Serotype and Number of Isolates | Antibiotic Resistance Pattern | No. of Isolates | Proportion of All Isolates (%) | Family of Antibiotic |
|---|---|---|---|---|---|
| 1 | S. Anatum (2) S. Braenderup (5) S. Carrau (4) S. diarizonae IIIb 60:r:z35 (1) S. enterica I H:y:- (2) S. Give (1) S. Miami (1) S. Montevideo (1) S. Newport (3) S. Nottingham (1) S. Panama (1) S. Poona (1) S. Saintpaul (3) S. Sandiego (1) S. Schwarzengrund (5) S. Soahanina (4) S. Typhimurium (1) S. Uganda (1) | AMK-GEN | 38 | 3.8 | Aminoglycoside |
| 2 | S. Typhimurium monophasic variant (7) S. Typhimurium (1) | AMK-AMP-CTX-CHL-GEN-TET-TGC | 8 | 6.9 | Aminoglycoside, beta-lactam, amphenicol, tetracycline |
| 3 | S. Typhimurium monophasic variant (4) S. Typhimurium (1) S. Kedougou (1) S. Saintpaul (1) | AMK-CHL-GEN-TET-TGC | 7 | 6.0 | Aminoglycoside, amphenicol, tetracycline |
| 4 | S. Typhimurium monophasic variant (7) | AMK-AMP-CHL-GEN-SMX-TET-TGC | 7 | 6.0 | Aminoglycoside, beta-lactam, amphenicol, folate pathway antagonist, tetracycline |
| 5 | S. Typhimurium monophasic variant (4) S. Muenster (1) | AMK-AMP-CTX-CHL-GEN-SMX-TET-TGC-TMP | 5 | 4.3 | Aminoglycoside, beta-lactam, amphenicol, folate pathway antagonist, tetracycline |
| 6 | S. Typhimurium monophasic variant (5) | AMK-AMP-CHL-GEN-NAL-SMX-TET | 5 | 4.3 | Aminoglycoside, beta-lactam, amphenicol, quinolone, folate pathway antagonist, tetracycline |
| 7 | S. Typhimurium monophasic variant (3) | AMK-CHL-GEN-NAL-TET-TGC | 3 | 2.6 | Aminoglycoside, amphenicol, quinolone, tetracycline |
| 8 | S. Typhimurium monophasic variant (3) | AMK-AMP-CTX-CHL-GEN-NAL-TET-TGC | 3 | 2.6 | Aminoglycoside, beta-lactam, amphenicol, quinolone, tetracycline |
| 9 | S. Amager (1) S. Isangi (2) S. Typhimurium monophasic variant (1) | AMK-CHL-CIP-GEN-NAL-TET-TGC | 4 | 3.4 | Aminoglycoside, amphenicol, quinolone, tetracycline |
| 10 | S. Meleagridis (1) S. Rubislaw (1) S. Anatum (1) | AMK-GEN-SMX | 3 | 2.6 | Aminoglycoside, folate pathway antagonist |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chamorro-Tobar, I.C.; Pulido-Villamarín, A.; Carrascal-Camacho, A.K.; Barrientos-Anzola, I.; Wiesner, M.; Hernández-Toro, I.; Alban, L.; Olsen, J.E.; Dalsgaard, A.; Hounmanou, Y.M.G. Phenotypic and Genotypic Characterization of Antimicrobial Resistance in Salmonella enterica Serovars from Colombian Pig Farms. Appl. Microbiol. 2024, 4, 1729-1744. https://doi.org/10.3390/applmicrobiol4040116
Chamorro-Tobar IC, Pulido-Villamarín A, Carrascal-Camacho AK, Barrientos-Anzola I, Wiesner M, Hernández-Toro I, Alban L, Olsen JE, Dalsgaard A, Hounmanou YMG. Phenotypic and Genotypic Characterization of Antimicrobial Resistance in Salmonella enterica Serovars from Colombian Pig Farms. Applied Microbiology. 2024; 4(4):1729-1744. https://doi.org/10.3390/applmicrobiol4040116
Chicago/Turabian StyleChamorro-Tobar, Iliana C., Adriana Pulido-Villamarín, Ana Karina Carrascal-Camacho, Irina Barrientos-Anzola, Magdalena Wiesner, Ivonne Hernández-Toro, Lis Alban, John Elmerdahl Olsen, Anders Dalsgaard, and Yaovi Mahuton Gildas Hounmanou. 2024. "Phenotypic and Genotypic Characterization of Antimicrobial Resistance in Salmonella enterica Serovars from Colombian Pig Farms" Applied Microbiology 4, no. 4: 1729-1744. https://doi.org/10.3390/applmicrobiol4040116
APA StyleChamorro-Tobar, I. C., Pulido-Villamarín, A., Carrascal-Camacho, A. K., Barrientos-Anzola, I., Wiesner, M., Hernández-Toro, I., Alban, L., Olsen, J. E., Dalsgaard, A., & Hounmanou, Y. M. G. (2024). Phenotypic and Genotypic Characterization of Antimicrobial Resistance in Salmonella enterica Serovars from Colombian Pig Farms. Applied Microbiology, 4(4), 1729-1744. https://doi.org/10.3390/applmicrobiol4040116






