Appendix A
This appendix contains details and explanations supplemental to the subsection, “4.1. Application of AI-based diagnostic tools in early detection, risk stratification, and monitoring of IFTA” of the main text. The explanations of the quality assessment of the included study articles [
1] through [
7] using the PROBAST and QUADAS-2 tools would disrupt the flow of the main text. However, this discussion is crucial to understanding the overall significance of these studies.
Table A1.
QUADAS-2 RoB and Applicability Assessment for Athvale et al., 2021 [
1].
| Domain | RoB | Judgment & Justification | Applicability Concern |
|---|
1. Patient
Selection | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High High | The study used a single-center, retrospective, consecutive sample (in = 352) of patients who underwent kidney biopsy at a tertiary hospital. This introduces potential selection bias, as patients were not randomly sampled and may not represent broader clinical populations. | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High, Participants are limited to a specific demographic (Cook County Hospital, Chicago), potentially limiting generalizability. High, Participants are limited to a specific demographic (Cook County Hospital, Chicago), potentially limiting generalizability. |
2. Index Test
(DL + XGBoost model) | ![Biomedinformatics 05 00067 i004 Biomedinformatics 05 00067 i004]() / /![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() Moderate to High Moderate to High | The model was trained and tested on the same institutional data, with no external validation. It’s unclear whether the index test was interpreted blinded to the reference standard. Deep learning feature extraction may vary across ultrasound machines. | ![Biomedinformatics 05 00067 i004 Biomedinformatics 05 00067 i004]() Moderate, Implementation in other settings could yield variable results due to equipment and imaging protocol differences. Moderate, Implementation in other settings could yield variable results due to equipment and imaging protocol differences. |
3. Reference
Standard
(Histopathologic IFTA grading) | ![Biomedinformatics 05 00067 i001 Biomedinformatics 05 00067 i001]() Low Low | Histopathology is the accepted gold standard for assessing interstitial fibrosis and tubular atrophy. It’s likely that assessments were performed by qualified nephropathologists. However, blinding between reference and index test evaluators was not explicitly mentioned. | ![Biomedinformatics 05 00067 i001 Biomedinformatics 05 00067 i001]() Low, The outcome is directly relevant to clinical practice. Low, The outcome is directly relevant to clinical practice. |
4. Flow and
Timing | ![Biomedinformatics 05 00067 i004 Biomedinformatics 05 00067 i004]() Moderate Moderate | All patients underwent both the index test (ultrasound) and the reference standard (biopsy) in the same period, but timing between the two was not specified. Missing data handling and patient exclusions were not detailed. | ![Biomedinformatics 05 00067 i001 Biomedinformatics 05 00067 i001]() Low, Likely applicable since both tests relate to the same diagnostic event. Low, Likely applicable since both tests relate to the same diagnostic event. |
Table A1 presents the application of the QUADAS-2 tool to the 1st paper, Athvale et al. 2021 [
1], which utilizes an AI-based diagnostic tool for early detection, risk stratification, and monitoring of IFTA. Using QUADAS-2, we evaluated the RoB and applicability concerns of this study across four domains: Patient Selection, Index Test, Reference Standard, and Flow and Timing.
Although the study demonstrates promising accuracy for noninvasive quantification of IFTA using DL on ultrasound images, its single-center design, lack of external validation, and small dataset relative to model complexity result in a high RoB and limited generalizability. Future multicenter validation and calibration studies are needed to strengthen certainty.
Overall QUADAS-2 judgment for this study is as follows: RoB, moderate to high, and applicability concerns, moderate to high.
Table A2.
QUADAS-2 RoB and applicability assessment for Trojani et al., 2024 [
2].
| Domain | RoB | Reasoning/Justification | Applicability Concern |
|---|
1. Patient
Selection | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High High | Retrospective, single-center study; patients were included only if they had both MRI and biopsy within six months, which introduces selection bias (enriched population, not consecutive diagnostic workflow). Excluded poor-quality MRIs and unsuitable biopsies, which further limits representativeness. | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High, The study population (transplant recipients in a tertiary center with available MRI) does not represent the full clinical spectrum of post-transplant patients. High, The study population (transplant recipients in a tertiary center with available MRI) does not represent the full clinical spectrum of post-transplant patients. |
| 2. Index Test (MRI-radiomic ML model) | ![Biomedinformatics 05 00067 i004 Biomedinformatics 05 00067 i004]() / /![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() Moderate to High Moderate to High | The MRI radiomics-based ML model was developed and validated on the same institutional data, using internal train/test splits (no external validation). The index test likely was not interpreted fully blinded to the biopsy results during feature selection and model tuning. Performance may be optimistically biased (AUC drop between training and test). | ![Biomedinformatics 05 00067 i004 Biomedinformatics 05 00067 i004]() Moderate, MRI protocols, scanners, and pre-processing steps are highly site-specific; generalization to other centers may be limited. Moderate, MRI protocols, scanners, and pre-processing steps are highly site-specific; generalization to other centers may be limited. |
| 3. Reference Standard (Histopathologic biopsy with Banff IFTA grading) | ![Biomedinformatics 05 00067 i001 Biomedinformatics 05 00067 i001]() Low Low | The biopsy-based Banff classification is a recognized gold standard for IFTA. Assessment was performed by an expert nephropathologist, though blinding to MRI results was not explicitly confirmed. | ![Biomedinformatics 05 00067 i001 Biomedinformatics 05 00067 i001]() Low, The biopsy grading directly answers the target condition and is appropriate for the study aim. Low, The biopsy grading directly answers the target condition and is appropriate for the study aim. |
| 4. Flow and Timing | ![Biomedinformatics 05 00067 i004 Biomedinformatics 05 00067 i004]() Moderate Moderate | MRI and biopsy were performed within six months, which may be long enough for histological changes in IFTA to progress, introducing potential misclassification. Only 70 MR-biopsy pairs analyzed out of 254 biopsies performed, indicating patient/exam attrition. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Low to Moderate, The six-month interval could affect diagnostic consistency, but within chronic IFTA context, it’s partly acceptable. Low to Moderate, The six-month interval could affect diagnostic consistency, but within chronic IFTA context, it’s partly acceptable. |
Table A2 presents the application of the QUADAS-2 tool to the 2nd paper, Trojani et al. 2024 [
2], which uses an AI-based diagnostic tool for early detection, risk stratification, and monitoring of IFTA. Using QUADAS-2, we evaluated the RoB and applicability concerns of this study across four domains: Patient Selection, Index Test, Reference Standard, and Flow and Timing.
This single-center, retrospective diagnostic accuracy study presents a moderate-to-high RoB, mainly due to non-representative patient selection, lack of external validation, and potential overfitting of the radiomics-based ML model. The reference standard (biopsy-based Banff IFTA grading) is appropriate and reliable, though blinding was not fully reported. The six-month window between MRI and biopsy could have introduced temporal bias, and missing data handling was not fully described. Applicability is limited by center-specific MRI acquisition protocols, pre-processing pipelines, and manual segmentation.
Overall QUADAS-2 judgment for this study is as follows: RoB, moderate to high, and applicability concerns, moderate to high.
Table A3.
QUADAS-2 RoB and applicability assessment for Ginley et al., 2021 [
3].
| Domain | RoB | Reasoning/Justification | Applicability Concern |
|---|
| 1. Patient Selection | ![Biomedinformatics 05 00067 i004 Biomedinformatics 05 00067 i004]() Moderate Moderate | The study used 116 whole-slide biopsy images, retrospectively selected from existing archives. No mention of consecutive or random sampling. Slides were chosen for image quality and completeness, introducing potential selection bias. However, inclusion appears broad across chronic kidney injuries, not limited to a narrow subset. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Low to Moderate, The sample (renal biopsies with chronic injury) represents the target clinical population, though external representativeness is uncertain. Low to Moderate, The sample (renal biopsies with chronic injury) represents the target clinical population, though external representativeness is uncertain. |
| 2. Index Test (CNN-based ML model, DeepLab v2) | ![Biomedinformatics 05 00067 i004 Biomedinformatics 05 00067 i004]() Moderate Moderate | The convolutional neural network (DeepLab v2) was trained on annotated WSIs and evaluated internally and externally. External testing included only 20 slides, from the same or similar institutional context, and blinding to the reference standard was not specified. Model tuning and performance optimization may have introduced optimistic bias. | ![Biomedinformatics 05 00067 i004 Biomedinformatics 05 00067 i004]() Moderate, Deep learning performance may vary with slide scanners, staining, and lab protocols, affecting real-world applicability. Moderate, Deep learning performance may vary with slide scanners, staining, and lab protocols, affecting real-world applicability. |
| 3. Reference Standard (Renal Pathologist Grading of IFTA and Glomerulosclerosis) | ![Biomedinformatics 05 00067 i001 Biomedinformatics 05 00067 i001]() Low Low | The ground truth labels were assigned by expert renal pathologists, the clinical gold standard. Multiple pathologists participated in validation, and performance was benchmarked against their agreement levels. There’s no explicit statement on blinding to model outputs, but the use of independent test slides suggests low bias. | ![Biomedinformatics 05 00067 i001 Biomedinformatics 05 00067 i001]() Low, Pathologist-based grading directly reflects the intended diagnostic construct. Low, Pathologist-based grading directly reflects the intended diagnostic construct. |
| 4. Flow and Timing | ![Biomedinformatics 05 00067 i001 Biomedinformatics 05 00067 i001]() Low Low | All slides underwent both the index test (ML assessment) and reference evaluation (pathologist grading) from the same biopsy samples. No participants or slides appear to have been excluded after inclusion. Flow is consistent and clearly reported. | ![Biomedinformatics 05 00067 i001 Biomedinformatics 05 00067 i001]() Low, The timing of analyses aligns with typical diagnostic workflows. Low, The timing of analyses aligns with typical diagnostic workflows. |
Table A3 presents the application of the QUADAS-2 tool for the 3rd paper, Ginley et al. 2021 [
3], which uses an AI-based diagnostic tool for the early detection, risk stratification, and monitoring of IFTA progression. Using QUADAS-2, we evaluated the RoB and applicability concerns of this study across four domains: Patient Selection, Index Test, Reference Standard, and Flow and Timing.
This diagnostic accuracy study demonstrates moderate overall RoB, mainly due to retrospective sampling and limited external validation of the CNN model. The reference standard (expert renal pathologist assessment) is robust, and the index test was applied appropriately, achieving pathologist-level performance.
In summary, the study shows low concern in the reference standard and flow, but moderate risk in patient selection and index test domains, yielding an overall QUADAS-2 judgment of moderate RoB and moderate concern for applicability.
Overall QUADAS-2 judgment for this study is as follows: RoB, moderate, and applicability concerns, low to moderate.
Table A4.
QUADAS-2 RoB and applicability assessment for Zheng et al., 2021 [
4].
| Domain | RoB | Reasoning/Justification | Applicability Concern |
|---|
| 1. Patient Selection | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Low-Moderate Low-Moderate | Patients included 64 from OSUWMC (67 WSIs) and 14 from Kidney Precision Medicine Project/KPMP (28 WSIs). WSIs underwent manual quality checks to exclude slides with artifacts. Some clinical data was missing (e.g., proteinuria, eGFR), but all eligible biopsies were included. The sample may not fully represent all renal biopsy populations, and KPMP had no severe IFTA cases. | ![Biomedinformatics 05 00067 i001 Biomedinformatics 05 00067 i001]() Low, The population represents patients undergoing renal biopsy, which matches the intended clinical target population for IFTA grading. Low, The population represents patients undergoing renal biopsy, which matches the intended clinical target population for IFTA grading. |
| 2. Index Test (DL Model, glpathnet) | ![Biomedinformatics 05 00067 i001 Biomedinformatics 05 00067 i001]() Low Low | The DL model combined local patch-level and global WSI-level features to predict IFTA grade. Model was trained on OSUWMC data with 5-fold cross-validation and tested externally on KPMP data. Patch-level probabilities were reviewed after reference grading to avoid bias. | ![Biomedinformatics 05 00067 i001 Biomedinformatics 05 00067 i001]() Low, The model directly addresses automated IFTA grading on digitized WSIs, the intended purpose of the index test. Low, The model directly addresses automated IFTA grading on digitized WSIs, the intended purpose of the index test. |
| 3. Reference Standard (Pathologist Consensus, Majority Vote) | ![Biomedinformatics 05 00067 i004 Biomedinformatics 05 00067 i004]() Moderate Moderate | IFTA grades were determined by majority vote of five nephropathologists (OSUWMC) and by study investigators (KPMP), with moderate interobserver agreement (κ = 0.31–0.50). Grading is inherently subjective and may vary between pathologists, though consensus aligns with standard clinical practice. | ![Biomedinformatics 05 00067 i001 Biomedinformatics 05 00067 i001]() Low, Reference standard is clinically appropriate, using expert nephropathologists’ evaluation of renal biopsy WSIs. Low, Reference standard is clinically appropriate, using expert nephropathologists’ evaluation of renal biopsy WSIs. |
| 4. Flow and Timing | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Low-Moderate Low-Moderate | All WSIs were digitized consistently at ×40 magnification. DL training and testing were separated between OSUWMC (training) and KPMP (external validation). Some KPMP cases lacked severe IFTA. No missing WSIs, and the same grading criteria were applied across datasets. | ![Biomedinformatics 05 00067 i001 Biomedinformatics 05 00067 i001]() Low, Data handling, timing, and grading criteria are consistent, reflecting intended workflow for WSI analysis. Low, Data handling, timing, and grading criteria are consistent, reflecting intended workflow for WSI analysis. |
Table A4 presents the application of the QUADAS-2 tool for the 4th paper, Zheng et al., 2021 [
4], which uses an AI-based diagnostic tool for the early detection, risk stratification, and monitoring of IFTA progression. Using QUADAS-2, we evaluated the RoB and applicability concerns of this study across four domains: Patient Selection, Index Test, Reference Standard, and Flow and Timing.
The study has an overall moderate RoB. This is because while the index test and flow/timing are low risk, moderate concerns arise from patient selection and the subjectivity of the reference standard. Applicability concerns across all domains are low, indicating that the study population, index test, and reference standard are relevant to the intended clinical context.
Overall QUADAS-2 judgment for this study is as follows: RoB, moderate, and applicability concerns, low.
Table A5.
QUADAS-2 RoB and applicability assessment for Athvale et al., 2020 [
5].
| Domain | RoB | Reasoning/Justification | Applicability Concern |
|---|
| 1. Patient Selection | ![Biomedinformatics 05 00067 i004 Biomedinformatics 05 00067 i004]() Moderate Moderate | Patients were included retrospectively from a single center (Cook County Health, Chicago, IL). Ultrasound images were obtained from 352 patients who underwent kidney biopsy. Potential selection bias exists as only patients with available biopsy-confirmed IFTA, and usable ultrasound images were included. | ![Biomedinformatics 05 00067 i004 Biomedinformatics 05 00067 i004]() Moderate, Population reflects patients undergoing biopsy but may not represent broader populations or those without biopsy, limiting generalizability to all kidney disease patients. Moderate, Population reflects patients undergoing biopsy but may not represent broader populations or those without biopsy, limiting generalizability to all kidney disease patients. |
| 2. Index Test (DL Ultrasound Classification) | ![Biomedinformatics 05 00067 i001 Biomedinformatics 05 00067 i001]() Low Low | The DL system classified IFTA from ultrasound images with masked kidneys based on a 91% accurate segmentation routine. The system was trained, validated, and tested on separate datasets, reducing bias. Performance metrics (accuracy, precision, recall, F1-score) were reported for all sets. | ![Biomedinformatics 05 00067 i001 Biomedinformatics 05 00067 i001]() Low, The index test is directly relevant to the clinical task of non-invasive IFTA assessment. Low, The index test is directly relevant to the clinical task of non-invasive IFTA assessment. |
| 3. Reference Standard (Biopsy IFTA by Nephropathologist) | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Low-Moderate Low-Moderate | Reference standard was histologic IFTA grading on trichrome-stained kidney biopsy by nephropathologists. While widely accepted, inter-observer variability in IFTA grading is known, but majority consensus or standardized scoring was not specified. | ![Biomedinformatics 05 00067 i001 Biomedinformatics 05 00067 i001]() Low, The reference standard is clinically appropriate and directly measures the target condition. Low, The reference standard is clinically appropriate and directly measures the target condition. |
| 4. Flow and Timing | ![Biomedinformatics 05 00067 i001 Biomedinformatics 05 00067 i001]() Low Low | Training, validation, and test datasets were clearly separated. No mention of missing images or exclusions post-acquisition, and all images were processed using the same protocol. Timing between ultrasound and biopsy was not specified but likely contemplate for clinical workflow. | ![Biomedinformatics 05 00067 i001 Biomedinformatics 05 00067 i001]() Low, Flow and timing are appropriate for evaluating the DL model against the biopsy reference standard. Low, Flow and timing are appropriate for evaluating the DL model against the biopsy reference standard. |
Table A5 presents the application of the QUADAS-2 tool for the 5th paper, Athvale et al., 2020 [
5], which uses an AI-based diagnostic tool for the early detection, risk stratification, and monitoring of IFTA progression. Using QUADAS-2, we evaluated the RoB and applicability concerns of this study across four domains: Patient Selection, Index Test, Reference Standard, and Flow and Timing.
The study demonstrates robust performance of the DL system for non-invasive IFTA assessment, with moderate caution due to selection bias and potential variability in biopsy grading.
Overall QUADAS-2 judgment for this study is as follows: RoB, moderate, and applicability concerns, low.
Table A6.
QUADAS-2 RoB and applicability assessment for Ginley et al., 2020 [
6].
| Domain | RoB | Reasoning/Justification | Applicability Concern |
|---|
| 1. Patient/Tissue Selection | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Low-Moderate Low-Moderate | The study used renal biopsy samples stained with periodic acid-Schiff (PAS). Data came from a single institution for intra-institutional holdout testing and an external institution for inter-institutional testing. Exact selection criteria and sample size were not fully described, introducing potential selection bias. | ![Biomedinformatics 05 00067 i001 Biomedinformatics 05 00067 i001]() Low, Study samples are representative of patients undergoing renal biopsy for glomerulosclerosis and IFTA assessment, the intended target population. Low, Study samples are representative of patients undergoing renal biopsy for glomerulosclerosis and IFTA assessment, the intended target population. |
| 2. Index Test (CNN Segmentation) | ![Biomedinformatics 05 00067 i001 Biomedinformatics 05 00067 i001]() Low Low | CNNs were trained to segment glomerulosclerosis and IFTA on PAS-stained biopsies. The model performance was evaluated on holdout intra- and inter-institutional datasets. Segmentation outputs were quantitatively compared to reference annotations, and high correlations were reported, indicating low bias in test conduct. | ![Biomedinformatics 05 00067 i001 Biomedinformatics 05 00067 i001]() Low, The index test directly addresses the clinical task of automated segmentation and quantitation of renal histologic injury. Low, The index test directly addresses the clinical task of automated segmentation and quantitation of renal histologic injury. |
| 3. Reference Standard (Pathologist Annotations) | ![Biomedinformatics 05 00067 i004 Biomedinformatics 05 00067 i004]() Moderate Moderate | Ground truth was based on manual segmentation by renal pathologists. The study notes that the CNN sometimes predicted regions “better than the ground truth,” indicating some subjectivity and potential variability in reference standard. Inter-observer variability of annotations was not formally reported. | ![Biomedinformatics 05 00067 i001 Biomedinformatics 05 00067 i001]() Low, Expert pathologist annotations are clinically relevant and appropriate for training and validating the model. Low, Expert pathologist annotations are clinically relevant and appropriate for training and validating the model. |
| 4. Flow and Timing | ![Biomedinformatics 05 00067 i001 Biomedinformatics 05 00067 i001]() Low Low | The training, intra-institutional holdout, and inter-institutional holdout testing datasets were clearly separated. No missing data issues were reported, and all images were analyzed according to the same protocol. | ![Biomedinformatics 05 00067 i001 Biomedinformatics 05 00067 i001]() Low, Flow and timing reflect intended use, with proper separation of training and test datasets. Low, Flow and timing reflect intended use, with proper separation of training and test datasets. |
Table A6 presents the application of the QUADAS-2 tool for the 6th paper, Ginley et al., 2020 [
6], which uses an AI-based diagnostic tool for the early detection, risk stratification, and monitoring of IFTA progression. Using QUADAS-2, we evaluated the RoB and applicability concerns of this study across four domains: Patient Selection, Index Test, Reference Standard, and Flow and Timing.
The QUADAS-2 assessment indicates that this study evaluating CNNs for segmentation of glomerulosclerosis and interstitial fibrosis/tubular atrophy (IFTA) in PAS-stained renal biopsies is generally robust. The index test (CNN-based segmentation) demonstrates low RoB, as it was trained and validated on holdout intra- and inter-institutional datasets with quantitative evaluation against reference annotations. The reference standard, based on pathologist manual annotations, carries a moderate RoB due to inherent subjectivity and potential variability, though it remains clinically appropriate. Patient and tissue selection present low-moderate risk because sample sizes and selection criteria were not fully described, and external validation data came from a single additional institution. The flow and timing domain has a low RoB, with consistent image handling and clear separation of training and test datasets.
Overall QUADAS-2 judgment for this study is as follows: RoB, moderate, and applicability concerns, low.
Table A7.
PROBAST assessment for Yin et al., 2023 [
7].
| Domain | Details | RoB/Concern |
|---|
| Population | Post-transplant kidney patients from five GEO datasets (GSE98320 [45], GSE76882 [46]: training; GSE22459 [47], GSE53605 [48]: validation; GSE21374 [49]: prognosis). Total sample sizes vary; cohorts selected based on ≥50 samples and availability of biopsy-confirmed IFTA or survival data. Heterogeneity is due to platform differences and batch effects. | ![Biomedinformatics 05 00067 i004 Biomedinformatics 05 00067 i004]() Moderate, selection bias possible; not fully representative of all transplant patients Moderate, selection bias possible; not fully representative of all transplant patients |
| Index Model | Stepglm[both] + RF diagnostic model based on 28 necroptosis-related genes. Developed from 114 combinations of 13 ML algorithms (LASSO, Ridge, Enet, Stepglm, SVM, glmboost, LDA, plsRglm, RF, Gradient Boosting Machine/GBM, XGBoost, Naive Bayes, ANN). High-dimensional data relative to sample size increases risk of overfitting. | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High, multiple model testing, risk of overfitting, small validation sets relative to training High, multiple model testing, risk of overfitting, small validation sets relative to training |
| Comparator/Reference Model | Biopsy-confirmed IFTA status (histopathological evaluation) and survival data (post-transplant graft loss) from GEO datasets. Used as reference standard to evaluate predictive performance (AUC, ROC). | ![Biomedinformatics 05 00067 i001 Biomedinformatics 05 00067 i001]() Low, clinically accepted standard; reference outcome is relevant and reliable Low, clinically accepted standard; reference outcome is relevant and reliable |
| Outcome | IFTA classification (binary or graded) and post-transplant graft survival. Modeled outcome includes differential gene expressions associated with necroptosis. Performance evaluated with AUC, Principal Component Analysis/PCA separation, and Kaplan–Meier curves for survival. | ![Biomedinformatics 05 00067 i004 Biomedinformatics 05 00067 i004]() Moderate, grading differences across cohorts and batch effects may introduce misclassification bias Moderate, grading differences across cohorts and batch effects may introduce misclassification bias |
| Timing | Gene expression data from biopsies collected at variable post-transplant time points. Prognostic evaluation uses longitudinal survival data. Model development used cross-sectional training/validation datasets; timing differences between datasets could affect predictive performance. | ![Biomedinformatics 05 00067 i004 Biomedinformatics 05 00067 i004]() Moderate, timing differences and cross-sectional data may limit prediction consistency Moderate, timing differences and cross-sectional data may limit prediction consistency |
| Setting | Publicly available GEO gene expression datasets from multiple kidney transplant cohorts. Laboratory and bioinformatics setting; no prospective or clinical trial validation. | ![Biomedinformatics 05 00067 i004 Biomedinformatics 05 00067 i004]() Moderate, datasets may not represent real-world clinical populations Moderate, datasets may not represent real-world clinical populations |
| Intended Use of Prediction Model | Early identification of IFTA and stratification of kidney transplant patients by risk of fibrosis progression or graft loss. Aims to support clinical decision-making and follow-up prioritization. Not yet validated for clinical deployment. | ![Biomedinformatics 05 00067 i004 Biomedinformatics 05 00067 i004]() Moderate, potential clinical use, but external validation and clinical implementation pending Moderate, potential clinical use, but external validation and clinical implementation pending |
Table A7 integrates PROBAST domains with the study-specific details (population, predictors, comparator, outcome, timing, setting, intended use) for a clear, structured assessment of the journal publication, Yin et al., 2023 [
7].
The overall PROBAST judgment for this study is:
![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]()
High RoB, with
![Biomedinformatics 05 00067 i004 Biomedinformatics 05 00067 i004]()
Moderate Concerns for Applicability.
Justification is as follows:
High RoB arises mainly from the model development and analysis domain, where extensive algorithm testing (114 model combinations) and limited validation increase the likelihood of overfitting.
The population is retrospectively selected from heterogeneous GEO datasets, adding potential selection and batch-related biases.
Applicability concerns are moderate because the predictors (necroptosis-related genes) and outcomes (biopsy-confirmed IFTA and graft survival) are clinically relevant, but external generalizability to broader or prospective clinical populations remains uncertain.
Appendix B
This appendix contains details and explanations supplemental to the subsection, “4.2. Application of AI-based models to different urinary biomarkers for early detection, risk stratification, and monitoring of CKD progression” of the main text. The explanations of the quality assessment of the included study articles [
26] through [
30] using the PROBAST and QUADAS-2 tools would disrupt the flow of the main text. However, this discussion is crucial to understanding the overall significance of these studies.
Table A8.
PROBAST assessment for Bienaime et al., 2023 [
26].
| Domain/Item | Details | RoB/Concern |
|---|
| Population | Participants were 229 adults with chronic kidney disease (mean age 61 years; 66% male; mean baseline mGFR 38 mL/min) from the prospective NephroTest cohort. Fast CKD progression was defined as >10% annual mGFR decline. The cohort is well-characterized, but the subsample size is modest and may not reflect the full CKD spectrum. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Low-Moderate, Prospective and clinically relevant, but limited sample and single cohort reduce representativeness. Low-Moderate, Prospective and clinically relevant, but limited sample and single cohort reduce representativeness. |
| Index Model | A LASSO logistic regression model combining five urinary biomarkers (CCL2, EGF, KIM1, NGAL, and TGF-α) with clinical variables (age, sex, mGFR, albuminuria) to predict fast CKD progression. Model selection used repeated resampling (100 iterations). | ![Biomedinformatics 05 00067 i004 Biomedinformatics 05 00067 i004]() Moderate, LASSO penalization reduces overfitting risk, but internal-only validation and data-driven selection of biomarkers may inflate performance estimates. Moderate, LASSO penalization reduces overfitting risk, but internal-only validation and data-driven selection of biomarkers may inflate performance estimates. |
| Comparator/Reference Model | The Kidney Failure Risk Equation (KFRE) variables (age, sex, mGFR, albuminuria) served as the baseline comparator for performance evaluation. | ![Biomedinformatics 05 00067 i001 Biomedinformatics 05 00067 i001]() Low, Comparator is appropriate, widely accepted, and clinically meaningful. Low, Comparator is appropriate, widely accepted, and clinically meaningful. |
| Outcome | Outcome was fast CKD progression, defined as >10% decline per year in measured GFR using ^51Cr-EDTA clearance-a gold standard assessment. | ![Biomedinformatics 05 00067 i001 Biomedinformatics 05 00067 i001]() Low, Objective and precise measurement of kidney function minimizes outcome misclassification. Low, Objective and precise measurement of kidney function minimizes outcome misclassification. |
| Timing | Predictor and outcome data came from a prospective follow-up design within the NephroTest cohort. Urine biomarkers and clinical variables were measured at baseline; outcomes were observed longitudinally. | ![Biomedinformatics 05 00067 i001 Biomedinformatics 05 00067 i001]() Low, Clear temporal sequence supports valid prediction; prospective data collection minimizes bias. Low, Clear temporal sequence supports valid prediction; prospective data collection minimizes bias. |
| Setting | Conducted in a clinical research cohort of CKD patients under nephrology care at French academic hospitals (NephroTest). Laboratory-based ELISA assays underwent rigorous FDA-standard validation prior to modeling. | ![Biomedinformatics 05 00067 i001 Biomedinformatics 05 00067 i001]() Low, Well-controlled research setting; consistent sample handling and assay validation. Low, Well-controlled research setting; consistent sample handling and assay validation. |
| Intended Use of Prediction Model | The model aims to improve risk stratification for CKD progression beyond standard clinical variables by adding validated urinary biomarkers, potentially guiding early intervention and follow-up intensity. | ![Biomedinformatics 05 00067 i001 Biomedinformatics 05 00067 i001]() Low, Intended use is clinically relevant and aligned with current CKD management goals. Low, Intended use is clinically relevant and aligned with current CKD management goals. |
Table A8 integrates PROBAST domains with the study-specific details (population, predictors, comparator, outcome, timing, setting, intended use) for a clear, structured assessment of the journal publication, Bienaime et al., 2023 [
26].
The overall PROBAST judgment for this study is:
![Biomedinformatics 05 00067 i004 Biomedinformatics 05 00067 i004]()
Moderate Overall RoB, with
![Biomedinformatics 05 00067 i001 Biomedinformatics 05 00067 i001]()
Low Applicability Concern
Justification is as follows:
The main limitation of this study lies in the modeling domain, where internal validation and data-driven biomarker selection raise a moderate risk of overfitting. Applicability is strong, the predictors, outcomes, and setting reflect real-world nephrology practice, making the findings highly relevant but needing external validation in independent CKD populations.
Table A9.
PROBAST assessment for Pizzini et al., 2017 [
27].
| Domain/Item | Details | RoB/Concern |
|---|
| Population | 118 adult CKD patients (mean age 62 ± 11 years; 59% male; mean eGFR ≈ 35 mL/min/1.73 m2) from a single nephrology center in Reggio Calabria, Italy. Follow-up: 3 years. Outcome: composite renal endpoint (eGFR decline > 30%, dialysis, or transplantation). Pilot cohort, relatively small sample size, and unclear recruitment method. | ![Biomedinformatics 05 00067 i004 Biomedinformatics 05 00067 i004]() Moderate, Clinically relevant CKD population but small, single-center, and possibly non-representative sample. Moderate, Clinically relevant CKD population but small, single-center, and possibly non-representative sample. |
| Index Model | Composite tubular risk score derived from urinary NGAL, Uromodulin, and KIM-1 (binary: above/below median). Developed via multiple Cox regression, combined later with eGFR in an integrated model. Internal performance assessed via Harrell’s C-index (0.79 vs. 0.77 for eGFR alone). | ![Biomedinformatics 05 00067 i004 Biomedinformatics 05 00067 i004]() Moderate, Simple derivation method, but internal-only validation, small sample, and data-driven thresholding raise overfitting risk. Moderate, Simple derivation method, but internal-only validation, small sample, and data-driven thresholding raise overfitting risk. |
| Comparator/Reference Model | eGFR-based model (single predictor) used as the reference comparator for assessing incremental prognostic value. | ![Biomedinformatics 05 00067 i001 Biomedinformatics 05 00067 i001]() Low, eGFR is a gold-standard clinical reference for kidney function. Low, eGFR is a gold-standard clinical reference for kidney function. |
| Outcome | Composite renal outcome: >30% eGFR decline, dialysis, or transplantation during 3 years. Objectively defined and clinically relevant. | ![Biomedinformatics 05 00067 i001 Biomedinformatics 05 00067 i001]() Low, Outcome is standardized, measurable, and clinically meaningful. Low, Outcome is standardized, measurable, and clinically meaningful. |
| Timing | Prospective follow-up of 3 years; predictors (urinary biomarkers) measured at baseline, outcome assessed longitudinally. | ![Biomedinformatics 05 00067 i001 Biomedinformatics 05 00067 i001]() Low, Appropriate temporal relationship between predictors and outcome. Low, Appropriate temporal relationship between predictors and outcome. |
| Setting | Academic nephrology and renal transplantation unit; research laboratory with validated urinary biomarker assays. | ![Biomedinformatics 05 00067 i001 Biomedinformatics 05 00067 i001]() Low, Controlled clinical and analytical environment ensures reliable data quality. Low, Controlled clinical and analytical environment ensures reliable data quality. |
| Intended Use of the Prediction Model | Early risk stratification of CKD patients for rapid progression or kidney failure, to complement eGFR-based clinical prediction tools. | ![Biomedinformatics 05 00067 i001 Biomedinformatics 05 00067 i001]() Low, Intended use aligns with nephrology practice and unmet clinical need. Low, Intended use aligns with nephrology practice and unmet clinical need. |
Table A9 integrates PROBAST domains with the study-specific details (population, predictors, comparator, outcome, timing, setting, intended use) for a clear, structured assessment of the journal publication, Pizzini et al., 2017 [
27].
The overall PROBAST judgment for this study is:
![Biomedinformatics 05 00067 i004 Biomedinformatics 05 00067 i004]()
Moderate Overall RoB with
![Biomedinformatics 05 00067 i001 Biomedinformatics 05 00067 i001]()
Low Applicability Concern
Justification is as follows:
The single-center design, small sample size, and lack of external validation introduce a moderate RoB, particularly in model development and analysis. Applicability concerns are low, as the predictors (urinary NGAL, Uromodulin, KIM-1) and outcomes reflect real-world CKD progression assessment.
Table A10.
PROBAST assessment for Qin et al., 2019 [
28].
| Domain | Description | RoB | Applicability Concern |
|---|
| Population | 1053 hospitalized adults with type 2 diabetes; after PSM, 500 (250 DKD, 250 non-DKD). All had eGFR ≥ 60 mL/min/1.73 m2. Excluded other kidney or systemic diseases. Hospital-based inpatient cohort, not representative of general outpatient T2DM populations. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate Moderate | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High, inpatient sample limits generalizability to screening or primary-care settings. High, inpatient sample limits generalizability to screening or primary-care settings. |
| Index Model/Predictors | Six urinary biomarkers measured once: transferrin (TF), immunoglobulin G (IgG), retinol-binding protein (RBP), β-galactosidase (GAL), N-acetyl-β-glucosaminidase (NAG), β2-microglobulin (β2MG). Each assessed individually with logistic regression and ROC AUC; no multivariable or externally validated model. | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High, simple univariable analysis; no validation or adjustment for overfitting. High, simple univariable analysis; no validation or adjustment for overfitting. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate, biomarkers clinically measurable but not yet standardized for DKD diagnosis. Moderate, biomarkers clinically measurable but not yet standardized for DKD diagnosis. |
| Comparator model/Reference Standard | 24 h urinary albumin excretion (UAE ≥ 30 mg/24 h) as gold standard for DKD. Overlaps mechanistically with some predictors, introducing incorporation bias. | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High, predictor-outcome dependency likely inflates AUCs. High, predictor-outcome dependency likely inflates AUCs. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate, UAE widely accepted but not ideal for early-stage DKD reference. Moderate, UAE widely accepted but not ideal for early-stage DKD reference. |
| Outcome | Presence of DKD (vs. normoalbuminuric) is defined cross-sectionally by 24 h UAE and eGFR ≥ 60. No longitudinal follow-up. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate, objective lab-based outcome but lacks temporal dimension. Moderate, objective lab-based outcome but lacks temporal dimension. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate, relevant to early DKD diagnosis but not progression prediction. Moderate, relevant to early DKD diagnosis but not progression prediction. |
| Timing | Cross-sectional; biomarkers and UAE measured concurrently during hospitalization (no temporal validation). | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High High | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High, not predictive; diagnostic only. High, not predictive; diagnostic only. |
| Setting | Single tertiary hospital (Tianjin Medical University Chu Hsien-I Memorial Hospital), China, 2018. All assays in hospital lab. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate Moderate | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High, single-center; potential institutional bias. High, single-center; potential institutional bias. |
| Intended Use of prediction model | Exploratory diagnostic discrimination to identify DKD among known T2DM inpatients with preserved eGFR. Not a prognostic or screening model. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate, appropriate for hypothesis generation only. Moderate, appropriate for hypothesis generation only. | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High, limited use beyond internal diagnostic context. High, limited use beyond internal diagnostic context. |
| Overall Judgment | Cross-sectional single-center study with internal ROC analysis only. High internal performance (RBP AUC 0.92) is likely optimistic. No calibration, temporal, or external validation performed. | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High overall RoB High overall RoB | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High applicability concern High applicability concern |
Table A10 integrates PROBAST domains with the study-specific details (population, predictors, comparator, outcome, timing, setting, intended use) for a clear, structured assessment of the journal publication, Qin et al., 2019 [
28].
This cross-sectional diagnostic study assessed six urinary biomarkers for detecting DKD among hospitalized adults with type 2 diabetes in Tianjin, China. Using 24 h urinary albumin excretion as the reference, RBP, TF, and IgG showed the best discrimination (AUCs of 0.92, 0.87, and 0.87, respectively). However, methodological appraisal with PROBAST indicates a high overall RoB due to the cross-sectional design, incorporation bias between biomarkers and outcome, and absence of external validation. Applicability is limited to hospital-based diagnostic research settings rather than predictive clinical screening or community-based use.
Table A11.
QUADAS-2 RoB and applicability concerns for Schanstra et al., 2015 [
29].
| Domain | Description | RoB | Applicability Concern |
|---|
| Patient Selection | Large multicenter cohort (n = 1990) including CKD patients across stages and healthy/at-risk controls; subset (n = 522) had longitudinal follow-up for progression. Inclusion criteria are broad and clinically relevant. Unclear if sampling was consecutive or random, though selection likely minimized spectrum bias by including multiple CKD etiologies. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Low-Moderate Low-Moderate | ![Biomedinformatics 05 00067 i001 Biomedinformatics 05 00067 i001]() Low, representative CKD and at-risk populations suitable for diagnostic and prognostic use. Low, representative CKD and at-risk populations suitable for diagnostic and prognostic use. |
| Index Test | Urinary multi-peptide biomarker classifier derived from proteomic analysis; algorithm validated internally and externally across centers. Index test interpreted blinded to reference measures (as implied). Uses objective mass-spectrometry-based quantification. | ![Biomedinformatics 05 00067 i001 Biomedinformatics 05 00067 i001]() Low, standardized proteomic measurement, objective analysis. Low, standardized proteomic measurement, objective analysis. | ![Biomedinformatics 05 00067 i001 Biomedinformatics 05 00067 i001]() Low, proteomic test applicable to intended CKD risk stratification setting. Low, proteomic test applicable to intended CKD risk stratification setting. |
| Reference Standard | Clinical CKD diagnosis and progression assessed by eGFR decline and/or albuminuria according to accepted criteria (Kidney Disease: Improving Global Outcome/KDIGO). Both objective and reproducible. However, albuminuria overlaps mechanistically with the index peptides, introducing some dependency. | ![Biomedinformatics 05 00067 i004 Biomedinformatics 05 00067 i004]() Moderate, potential incorporation bias with overlapping filtration markers. Moderate, potential incorporation bias with overlapping filtration markers. | ![Biomedinformatics 05 00067 i001 Biomedinformatics 05 00067 i001]() Low, consistent with clinical standards for CKD staging and progression. Low, consistent with clinical standards for CKD staging and progression. |
| Flow and Timing | Cross-sectional design for detection with a subset followed longitudinally (n = 522) for progression; uniform application of index and reference tests at baseline; consistent follow-up for outcome. | ![Biomedinformatics 05 00067 i001 Biomedinformatics 05 00067 i001]() Low, flow appropriate and timing consistent. Low, flow appropriate and timing consistent. | ![Biomedinformatics 05 00067 i001 Biomedinformatics 05 00067 i001]() Low, progression analysis based on follow-up data aligns with intended use. Low, progression analysis based on follow-up data aligns with intended use. |
| Overall RoB | Generally robust multicenter design with standardized proteomic measurement and appropriate statistical validation. Minor risk from partial overlap between index and reference measures. | ![Biomedinformatics 05 00067 i001 Biomedinformatics 05 00067 i001]() Low overall Low overall | ![Biomedinformatics 05 00067 i001 Biomedinformatics 05 00067 i001]() Low overall Low overall |
Table A11 presents the application of the QUADAS-2 tool for the RoB and applicability concerns assessment of one of the latter two studies [
29], utilizing various urinary biomarkers as input variables for various AI-based diagnostic tools for the early detection, risk stratification, and monitoring of CKD.
QUADAS-2 appraisal indicates an overall low RoB and good applicability, supported by rigorous proteomic quantification and validation across multiple centers. Minor concerns remain about partial incorporation bias since the reference standard (albuminuria, eGFR) overlaps biologically with some peptides. The study provides strong diagnostic and prognostic evidence for urinary proteome classifiers as complementary CKD risk stratification tools.
Table A12.
QUADAS-2 for RoB and applicability assessment for Muiru et al., 2021 [
30].
| Domain | Description | RoB | Applicability Concerns |
|---|
| 1. Patient Selection | Participants were drawn from the WIHS prospective cohort of women with HIV; inclusion required preserved kidney function (eGFR ≥ 60 mL/min/1.73 m2) and paired urine samples. | ![Biomedinformatics 05 00067 i005 Biomedinformatics 05 00067 i005]() Low-Moderate, selection limited to relatively healthy women, potentially introducing bias. Low-Moderate, selection limited to relatively healthy women, potentially introducing bias. | ![Biomedinformatics 05 00067 i005 Biomedinformatics 05 00067 i005]() Moderate, mostly middle-aged Black women with HIV; may not represent general CKD or HIV-positive male populations. Moderate, mostly middle-aged Black women with HIV; may not represent general CKD or HIV-positive male populations. |
| 2. Index Test (Urine Biomarkers) | 14 urine biomarkers measured in duplicate using standardized multiplex assays; results analyzed as continuous standardized values without diagnostic thresholds. | ![Biomedinformatics 05 00067 i005 Biomedinformatics 05 00067 i005]() Low-Moderate, laboratory methods robust, but no pre-specified diagnostic cut-offs. Low-Moderate, laboratory methods robust, but no pre-specified diagnostic cut-offs. | ![Biomedinformatics 05 00067 i005 Biomedinformatics 05 00067 i005]() Some concern, biomarkers used as exploratory indicators, not validated diagnostic tests. Some concern, biomarkers used as exploratory indicators, not validated diagnostic tests. |
| 3. Reference Standard | No true diagnostic “gold standard” for CKD; comparisons made to CKD risk factors (HbA1c, BP, viral load, etc.) rather than confirmed CKD diagnosis. | ![Biomedinformatics 05 00067 i006 Biomedinformatics 05 00067 i006]() High, absence of a defined reference standard limits diagnostic accuracy assessment. High, absence of a defined reference standard limits diagnostic accuracy assessment. | ![Biomedinformatics 05 00067 i006 Biomedinformatics 05 00067 i006]() High, reference variables do not constitute a diagnostic criterion for CKD. High, reference variables do not constitute a diagnostic criterion for CKD. |
| 4. Flow and Timing | Baseline and follow-up urine and serum specimens obtained 2.5 years apart for all 647 participants; consistent measurements across time points. | ![Biomedinformatics 05 00067 i007 Biomedinformatics 05 00067 i007]() Low, clear temporal structure and uniform application of tests. Low, clear temporal structure and uniform application of tests. | ![Biomedinformatics 05 00067 i007 Biomedinformatics 05 00067 i007]() Low, appropriate interval and consistent follow-up across participants. Low, appropriate interval and consistent follow-up across participants. |
The study demonstrates rigorous biomarker measurement and statistical modeling, but it is more exploratory (prognostic/associative) than diagnostic, thus QUADAS-2 partially applies. We applied QUADAS-2 principles to evaluate quality in diagnostic/biomarker inference. Since the QUADAS-2 framework evaluates the RoB and applicability concerns across four key domains, we explored how this study aligns with each domain:
Overall RoB for Domain 1: Low to moderate
![Biomedinformatics 05 00067 i005 Biomedinformatics 05 00067 i005]()
Overall RoB for Domain 2: Low to moderate
![Biomedinformatics 05 00067 i005 Biomedinformatics 05 00067 i005]()
Table A15.
QUADAS-2 for RoB and applicability assessment for Muiru et al., 2021 [
30], Domain 3: Reference Standard.
| Criterion | Assessment | Justification |
|---|
| Is the reference standard likely to correctly classify the target condition? | ![Biomedinformatics 05 00067 i009 Biomedinformatics 05 00067 i009]() High risk High risk | The study does not use a clinical diagnosis or gold-standard CKD outcome, only risk factors and biomarker change correlations. |
| Were the reference standard results interpreted without knowledge of the index test results? | ![Biomedinformatics 05 00067 i008 Biomedinformatics 05 00067 i008]() Low risk Low risk | Risk factors (A1c, blood pressure, HIV viral load, etc.) were measured independently of biomarkers. |
| Applicability concern | ![Biomedinformatics 05 00067 i009 Biomedinformatics 05 00067 i009]() High High | The “reference standard” here is not a diagnostic truth measure, so diagnostic accuracy cannot be directly evaluated. |
Overall RoB for Domain 3: High
![Biomedinformatics 05 00067 i006 Biomedinformatics 05 00067 i006]()
(not applicable as a true diagnostic accuracy study)
Overall RoB for Domain 4: Low
Overall RoB for this study: Moderate to High
Overall Applicability of this study: Moderate
Since this study is methodologically sound for associative/prognostic biomarker research but not a diagnostic accuracy study, and QUADAS-2 applies only partially, we also used the PROBAST tool to assess RoB and the applicability of this study.
Table A17.
PROBAST quality assessment for Muiru et al., 2021 [
30].
| Domain | Description | RoB | Applicability Concerns |
|---|
| Participants/Population | 647 women living with HIV from the U.S. Women’s Interagency HIV Study (WIHS). Inclusion required two urine samples and preserved kidney function (eGFR ≥ 60 mL/min/1.73 m2). Majority were middle-aged and Black (67%). | ![Biomedinformatics 05 00067 i005 Biomedinformatics 05 00067 i005]() Some concern, selection limited to women with preserved renal function; may not represent patients with advanced CKD or male populations. Some concern, selection limited to women with preserved renal function; may not represent patients with advanced CKD or male populations. | ![Biomedinformatics 05 00067 i005 Biomedinformatics 05 00067 i005]() Moderate, findings apply mainly to women with HIV and may not generalize to all HIV or CKD populations. Moderate, findings apply mainly to women with HIV and may not generalize to all HIV or CKD populations. |
| Index Model | Multivariable penalized regression model (MSG-LASSO) and simultaneous linear equations assessing associations between CKD risk factors and longitudinal changes in 14 urine biomarkers. | ![Biomedinformatics 05 00067 i005 Biomedinformatics 05 00067 i005]() Some concern, robust internal modeling, but no external or temporal validation; unclear internal validation (e.g., bootstrapping). Some concern, robust internal modeling, but no external or temporal validation; unclear internal validation (e.g., bootstrapping). | ![Biomedinformatics 05 00067 i005 Biomedinformatics 05 00067 i005]() Moderate, model exploratory, not clinically implemented; predictive performance not reported. Moderate, model exploratory, not clinically implemented; predictive performance not reported. |
| Comparator Model | None, no existing or alternative predictive model used for comparison; focus was on evaluating associations, not on model performance metrics. | ![Biomedinformatics 05 00067 i006 Biomedinformatics 05 00067 i006]() High, absence of comparator limits interpretation of predictive improvement or incremental value. High, absence of comparator limits interpretation of predictive improvement or incremental value. | ![Biomedinformatics 05 00067 i005 Biomedinformatics 05 00067 i005]() Some concern, not designed for model comparison or validation. Some concern, not designed for model comparison or validation. |
| Outcome | Longitudinal changes in kidney tubular and glomerular biomarkers (e.g., KIM-1, IL-18, UMOD, α1m, β2m). No hard kidney outcome (CKD progression, eGFR decline) assessed. | ![Biomedinformatics 05 00067 i005 Biomedinformatics 05 00067 i005]() Some concern, surrogate outcomes, not direct measures of kidney disease progression or patient-level endpoints. Some concern, surrogate outcomes, not direct measures of kidney disease progression or patient-level endpoints. | ![Biomedinformatics 05 00067 i005 Biomedinformatics 05 00067 i005]() Moderate, outcome biologically meaningful but limited for clinical prediction utility. Moderate, outcome biologically meaningful but limited for clinical prediction utility. |
| Timing | Prospective longitudinal cohort; baseline biomarker and clinical data collected in 2009–2011 and repeated ~2.5 years later. | ![Biomedinformatics 05 00067 i007 Biomedinformatics 05 00067 i007]() Low, consistent and appropriate timing for longitudinal biomarker change evaluation. Low, consistent and appropriate timing for longitudinal biomarker change evaluation. | ![Biomedinformatics 05 00067 i007 Biomedinformatics 05 00067 i007]() Low, timing consistent with biological plausibility for kidney biomarker change. Low, timing consistent with biological plausibility for kidney biomarker change. |
| Setting | Multi-center U.S. observational cohort study (academic research settings). | ![Biomedinformatics 05 00067 i007 Biomedinformatics 05 00067 i007]() Low, standardized data collection and laboratory protocols reduce bias. Low, standardized data collection and laboratory protocols reduce bias. | ![Biomedinformatics 05 00067 i005 Biomedinformatics 05 00067 i005]() Some concern, research setting may differ from clinical practice environments. Some concern, research setting may differ from clinical practice environments. |
| Intended Use of Predictive Model | Exploratory, to identify CKD risk factors associated with biomarker changes and to inform future development of kidney disease detection algorithms in HIV. | ![Biomedinformatics 05 00067 i005 Biomedinformatics 05 00067 i005]() Some concern, model not yet developed for clinical prediction; exploratory by design. Some concern, model not yet developed for clinical prediction; exploratory by design. | ![Biomedinformatics 05 00067 i005 Biomedinformatics 05 00067 i005]() Moderate, informative for biomarker research, but not directly applicable to clinical prediction or screening. Moderate, informative for biomarker research, but not directly applicable to clinical prediction or screening. |
The study demonstrates strong internal validity and robust measurement methods but is limited by a lack of external validation, a restricted population, and the use of biomarkers as surrogate outcomes. It is best viewed as a hypothesis-generating prognostic analysis rather than a finalized predictive model.
Overall RoB: Moderate
Overall Applicability: Moderate
Appendix F
This appendix contains details and explanations supplemental to the subsection, “4.6. Application of AI and ML-based algorithms for identifying and classifying existing diseases and subtypes, and forecasting disease progression and risk stratification” of the main text. The explanations of the quality assessment of the included study articles [
16] through [
23] using the PROBAST and QUADAS-2 tools would disrupt the flow of the main text. However, this discussion is crucial to understanding the overall significance of these studies.
Table A37.
QUADAS-2 RoB and applicability assessment for Lucarelli et al., 2023 [
16].
| Domain | RoB | Applicability Concerns |
|---|
| Patient Selection | ![Biomedinformatics 05 00067 i006 Biomedinformatics 05 00067 i006]() High/Unclear, Selective recruitment (urine proteomics + pathology cohort) with limited reporting on inclusion/exclusion; possible spectrum bias. High/Unclear, Selective recruitment (urine proteomics + pathology cohort) with limited reporting on inclusion/exclusion; possible spectrum bias. | ![Biomedinformatics 05 00067 i010 Biomedinformatics 05 00067 i010]() Moderate, Participants unlikely to reflect general T2DM population; community-clinic applicability uncertain. Moderate, Participants unlikely to reflect general T2DM population; community-clinic applicability uncertain. |
| Index Test (digital biomarkers via urinary proteomics + pathology) | ![Biomedinformatics 05 00067 i010 Biomedinformatics 05 00067 i010]() Moderate/Unclear, Unclear blinding; thresholds not pre-specified; same dataset used for discovery and model training → overfitting risk. Moderate/Unclear, Unclear blinding; thresholds not pre-specified; same dataset used for discovery and model training → overfitting risk. | ![Biomedinformatics 05 00067 i010 Biomedinformatics 05 00067 i010]() Moderate, Uses specialized proteomics pipeline; external platforms may differ → limited generalizability. Moderate, Uses specialized proteomics pipeline; external platforms may differ → limited generalizability. |
| Reference Standard | ![Biomedinformatics 05 00067 i005 Biomedinformatics 05 00067 i005]() Low → Moderate, Pathology-based reference appropriate but possibly heterogeneous (biopsy vs. clinical classification); unclear if consistent across subjects. Low → Moderate, Pathology-based reference appropriate but possibly heterogeneous (biopsy vs. clinical classification); unclear if consistent across subjects. | ![Biomedinformatics 05 00067 i010 Biomedinformatics 05 00067 i010]() Moderate, Standard relevant to DKD but not necessarily identical to routine clinical endpoints. Moderate, Standard relevant to DKD but not necessarily identical to routine clinical endpoints. |
| Flow & Timing | ![Biomedinformatics 05 00067 i006 Biomedinformatics 05 00067 i006]() High, Incomplete information on timing between index and reference tests; unclear follow-up; potential verification bias. High, Incomplete information on timing between index and reference tests; unclear follow-up; potential verification bias. | ![Biomedinformatics 05 00067 i010 Biomedinformatics 05 00067 i010]() Moderate, Specialized research workflow limits applicability to routine timelines and sampling logistics. Moderate, Specialized research workflow limits applicability to routine timelines and sampling logistics. |
| Overall Judgment | ![Biomedinformatics 05 00067 i006 Biomedinformatics 05 00067 i006]() Overall RoB: High, Unclear patient selection + limited reporting on timing increase risk. Overall RoB: High, Unclear patient selection + limited reporting on timing increase risk. | ![Biomedinformatics 05 00067 i010 Biomedinformatics 05 00067 i010]() Overall Applicability: Moderate, Study setting and assay platform differ from standard clinical practice; external validation needed. Overall Applicability: Moderate, Study setting and assay platform differ from standard clinical practice; external validation needed. |
Table A37 is a color-coded QUADAS-2 table for the study: Lucarelli et al., (2023) [
16]; “Discovery of Novel Digital Biomarkers for Type 2 Diabetic Nephropathy Classification via Integration of Urinary Proteomics and Pathology.”
This study represents an innovative early-phase effort to merge urinary proteomics with pathology for digital biomarker discovery in diabetic nephropathy. However, methodological transparency (especially around cohort definition, temporal sequence, and blinding) is limited. The findings are promising but not yet generalizable to multi-site or community settings without external validation and standardized assay calibration.
Table A38.
QUADAS-2 RoB and applicability assessment for Yan et al., 2024 [
17].
| Domain | RoB | Applicability Concerns |
|---|
| Patient Selection | ![Biomedinformatics 05 00067 i005 Biomedinformatics 05 00067 i005]() Low → Moderate, Participants included clear diagnostic categories (T2DM with/without DKD), but sampling method and exclusion criteria were not fully described; possible selection bias if matched retrospectively. Low → Moderate, Participants included clear diagnostic categories (T2DM with/without DKD), but sampling method and exclusion criteria were not fully described; possible selection bias if matched retrospectively. | ![Biomedinformatics 05 00067 i007 Biomedinformatics 05 00067 i007]() Low, DKD and control populations clinically relevant; findings likely applicable to standard T2DM cohorts with albuminuria-based classification. Low, DKD and control populations clinically relevant; findings likely applicable to standard T2DM cohorts with albuminuria-based classification. |
| Index Test (Urinary proteomics + ML classifiers) | ![Biomedinformatics 05 00067 i010 Biomedinformatics 05 00067 i010]() Moderate, ML approach (e.g., SVM, RF) applied without fully independent test set; unclear blinding to reference standard; internal validation only. Moderate, ML approach (e.g., SVM, RF) applied without fully independent test set; unclear blinding to reference standard; internal validation only. | ![Biomedinformatics 05 00067 i010 Biomedinformatics 05 00067 i010]() Moderate, Omics workflows and normalization pipelines may not generalize across assay platforms; batch effects could limit external use. Moderate, Omics workflows and normalization pipelines may not generalize across assay platforms; batch effects could limit external use. |
| Reference Standard | ![Biomedinformatics 05 00067 i007 Biomedinformatics 05 00067 i007]() Low, Diagnostic definitions followed established DKD criteria (albuminuria, eGFR decline); reference standard appropriate and consistently applied. Low, Diagnostic definitions followed established DKD criteria (albuminuria, eGFR decline); reference standard appropriate and consistently applied. | ![Biomedinformatics 05 00067 i007 Biomedinformatics 05 00067 i007]() Low, Reference outcomes align well with clinical practice and guidelines. Low, Reference outcomes align well with clinical practice and guidelines. |
| Flow & Timing | ![Biomedinformatics 05 00067 i010 Biomedinformatics 05 00067 i010]() Moderate, Timing between urine collection and DKD classification not explicitly stated; unclear if temporal gaps could bias associations. Moderate, Timing between urine collection and DKD classification not explicitly stated; unclear if temporal gaps could bias associations. | ![Biomedinformatics 05 00067 i007 Biomedinformatics 05 00067 i007]() Low → Moderate, Acceptable for cross-sectional biomarker screening but limited for longitudinal prediction. Low → Moderate, Acceptable for cross-sectional biomarker screening but limited for longitudinal prediction. |
| Overall Judgment | ![Biomedinformatics 05 00067 i010 Biomedinformatics 05 00067 i010]() Overall RoB: Moderate, Limited external validation and potential for overfitting; otherwise methodologically reasonable. Overall RoB: Moderate, Limited external validation and potential for overfitting; otherwise methodologically reasonable. | ![Biomedinformatics 05 00067 i007 Biomedinformatics 05 00067 i007]() Overall Applicability: Low Concern, Results relevant to typical DKD diagnostic settings, but omics reproducibility remains a constraint. Overall Applicability: Low Concern, Results relevant to typical DKD diagnostic settings, but omics reproducibility remains a constraint. |
Table A38 is a color-coded QUADAS-2 evaluation for the study: Yan et al., (2024) [
17]; “Application of Proteomics and Machine Learning Methods to Study the Pathogenesis of Diabetic Nephropathy and Screen Urinary Biomarkers.”
This study follows a single-center cohort. External validation and transparent reporting of temporal design are still needed to ensure trustworthy real-world deployment. The overall evidence is methodologically moderate with good clinical applicability.
Table A39.
PROBAST assessment for Dong et al., 2022 [
18].
| Domain | Description | RoB | Applicability Concern |
|---|
| Population/Participants | Adults with type 2 diabetes extracted from a large hospital EMR database in China; patients were followed for up to 3 years for DKD onset. Clear inclusion/exclusion criteria with sufficient baseline data. | ![Biomedinformatics 05 00067 i007 Biomedinformatics 05 00067 i007]() Low Low | ![Biomedinformatics 05 00067 i007 Biomedinformatics 05 00067 i007]() Low Low |
| Index Model | ML-based predictive models (RF, XGBoost, Logistic Regression) using routine EMR data (labs, demographics, comorbidities, medication, BP, BMI, eGFR, etc.) to predict 3-year risk of diabetic kidney disease. | ![Biomedinformatics 05 00067 i005 Biomedinformatics 05 00067 i005]() Moderate Moderate | ![Biomedinformatics 05 00067 i007 Biomedinformatics 05 00067 i007]() Low Low |
| Comparator Model | Traditional logistic regression models used as baseline comparators for performance benchmarking. | ![Biomedinformatics 05 00067 i007 Biomedinformatics 05 00067 i007]() Low Low | ![Biomedinformatics 05 00067 i007 Biomedinformatics 05 00067 i007]() Low Low |
| Outcomes | Onset of diabetic kidney disease (DKD) within 3 years, defined by KDIGO criteria (eGFR < 60 mL/min/1.73 m2 or UACR >30 mg/g). | ![Biomedinformatics 05 00067 i007 Biomedinformatics 05 00067 i007]() Low Low | ![Biomedinformatics 05 00067 i007 Biomedinformatics 05 00067 i007]() Low Low |
| Timing | Predictors measured at baseline; 3-year prediction horizon. However, it is unclear if temporal data splits (e.g., patient-level chronological separation) were strictly applied. | ![Biomedinformatics 05 00067 i005 Biomedinformatics 05 00067 i005]() Moderate Moderate | ![Biomedinformatics 05 00067 i005 Biomedinformatics 05 00067 i005]() Moderate Moderate |
| Setting | Single tertiary hospital EMR database (China); retrospective design. No external validation or community-level testing reported. | ![Biomedinformatics 05 00067 i006 Biomedinformatics 05 00067 i006]() High High | ![Biomedinformatics 05 00067 i005 Biomedinformatics 05 00067 i005]() Moderate Moderate |
| Intended Use of Predictive Model | Clinical decision support for early identification of high-risk DKD patients among those with type 2 diabetes; potentially guiding earlier interventions or referrals. | ![Biomedinformatics 05 00067 i007 Biomedinformatics 05 00067 i007]() Low Low | ![Biomedinformatics 05 00067 i007 Biomedinformatics 05 00067 i007]() Low Low |
| Statistical Analysis | Multiple ML algorithms compared. Data randomly split into training/testing sets; AUROC ≈ 0.86 reported. No external validation. No calibration curve, intercept, or DCA. Imputation and feature selection methods are not fully detailed, increasing overfitting risk. | ![Biomedinformatics 05 00067 i006 Biomedinformatics 05 00067 i006]() High High | ![Biomedinformatics 05 00067 i005 Biomedinformatics 05 00067 i005]() Moderate Moderate |
| Overall Judgment | Moderate-to-High RoB, Model trained and tested internally with strong discrimination but limited evidence of calibration, generalizability, and robustness to domain shift. Applicability concerns low, as EMR-based predictors are clinically relevant and reproducible. | ![Biomedinformatics 05 00067 i010 Biomedinformatics 05 00067 i010]() Moderate-High Moderate-High | ![Biomedinformatics 05 00067 i007 Biomedinformatics 05 00067 i007]() Low Low |
Table A39 is a structured PROBAST-style summary table for Dong et al. (2022) [
18].
Key Limitations of this study include a lack of external validation, unclear handling of temporal dependencies, and incomplete reporting of calibration.
Key Strengths of this study include a large, representative EMR cohort with a clinically meaningful 3-year DKD outcome.
Table A40.
PROBAST assessment for Hsu et al. 2023 [
19].
| Domain | Description | RoB | Applicability Concern |
|---|
| Population/Participants | Adult patients with type 2 diabetes from a large hospital network in Taiwan. Dataset derived from electronic medical records (EMRs) between 2012–2021. Exclusions applied for pre-existing ESRD or missing baseline renal data. | ![Biomedinformatics 05 00067 i007 Biomedinformatics 05 00067 i007]() Low Low | ![Biomedinformatics 05 00067 i007 Biomedinformatics 05 00067 i007]() Low Low |
| Index Model | Ensemble ML models (RF, XGBoost, Light GBM) built to predict risk of rapidly progressive kidney disease (RPKD), defined as ≥30% eGFR decline within 2 years, and identify patients needing nephrology referral. | ![Biomedinformatics 05 00067 i007 Biomedinformatics 05 00067 i007]() Low Low | ![Biomedinformatics 05 00067 i007 Biomedinformatics 05 00067 i007]() Low Low |
| Comparator Model | Traditional logistic regression and Cox regression are used as comparators. ML models outperformed conventional methods with higher AUROC values. | ![Biomedinformatics 05 00067 i007 Biomedinformatics 05 00067 i007]() Low Low | ![Biomedinformatics 05 00067 i007 Biomedinformatics 05 00067 i007]() Low Low |
| Outcomes | RPKD and nephrology referral within a 2-year period, defined using KDIGO-based eGFR decline thresholds and clinician referrals. Clear, clinically relevant endpoint definitions. | ![Biomedinformatics 05 00067 i007 Biomedinformatics 05 00067 i007]() Low Low | ![Biomedinformatics 05 00067 i007 Biomedinformatics 05 00067 i007]() Low Low |
| Timing | Predictors collected at baseline; follow-up period of up to 2 years. However, it is unclear whether patient-level temporal splits were enforced, or random sampling used for validation. | ![Biomedinformatics 05 00067 i005 Biomedinformatics 05 00067 i005]() Moderate Moderate | ![Biomedinformatics 05 00067 i005 Biomedinformatics 05 00067 i005]() Moderate Moderate |
| Setting | Retrospective single-center EMR dataset from a tertiary care hospital. While data volume was high, external validation or community-level generalizability testing was not reported. | ![Biomedinformatics 05 00067 i006 Biomedinformatics 05 00067 i006]() High High | ![Biomedinformatics 05 00067 i005 Biomedinformatics 05 00067 i005]() Moderate Moderate |
| Intended Use of Predictive Model | Designed to assist clinicians in early identification of diabetic patients at high risk for RPKD, to optimize referral timing and monitoring intensity. Potential clinical decision-support tool. | ![Biomedinformatics 05 00067 i007 Biomedinformatics 05 00067 i007]() Low Low | ![Biomedinformatics 05 00067 i007 Biomedinformatics 05 00067 i007]() Low Low |
| Statistical Analysis | Compared multiple ML models; best AUROC ≈ 0.89 (XGBoost). Used cross-validation for internal evaluation. Calibration metrics not clearly reported, and decision-curve analysis absent. Missing data handling and normalization is not fully described. | ![Biomedinformatics 05 00067 i005 Biomedinformatics 05 00067 i005]() Moderate Moderate | ![Biomedinformatics 05 00067 i005 Biomedinformatics 05 00067 i005]() Moderate Moderate |
| Overall Judgment | Moderate RoB. Model performance was strong (AUROC ≈ 0.89), but limited transparency on calibration, imputation, and temporal validation reduces real-world reliability. Applicability concerns are low, given the model’s clinically relevant predictors and outcomes, but external validation remains a key gap. | ![Biomedinformatics 05 00067 i005 Biomedinformatics 05 00067 i005]() Moderate Moderate | ![Biomedinformatics 05 00067 i007 Biomedinformatics 05 00067 i007]() Low Low |
Table A40 is a PROBAST evaluation of the study, Hsu et al., 2023 [
19].
The real-world deployment of the Hsu et al. 2023 [
19] model is limited by a lack of external validation and by incomplete reporting of calibration and data hygiene practices. Although the study demonstrates promise for supporting physicians’ clinical decisions, it still needs broader testing across diverse clinical settings, from tertiary-level hospitals to local clinics.
Table A41.
QUADAS-2 RoB and applicability concerns assessment for Paranjpe et al., 2023 [
20].
| Domain | Description/Key Details | RoB | Applicability Concerns |
|---|
| Patient Selection | Retrospective EMR data from large academic centers; inclusion criteria focused on diabetic kidney disease (DKD) with genotyping data available. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate risk, retrospective design may introduce selection bias and missingness in clinical-genetic linkage. Moderate risk, retrospective design may introduce selection bias and missingness in clinical-genetic linkage. | ![Biomedinformatics 05 00067 i001 Biomedinformatics 05 00067 i001]() Low concern, representative of tertiary diabetic populations. Low concern, representative of tertiary diabetic populations. |
| Index Test (Deep Learning Model) | DL model trained on multimodal EMR data with genomic integration to identify DKD sub-phenotypes linked to the Rho pathway. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate risk, details of model tuning and cross-validation split not fully reported; unclear if feature leakage was avoided. Moderate risk, details of model tuning and cross-validation split not fully reported; unclear if feature leakage was avoided. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate concern, algorithm may overfit tertiary-center EMR data, limiting community translation. Moderate concern, algorithm may overfit tertiary-center EMR data, limiting community translation. |
| Reference Standard | Genetic and clinical phenotyping used to define DKD subtypes; no independent gold-standard confirmation (e.g., biopsy). | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High risk, absence of external biological validation may weaken subtype credibility. High risk, absence of external biological validation may weaken subtype credibility. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate concern, relies on EMR and genetic proxies rather than clinical pathology confirmation. Moderate concern, relies on EMR and genetic proxies rather than clinical pathology confirmation. |
| Flow and Timing | Cross-sectional integration of EMR and genomic data; unclear timing alignment between phenotype and genotype capture. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate risk, potential temporal mismatch between clinical and genomic data points. Moderate risk, potential temporal mismatch between clinical and genomic data points. | ![Biomedinformatics 05 00067 i001 Biomedinformatics 05 00067 i001]() Low concern, overall timing reasonable for computational phenotyping. Low concern, overall timing reasonable for computational phenotyping. |
| Statistical Analysis | DL interpretability limited; performance metrics (e.g., AUC or clustering validity indices) partially reported; no external validation cohort. | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High risk, limited transparency and missing calibration or generalizability metrics. High risk, limited transparency and missing calibration or generalizability metrics. | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High concern, lack of independent validation and reproducibility assessment. High concern, lack of independent validation and reproducibility assessment. |
| Overall Judgment | The study offers valuable biological insight into DKD subtypes through EMR-genomic integration, but incomplete reporting and lack of external validation raise serious concerns about reproducibility and clinical generalizability. | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High overall RoB High overall RoB | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate applicability concern Moderate applicability concern |
Table A41 is a color-coded QUADAS-2 assessment table for the study by Paranjpe et al. (2023) [
20], designed for clarity.
Paranjpe et al. (2023) [
20] introduce an ambitious DL approach that links EMR and genomic data to uncover DKD subtypes, but transparency gaps and a lack of external validation limit its current clinical reliability. The findings are hypothesis-generating rather than ready for real-world application.
Table A42.
PROBAST assessment for Xu et al., 2020 [
21].
| PROBAST Item | Description/Assessment | RoB | Applicability Concerns |
|---|
| Study Type | Systematic literature review of ML models predicting diabetic microvascular complications (retinopathy, nephropathy, neuropathy) in Type 1 Diabetes Mellitus (T1DM). | - | - |
| Population/Participants | Participants were individuals with T1DM, from heterogeneous study cohorts (pediatric/adult, different disease durations, varying inclusion criteria). Lack of uniformity and representativeness. | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High High | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High High |
| Index Model(s) | Various ML algorithms, SVM, RF, Artificial Neural Networks (ANN), Decision Trees, etc. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Unclear Unclear | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Unclear Unclear |
| Comparator Model(s) | Some studies compared ML models with logistic regression or classical statistical methods; no consistent comparator and no meta-analysis. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Unclear Unclear | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Unclear Unclear |
| Outcome(s) | Diabetic retinopathy, nephropathy, and neuropathy. Outcome definitions and diagnostic criteria varied across studies. Limited reporting on outcome ascertainment. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Unclear Unclear | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Unclear Unclear |
| Timing | Prediction horizons varied (cross-sectional, retrospective cohort). No standardized follow-up intervals or prediction windows. | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High High | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate Moderate |
| Setting | Studies conducted in mixed clinical and research settings (hospital registries, EMR datasets). Contextual details are often missing. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Unclear Unclear | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Unclear Unclear |
| Intended Use | To support early detection and risk stratification for diabetic microvascular complications in T1DM, potentially informing personalized clinical management. | - | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate Moderate |
| Predictors | Predictors included demographic (age, sex), clinical (HbA1c, BP, duration of diabetes), lab (lipids, creatinine), and imaging (retinal images). Predictor handling poorly described and inconsistent. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Unclear Unclear | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate Moderate |
| Statistical/Modeling Analysis | Qualitative synthesis only. Most studies reported accuracy and AUC, but few addressed calibration, missing data, or validation. No pooled statistics or sensitivity analyses. | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High High | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High High |
| Domain 1, Participants | Populations varied; inclusion criteria and recruitment are often unclear; potential selection bias. | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High High | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High High |
| Domain 2, Predictors | Inconsistent predictor definitions and processing; limited reporting of feature selection and handling of missing data. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Unclear Unclear | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate Moderate |
| Domain 3, Outcome | Outcomes inconsistently defined; validation methods not standardized; blinding rarely reported. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Unclear Unclear | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate Moderate |
| Domain 4, Analysis | No formal validation in many studies; high risk of overfitting; small datasets; selective reporting of favorable results. | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High High | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High High |
| Overall RoB | Methodological transparency limited; heterogeneity high; no structured bias tool (like PROBAST) used in the review. | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High High | - |
| Overall Applicability | Useful overview of ML research trends but limited generalizability or clinical utility due to lack of validation and standardization. | - | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate Moderate |
| Overall PROBAST Judgment | High RoB; moderate applicability concerns. Review descriptive but insufficient for clinical adoption of ML models in T1DM complications. | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High High | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate Moderate |
Table A42 is a structured PROBAST summary table including both methodological domains and key study characteristics for the study, Xu et al., 2020 [
21].
Xu et al. (2020) [
21] provide a broad overview of ML applications in diabetic complication prediction. But the overall PROBAST rating is high RoB, with moderate concerns about applicability. The real-world deployment of this study is limited by a lack of structured RoB assessment, poor reporting of model development and validation across included studies, and substantial heterogeneity in predictors, outcomes, and data sources.
Table A43.
QUADAS-2 RoB and applicability assessment for Dong et al., 2024 [
22].
| Domain | Key Questions/Description | RoB | Applicability Concerns |
|---|
| 1. Patient Selection | Did the review include studies enrolling representative participants? Were inclusion/exclusion criteria clearly defined and appropriate for DN biomarker discovery? | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Unclear risk, The review included studies of diabetic patients with and without nephropathy, but inclusion criteria across studies varied widely (different stages of DN, different diabetes types, and demographic variability). No explicit description of how participants were selected in included studies. Unclear risk, The review included studies of diabetic patients with and without nephropathy, but inclusion criteria across studies varied widely (different stages of DN, different diabetes types, and demographic variability). No explicit description of how participants were selected in included studies. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Unclear concern, Applicability limited by heterogeneity in populations (T1DM vs. T2DM, ethnic differences, sample sources). Unclear concern, Applicability limited by heterogeneity in populations (T1DM vs. T2DM, ethnic differences, sample sources). |
| 2. Index Test (MLModels/Biomarker Identification Methods) | Were ML methods described in sufficient detail to permit replication? Was model performance validated appropriately? | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High risk, The review summarized various ML approaches (e.g., RF, SVM, LASSO), but many primary studies lacked validation procedures or transparent performance metrics. The review did not critically appraise algorithm robustness or validation quality. High risk, The review summarized various ML approaches (e.g., RF, SVM, LASSO), but many primary studies lacked validation procedures or transparent performance metrics. The review did not critically appraise algorithm robustness or validation quality. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate concern, Some ML models aimed to identify candidate biomarkers (not diagnostic tools), so their direct clinical applicability is limited. Moderate concern, Some ML models aimed to identify candidate biomarkers (not diagnostic tools), so their direct clinical applicability is limited. |
| 3. Reference Standard (Diagnosis of Diabetic Nephropathy) | Was DN defined and confirmed consistently across included studies? Was the reference standard likely to correctly classify the condition? | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Unclear risk, DN definitions varied (e.g., based on eGFR, albuminuria, biopsy, or clinical diagnosis). The review did not standardize or stratify based on diagnostic criteria. Unclear risk, DN definitions varied (e.g., based on eGFR, albuminuria, biopsy, or clinical diagnosis). The review did not standardize or stratify based on diagnostic criteria. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Unclear concern, Variability in diagnostic definitions affects generalizability of biomarker findings. Unclear concern, Variability in diagnostic definitions affects generalizability of biomarker findings. |
| 4. Flow and Timing | Were all patients included in the analysis? Was there an appropriate interval between biomarker testing and reference standard assessment? | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High risk, The review did not evaluate timing consistency or completeness of datasets in primary studies. Missing data handling and participant flow were not discussed. High risk, The review did not evaluate timing consistency or completeness of datasets in primary studies. Missing data handling and participant flow were not discussed. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate concern, Potential temporal mismatch between biomarker sampling and DN diagnosis may bias interpretation. Moderate concern, Potential temporal mismatch between biomarker sampling and DN diagnosis may bias interpretation. |
| Overall Judgment | General methodological and reporting quality of the review and the included studies. | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High RoB, No formal quality or bias assessment (e.g., QUADAS-2, PROBAST) applied; heterogeneous populations, methods, and validation standards. High RoB, No formal quality or bias assessment (e.g., QUADAS-2, PROBAST) applied; heterogeneous populations, methods, and validation standards. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate applicability concern, Review useful for identifying research trends but limited clinical translation due to poor standardization and unclear diagnostic validity. Moderate applicability concern, Review useful for identifying research trends but limited clinical translation due to poor standardization and unclear diagnostic validity. |
Table A43 is an adapted QUADAS-2 tool evaluation of the study Dong et al., 2024 [
22] to assess the quality and bias of the review’s approach and of the studies it summarized.
Across the included literature, common issues persist: data leakage arising from sloppy time windows between biomarker measurement and outcome ascertainment, undefined handling of missing data, and outcome labels that shift across sites or depend on inconsistent diagnostic criteria for diabetic nephropathy. Follow-up intervals were often too short to capture clinically meaningful disease progression, further limiting interpretability.
Most primary studies used small, retrospective datasets and reported impressive accuracy metrics without adequate external validation or calibration. These eye-catching numbers are likely inflated by methodological weaknesses, which limit the study’s generalizability to new populations and real-world deployment.
Table A44.
QUADAS-2 RoB and applicability concerns assessment for the study, Nagaraj et al., 2021 [
23].
| Domain | Key Questions/Description | RoB | Applicability Concerns |
|---|
| 1. Patient Selection | Participants were individuals with DKD from observational cohorts. Inclusion and exclusion criteria were clearly defined, and baseline characteristics were described. However, participants were drawn from limited cohorts, potentially leading to selection bias. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Unclear risk, Sampling may not reflect broader DKD populations (mostly moderate to severe stages). Unclear risk, Sampling may not reflect broader DKD populations (mostly moderate to severe stages). | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Unclear concern, Applicability may be limited to similar hospital-based populations. Unclear concern, Applicability may be limited to similar hospital-based populations. |
| 2. Index Test (Kidney Age Index, ML Model) | The KAI framework was developed using ML algorithm to estimate biological kidney age from clinical and biochemical parameters. Model development and internal validation were described, but external validation was not performed. There is potential for data leakage if temporal splits are not strictly enforced. Handling of missing data and feature selection steps were insufficiently detailed. | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High risk, Possible overfitting and unclear missing data handling. High risk, Possible overfitting and unclear missing data handling. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate concern, KAI may not generalize to settings with different patient characteristics or lab standards. Moderate concern, KAI may not generalize to settings with different patient characteristics or lab standards. |
| 3. Reference Standard (Measured Kidney Function) | The reference standard was eGFR and albuminuria-based diagnosis of DKD, which are accepted clinical measures. However, measurement variability and assay calibration differences were not discussed. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Unclear risk, Reference measures acceptable but not standardized across datasets. Unclear risk, Reference measures acceptable but not standardized across datasets. | ![Biomedinformatics 05 00067 i008 Biomedinformatics 05 00067 i008]() Low concern, Consistent with clinical definitions of kidney function. Low concern, Consistent with clinical definitions of kidney function. |
| 4. Flow and Timing | The study used retrospective data; timing of biomarker and outcome measurements was not always synchronized. Follow-up duration for assessing kidney decline was limited, making it difficult to assess long-term predictive validity. | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High risk, Potential bias due to inconsistent timing and incomplete follow-up data. High risk, Potential bias due to inconsistent timing and incomplete follow-up data. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate concern, Limited longitudinal data reduce applicability for real-world progression prediction. Moderate concern, Limited longitudinal data reduce applicability for real-world progression prediction. |
| Overall Judgment | Innovative and well-conceptualized approach, but limited by internal-only validation, potential data leakage, and incomplete transparency on data preprocessing. | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High overall RoB High overall RoB | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate applicability concern Moderate applicability concern |
Table A44 is a structured QUADAS-2 evaluation summary table including both methodological domains and key structures of the study, Nagaraj et al., 2021 [
23].
This study introduces KAI, an ML-derived biomarker estimating kidney function in DKD by mapping clinical variables to an “age-equivalent” kidney function score. While conceptually strong, the diagnostic accuracy framework is limited by several common pitfalls in ML-driven biomarker development:
Possible leakage between training and testing data due to unclear temporal separation.
Undefined handling of missing data, imputation, and variable selection.
Outcome labels (e.g., eGFR thresholds, albuminuria cutoffs) may shift across sites or datasets, affecting comparability.
Follow-up periods are too short to meaningfully assess the decline in kidney function or validate clinical relevance.
Despite these issues, the KAI framework represents a promising step toward personalized nephrology, and its methodological transparency and external validation should determine its eventual utility.
Appendix G
This appendix contains details and explanations supplemental to the subsection, “4.7. Predicting future outcomes, such as mortality or cardiovascular events, using ML algorithms or patients’ biomarkers” of the main text. The explanations of the quality assessment of the included study articles [
34] through [
40] using the PROBAST and QUADAS-2 tools would disrupt the flow of the main text. However, this discussion is crucial to understanding the overall significance of these studies.
Table A45.
PROBAST assessment for Ma et al., 2023 [
34].
| Domain | Description/Assessment | RoB | Applicability Concerns |
|---|
| Population/Participants | 656 peritoneal dialysis patients with 13,091 visits; external testing on 1363 hemodialysis patients. Inclusion criteria were clear, but single center (or limited centers) may not represent broader PD populations globally. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Unclear Unclear | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Unclear Unclear |
| Index Model | “AICare” ML model uses adaptive feature-importance recalibration and multi-channel feature extraction from longitudinal EMR data. Sophisticated model, but missing data handling, temporal separation, and feature selection processes are not fully described. | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High High | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate Moderate |
| Comparator Model | Conventional ML baselines (RF, logistic regression) and ablation versions of AICare were compared. Comparisons performed internally; no external peritoneal dialysis comparators beyond the hemodialysis dataset. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Unclear Unclear | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate Moderate |
| Outcome | 1-year mortality following each clinical visit (binary). Mortality is a hard endpoint; cause-specific mortality is not considered. Outcome timing per visit introduces potential inconsistencies. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Unclear Unclear | ![Biomedinformatics 05 00067 i008 Biomedinformatics 05 00067 i008]() Low Low |
| Timing | Prediction horizon = 1-year post-visit; dataset spans ~12 years. Potential data leakage if future visits influence predictors. Follow-up per patient variable; censoring not fully detailed. | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High High | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate Moderate |
| Setting | Single-center (or few-center) tertiary care peritoneal dialysis patients; external hemodialysis dataset for testing. Retrospective EMR data. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Unclear Unclear | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate Moderate |
| Intended Use of Predictive Model | Early risk stratification for peritoneal dialysis patients to guide clinical interventions and prioritize high-risk patients for monitoring or treatment adjustments. | - | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate Moderate |
| Statistical Analysis | Multi-channel ML with adaptive recalibration, internal cross-validation, and testing on external hemodialysis cohort. Metrics: AUROC, AUPRC. Limited reporting on calibration, missingness handling, or temporal data leakage mitigation. | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High High | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate Moderate |
| Overall Judgment | Innovative ML approach with interpretability, large longitudinal dataset. High RoB due to potential leakage, unclear missing data handling, and limited external validation in PD population. Applicability moderate; promising method but caution needed in generalizing results. | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High High | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate Moderate |
Table A45 is a structured PROBAST evaluation summary table with both methodological domains and key features of the study, Ma et al., 2023 [
34].
This study has common pitfalls in predictive modeling that are evident:
Potential data leakage: Because predictions are made at each visit, there is a risk that future information or visit timing may inadvertently inform earlier predictions if not strictly excluded.
Handling of missingness: Although many longitudinal features are used, the report gives limited detail on how missing values, irregular visit intervals, or drop-outs were handled.
Outcome labels and timing consistency: While mortality is a robust endpoint, the consistency of “1-year ahead from visit” across all visits may vary; also, differences between peritoneal dialysis and hemodialysis cohorts (for external testing) may limit applicability.
Follow-up and generalizability: Although the dataset spans ~12 years, the actual follow-up per individual and event rate (39.8% died in the peritoneal dialysis cohort) may limit how well the model predicts longer-term outcomes beyond 1 year. Also, the population appears from one tertiary center (or limited centers) in China, which may differ from other geographic settings.
Given these limitations, when this study reports high performance (e.g., AUROC ~ 0.816 in peritoneal dialysis dataset, AUPRC ~ 0.472), one must interpret the results cautiously: these numbers may not transfer to other centers, populations, or real-world deployment without further external validation and scrutiny of the modeling pipeline.
Table A46.
PROBAST assessment for Chen et al., 2025 [
35].
| Domain | Description/Assessment | RoB | Applicability Concerns |
|---|
| Population/Participants | Retrospective cohort of 359 hemodialysis patients from one hospital (January 2017–June 2023) in China. Inclusion appears clear, but limited to one center, limited external validation. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Unclear Unclear | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate Moderate |
| Index Model | Two ML models: Model A (85 variables) and Model B (22 variables), using RF/SVM/logistic regression, with SHAP for interpretability. Model description is good, but detail on missing data, temporal split, feature engineering is limited. | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High High | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate Moderate |
| Comparator Model | Comparisons among ML methods (RF/SVM/Logistic), but no strong external reference standard model (e.g., purely conventional risk model) reported. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Unclear Unclear | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate Moderate |
| Outcome | Two outcomes: (1) all-cause mortality; (2) time to death (regression) for hemodialysis patients. Mortality is meaningful, but follow-up time, censoring, competing risks are not fully described. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Unclear Unclear | ![Biomedinformatics 05 00067 i008 Biomedinformatics 05 00067 i008]() Low Low |
| Timing | Data span about 6.5 years; models are built on retrospective data. Unclear if proper temporal separation between training and validation; possible leakage if later data influence earlier predictions. | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High High | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate Moderate |
| Setting | Single tertiary-care hemodialysis center in China; hospital setting. Raises concern about generalizability to other geographic/health-system settings. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Unclear Unclear | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate Moderate |
| Intended Use of Predictive Model | To support clinical decision-making by predicting mortality risk and time to death in hemodialysis patients, presumably to identify high-risk individuals for intervention. | - | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate Moderate |
| Statistical Analysis | Performance metrics: for Model A AU-ROC ~ 0.86 ± 0.07; for Model B AU-ROC ~ 0.80 ± 0.06. Regression (R2) for time to death reported. But calibration, missing data handling, external validation, temporal validation poorly described. | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High High | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate Moderate |
| Overall Judgment | Innovative and interpretable ML modeling, but significant methodological concerns: single-center data, risk of leakage, limited reporting of missingness/temporal splits, no strong external validation in hemodialysis populations. | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High High | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate Moderate |
Table A46 is a PROBAST evaluation summary table with both methodological domains and key structures of the study, Chen et al., 2025 [
35].
The study by Chen et al. (2025) [
35] proposes two interpretable ML tools for predicting all-cause mortality and time to death among hemodialysis patients. Using a retrospective cohort from a single center in China, the authors achieved relatively high discrimination (AU-ROC ~ 0.86 for their more complex model). However, from a PROBAST perspective, several shortcomings reduce confidence in generalizability:
Data leakage risk: It is not clear whether temporal separation was strictly maintained between training and validation sets (e.g., avoiding future visits influencing predictions).
Handling of missing data and preprocessing is insufficiently described, increasing bias risk.
Outcome definitions and follow-up: While mortality is a robust endpoint, details on censoring, competing risks, and follow-up duration are sparse, raising questions about validity over time.
Setting and sample: Single-center Chinese hemodialysis population limits applicability to other regions, dialysis modalities, or health systems.
Model evaluation: While discrimination is reported, calibration and external validation are absent, meaning the “eye-catching” performance may not travel to other settings.
Table A47.
PROBAST assessment for Hung et al., 2022 [
36].
| Domain | Description/Assessment | RoB | Applicability Concerns |
|---|
| Population/Participants | Retrospective cohort of 2932 ICU patients who received CRRT at a single tertiary center (Changhua Christian Hospital, Taiwan) from January 2010 to April 2021. Excluded ESRD on dialysis (n = 283), <20 yrs (n = 15), missing lab data (n = 73). While large cohort, single-center setting may limit representativeness of other ICU/CRRT populations. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Unclear risk Unclear risk | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate concern Moderate concern |
| Index Model | ML algorithms (GBM, XGBoost, RF, SVM) with feature selection (recursive feature elimination) and cross-validation; explainability via SHAP (global and local). However, details on missing data handling, temporal data splits (future leakage), and predictor measurement timing are limited. | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High risk High risk | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate concern Moderate concern |
| Comparator Model | Several ML models compared among themselves; no standard external clinical risk score comparator (or head-to-head with established CRRT mortality score) reported. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Unclear risk Unclear risk | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate concern Moderate concern |
| Outcome | Primary outcome: in-hospital mortality after CRRT initiation. Secondary endpoints: 28-day and 90-day mortality. Outcome is clinically meaningful and well defined (death during hospitalization). | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Unclear risk Unclear risk | ![Biomedinformatics 05 00067 i008 Biomedinformatics 05 00067 i008]() Low concern Low concern |
| Timing | Cohort spans ~11 years (2010-2021). The split: 80% training (n = 2345), 20% test (n = 587). But the temporal validation (e.g., future data) is not clearly described; potential for data leakage if later-visit data contributed to earlier predictions; handling of censoring/competing risks not deeply addressed. | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High risk High risk | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate concern Moderate concern |
| Setting | Single tertiary university hospital ICU and CRRT dataset in Taiwan. Retrospective EMR. Limits generalizability to other geographies, types of ICUs, CRRT protocols, and patient populations. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Unclear risk Unclear risk | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate concern Moderate concern |
| Intended Use of Predictive Model | To provide interpretable, personalized risk predictions of in-hospital mortality for CRRT patients, aiding clinicians in decision-making, family discussions, possibly guiding care strategy. | - | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate concern Moderate concern |
| Statistical/Modeling Analysis | The authors used RFE feature selection, 10-fold cross-validation (repeated 5 times), multiple ML algorithms, calibration belts, SHAP interpretability. Performance: AUC ~ 0.806 (XGBoost) to ~ 0.823 (GBM) in test set. Nonetheless, external validation not performed, unknown missing-data imputation process, potential overfitting risk. | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High risk High risk | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate concern Moderate concern |
| Overall Judgment | The study is well-designed in terms of sample size and use of modern ML + interpretability tools. However, key methodological uncertainties (data leakage, missingness handling, single-center dataset, no external validation) raise high RoB. Applicability is moderate: the model might work in similar ICU/CRRT settings but caution in broader settings. | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High risk High risk | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate concern Moderate concern |
Table A47 is a PROBAST evaluation summary table with both methodological domains and key structures of the study, Hung et al., 2022 [
36].
This study presents a robust attempt at building an explainable ML model to predict in-hospital mortality among ICU patients receiving CRRT. The dataset is relatively large (n ≈ 2932) and the authors employ modern tools like SHAP to improve transparency. Yet, common problems in this space remain: potential leakage (since predictor data may span the CRRT initiation window without strict temporal partitioning), missing data/irregular time windows (the paper excludes patients with missing labs but does not fully describe handling or impact), outcome definitions are consistent for in-hospital death but may vary in timing and discharge practices across centers, and the follow-up window is short (in-hospital death) rather than long-term survival. When a model reports an AUC ~ 0.82, the eye-catching number here, one must remember it is built in one center with retrospective data and no external validation, all of which may limit its transportability. More weight should be given to studies that externally validate, transparently report missing data/imputation, enforce temporal separation, and calibrate properly across populations.
Table A48.
PROBAST assessment for Lin et al., 2023 [
37].
| Domain | Description/Assessment | RoB | Applicability Concerns |
|---|
| Population/Participants | 103 hemodialysis patients (age > 20, hemodialysis > 3 months) at a single center in Taiwan; followed for 36 months; 26 deaths (25.2%) occurred. While inclusion criteria are defined, the small sample size and single-center setting limit representativeness. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Unclear Unclear | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate Moderate |
| Index Test (Biomarker: Endocan) | Serum endocan levels measured at baseline; the authors explored association with all-cause mortality in hemodialysis patients. The biomarker assay is specified, but details on timing of measurement in relation to hemodialysis initiation, repeated measures, or variability are limited. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Unclear Unclear | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate Moderate |
| Comparator/Other Predictors | The study adjusted for prognostic variables (age, diabetes, creatinine, albumin) in multivariable analysis; but no formal predictive model comprehensively compared to endocan alone or other biomarker panels. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Unclear Unclear | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate Moderate |
| Outcome | Outcome is all-cause mortality over 36 months. Hard endpoint; well-defined. | ![Biomedinformatics 05 00067 i008 Biomedinformatics 05 00067 i008]() Low Low | ![Biomedinformatics 05 00067 i008 Biomedinformatics 05 00067 i008]() Low Low |
| Timing | Baseline endocan measured, then followed for up to 36 months. However, the study does not fully specify whether visits/predictor measurement preceded outcome uniformly, or how missing follow-up or censoring was handled. | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High High | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate Moderate |
| Setting | Single tertiary hospital dialysis center in Taiwan; hemodialysis patients. This may restrict applicability to other populations, geographies, or dialysis practices. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Unclear Unclear | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate Moderate |
| Intended Use of Predictive Model/Biomarker | The authors propose serum endocan as a biomarker for mortality risk stratification among hemodialysis patients. The intended use is prognostic rather than interventional. | - | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate Moderate |
| Statistical Analysis | Kaplan–Meier analysis by endocan median group; ROC curve for endocan (AUC reported); multivariable Cox regression adjusting for select covariates (endocan p = 0.010; creatinine p = 0.034). However, calibration, missing data handling, external validation, temporal separation, and model discrimination beyond biomarker association are not detailed. | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High High | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate Moderate |
| Overall Judgment | The study presents an interesting candidate biomarker (endocan) for mortality risk in hemodialysis patients, but methodological limitations (small sample, single center, limited timing/validation detail) raise substantial concerns about bias and generalizability. | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High High | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate Moderate |
Table A48 is a PROBAST evaluation summary table with both methodological domains and key structures of the study, Lin et al., 2023 [
37].
This study investigates whether serum endocan, a marker of endothelial dysfunction, predicts all-cause mortality in a cohort of 103 hemodialysis patients over a 36-month follow-up. The key strength is the use of a hard clinical endpoint (death) and the identification of a statistically significant association (higher endocan → higher mortality). However, several methodological problems reduce confidence in the robustness and transportability of the findings:
Timing and follow-up: While baseline measurement and 36-month follow-up are reported, the study does not fully elucidate whether predictor measurement preceded outcome in all cases, how censoring was handled, or how missing follow-up impacted results, raising risk of, in effect, non-uniform timing windows.
Small sample size & setting: With only 103 patients (26 events) and data from a single center, results may be prone to overfitting or idiosyncratic to this environment. External generalizability is modest.
Limited model development/validation: The study essentially reports a biomarker-mortality association rather than a fully developed predictive model with performance metrics (calibration, discrimination in an independent cohort). As such, the “eye-catching” association may not hold in other populations.
Missing or variable data handling: The report does not deeply describe how missing biomarker/clinical values, variability in hemodialysis practices, or shifting outcome definitions (e.g., timing of death, cause of death) were addressed, potential sources of bias.
Table A49.
PROBAST assessment of the study, Tran et al., 2024 [
38].
| Domain | Description/Assessment | RoB | Applicability Concerns |
|---|
| Population/Participants | External validation dataset of 527 outpatients with stage 4 or 5 CKD (non-dialysis) from a French regional cohort; 91 of 527 died within 2 years. The validation cohort differed from the development cohort (younger age, lower death rate). | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Unclear, while external, differences in cohort characteristics suggest possible selection or spectrum bias. Unclear, while external, differences in cohort characteristics suggest possible selection or spectrum bias. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Unclear, setting (French outpatient CKD stage 4–5) may limit generalizability to other countries, dialysis cohorts, or more heterogeneous CKD populations. Unclear, setting (French outpatient CKD stage 4–5) may limit generalizability to other countries, dialysis cohorts, or more heterogeneous CKD populations. |
| Index Model | A previously developed ML-based 2-year all-cause mortality prediction tool (7 variables: age, ESA use, cardiovascular history, smoking status, 25-OH vitamin D, PTH, ferritin) from an earlier cohort. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Unclear, The validation uses an existing model, but details of model adaptation, calibration and predictor measurement in the new cohort are limited. Unclear, The validation uses an existing model, but details of model adaptation, calibration and predictor measurement in the new cohort are limited. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Unclear, Model developed in one dataset may perform differently in new populations with different baseline risks and features. Unclear, Model developed in one dataset may perform differently in new populations with different baseline risks and features. |
| Comparator Model | The study does not present a head-to-head comparison against a standard clinical risk score or alternative predictive model in the validation cohort; rather, it applies to the existing tool. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Unclear, absence of comparator limits assessment of relative performance. Unclear, absence of comparator limits assessment of relative performance. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Unclear, Without benchmark models, it’s hard to evaluate added value in this setting. Unclear, Without benchmark models, it’s hard to evaluate added value in this setting. |
| Outcome | All-cause mortality at 2 years follow-up. Hard clinical endpoint clearly defined. In the validation dataset, 91/527 died within 2 years. | ![Biomedinformatics 05 00067 i008 Biomedinformatics 05 00067 i008]() Low, Outcome is well-defined and appropriate. Low, Outcome is well-defined and appropriate. | ![Biomedinformatics 05 00067 i008 Biomedinformatics 05 00067 i008]() Low, Applicability of outcome is good for clinical mortality prediction in CKD stage 4–5. Low, Applicability of outcome is good for clinical mortality prediction in CKD stage 4–5. |
| Timing | Predictor variables measured at baseline; follow-up period = 2 years. However, there is limited information on timing of predictor ascertainment relative to baseline, censoring and missing follow-up, and whether temporal effects or secular changes were accounted for. | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High, The potential for bias is elevated because of insufficient reporting of timing, missing follow-up handling, and potential changes in care over time. High, The potential for bias is elevated because of insufficient reporting of timing, missing follow-up handling, and potential changes in care over time. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate concern, The 2-year horizon is clinically relevant, but differences in cohorts and treatment eras may affect transportability. Moderate concern, The 2-year horizon is clinically relevant, but differences in cohorts and treatment eras may affect transportability. |
| Setting | Outpatient nephrology settings in France (stage 4–5 CKD non-dialysis). The model was externally validated in this setting. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Unclear, Single region may limit heterogeneity; applicability to broader settings uncertain. Unclear, Single region may limit heterogeneity; applicability to broader settings uncertain. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate concern, The setting is relevant for outpatient CKD patients, but generalization to other geographies, healthcare systems or dialysis populations is uncertain. Moderate concern, The setting is relevant for outpatient CKD patients, but generalization to other geographies, healthcare systems or dialysis populations is uncertain. |
| Intended Use of Predictive Model | To predict 2-year all-cause mortality in stage 4–5 CKD patients to support risk stratification and potentially inform monitoring or early intervention. | - | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate concern, Intended use is compatible with the validation setting, but the lack of broad transportability reduces practical applicability. Moderate concern, Intended use is compatible with the validation setting, but the lack of broad transportability reduces practical applicability. |
| Statistical/Modeling Analysis | Validation reported AUC-ROC = 0.72, accuracy = 63.6%, sensitivity = 72.5%, specificity = 61.7%. The model showed significant separation of survival curves (p < 0.001). However, calibration metrics, handling of missing data, sample size adequacy for external validation, and robustness of performance across subgroups are not fully detailed. | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High, Key modeling aspects (calibration, missing data, model updating) are inadequately reported, increasing bias risk. High, Key modeling aspects (calibration, missing data, model updating) are inadequately reported, increasing bias risk. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate concern, The model shows reasonable discrimination, but limited detail and moderate specificity raise concerns about real-world performance. Moderate concern, The model shows reasonable discrimination, but limited detail and moderate specificity raise concerns about real-world performance. |
| Overall Judgment | While this is a genuine external validation of a predictive model (which is a strength), the limitations around timing, missing data/reporting, sample representativeness, and limited reporting of calibration mean the RoB is high. Applicability is moderate: the tool may work in similar outpatient CKD stage 4–5 populations in France but transfer to other settings is uncertain. | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High RoB High RoB | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate applicability concern Moderate applicability concern |
Table A49 is a PROBAST evaluation summary table that includes both methodological domains and key structures of the study, Tran et al., 2024 [
38].
This study by Tran and colleagues conducts an external validation of a 2-year all-cause mortality prediction tool developed via ML in patients with stage 4–5 CKD [
38]. The validation cohort of 527 patients, with 91 deaths in 2 years, demonstrates modest performance (AUC 0.72), which is commendable for external validation. However, several methodological issues raise caution:
Measurement or selection bias: the validation cohort differed significantly from the development cohort (younger age, lower event rate), which may affect calibration or transportability.
Timing and missing-data concerns: the study provides limited detail on how baseline predictor data were timed in relation to baseline, how missing values were handled, or how secular changes in care were accounted for. This opens the possibility of bias or reduced reliability.
Model transportability: though validated externally, it is still within a French outpatient nephrology context; applicability to other healthcare systems, patient populations (e.g., dialysis, other countries), or treatment eras is untested.
Limited statistical reporting: Calibration metrics (e.g., calibration slope/intercept), decision-curve analysis, or subgroup performance are not fully reported, reducing confidence in clinical implementation despite the “eye-catching” AUC of 0.72.
Table A50.
PROBAST assessment for the study, Kim et al., 2020 [
39].
| Domain | Description/Assessment | RoB | Applicability Concerns |
|---|
| Population/Participants | Hemodialysis patients from a prospective multi-center Korean ESRD cohort (“K-cohort”), of which 354 of 452 eligible had plasma endocan measured and were followed for ~34.6 months. Selection of those with available endocan data may introduce selection bias; cohort limited to Korean center(s). | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Unclear Unclear | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate Moderate |
| Index Test (Biomarker: Plasma Endocan) | Baseline plasma endocan measured once (EDTA tube, fasting, mid-week dialysis). The biomarker is under investigation as predictor of cardiovascular events. Single measurement only; no repeated measures or longitudinal biomarker changes assessed. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Unclear Unclear | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate Moderate |
| Comparator/Other Predictors | The study uses multivariable Cox regression adjusting for prior cardiovascular events, albumin, BMI, TG and other covariates; but no formal standard risk-score model benchmark is reported. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Unclear Unclear | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate Moderate |
| Outcome | Composite cardiovascular event (acute coronary syndrome, stable angina requiring PCI/CABG, heart failure, ventricular arrhythmia, cardiac arrest, sudden death) and non-cardiac death. Outcome clearly defined, measured over follow-up. | ![Biomedinformatics 05 00067 i008 Biomedinformatics 05 00067 i008]() Low Low | ![Biomedinformatics 05 00067 i008 Biomedinformatics 05 00067 i008]() Low Low |
| Timing/Flow | Baseline biomarker measured, then follow-up (mean ~34.56 months). However: the study excludes patients missing endocan data, does not clearly describe censoring, handling of missing follow-up, or whether timing of predictor measurement relative to baseline events might introduce bias. | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High High | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate Moderate |
| Setting | Multi-center (six hospitals in South Korea) ESRD patients on hemodialysis, three times/week, >3 months vintage. While multi-center, all in one country/region; ESRD hemodialysis population specific. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Unclear Unclear | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate Moderate |
| Statistical/Modeling Analysis | Kaplan–Meier survival, determination of optimal cut-off for endocan via MaxStat, univariate and multivariable Cox regression (HR ~ 1.949 for high vs. low endocan, 95% CI 1.144–3.319, p = 0.014) for cardiovascular events. But limited reporting of calibration, no external validation, no missing data imputation described, risk of over-fitting given moderate sample size/events. | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High High | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate Moderate |
| Overall Judgment | The study provides a suggestive biomarker association of plasma endocan with cardiovascular events in hemodialysis patients, but limited by single measurement, missing data may bias results, modest sample size, no external validation, and uncertain handling of timing/censoring. | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High RoB High RoB | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate applicability concern Moderate applicability concern |
Table A50 is a PROBAST evaluation summary table that includes both methodological domains and key structures of the study, Kim et al., 2020 [
40].
This study explores the prognostic value of plasma endocan, a marker of endothelial dysfunction, in predicting cardiovascular events in ESRD patients on CRRT. Strengths include a well-defined cohort, clear outcome definition, and finding a significant association (higher endocan → higher risk of cardiovascular events).
However, from a prediction-modeling evaluation (via PROBAST lens), several key limitations emerge:
Timing/flow leakage risk: Although baseline biomarker measurement and ~34-month follow-up are reported, the exclusion of ~100 eligible patients (354/452), unclear censoring, and single biomarker measurement raise concerns of selection bias and missingness.
Missing data/measurement inconsistency: The biomarker was measured once; no repeated measures to capture dynamic risk changes. Handling of missing covariate data is not detailed.
Outcome label consistency and generalizability: The composite cardiovascular events definition is broad (includes arrhythmia, sudden death, heart failure), which may vary across settings; the cohort consists of Korean hemodialysis patients, which may limit transportability.
Statistical modeling limitations: Although survival analysis was used, the study lacks external validation, calibration metrics, and comprehensive missing data/imputation strategies. The cut-off determination via MaxStat is data-driven and may overestimate effect size (cut-off bias).
Follow-up may be adequate (~3 years) but no comment on competing risks (e.g., non-cardiovascular deaths) or censoring due to transplantation, modality shift, or loss to follow-up.
Thus, while the “eye-catching” hazard ratio (~1.95) is encouraging, given the high RoB and moderate applicability concerns, these findings should be interpreted cautiously. The results may not travel well to other hemodialysis populations, regions, or settings without further validation.
Table A51.
PROBAST assessment for the study, Zhu et al., 2024 [
40].
| Domain | Description/Assessment | RoB | Applicability Concerns |
|---|
| Population/Participants | The study used electronic medical records from a single center (Chinese PLA General Hospital) from 2015 to 2020, enrolling 8894 CKD patients (incident or ongoing) and followed them for composite CVD events. Inclusion criteria, exclusion criteria, and representativeness beyond that center are not fully elaborated. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Unclear risk Unclear risk | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate concern, single-center data may not generalize to other populations or health systems Moderate concern, single-center data may not generalize to other populations or health systems |
| Index Model | They selected predictors via LASSO regression, then developed seven ML classification algorithms, with XGBoost being the top performer. They used SHAP for interpretability. However, descriptions of how missing data were handled, how predictor timing was controlled (to avoid leakage), and feature engineering are limited. | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High risk High risk | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate concern, model may be overfit to center-specific patterns Moderate concern, model may be overfit to center-specific patterns |
| Comparator Model | They compare across ML algorithms (e.g., XGBoost vs. RF, SVM, etc.), and contrast with baseline logistic regression/simpler models. But no strong external benchmark or well-established clinical risk score is used for comparison. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Unclear risk Unclear risk | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate concern Moderate concern |
| Outcome | The outcome is a composite CVD event (broad definition) including coronary, cerebrovascular, peripheral vascular disease, heart failure, and death. The composite definition is broad, which may dilute specificity, and the inclusion of “deaths from all causes (cardiovascular, non-cardiovascular, unknown)” further complicates interpretation. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Unclear risk Unclear risk | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate concern Moderate concern |
| Timing/Flow | Predictor data from 2015–2020; models evaluated on held-out (test) set. However, the paper does not strongly detail temporal separation (i.e., using earlier data to predict future), risk of data leakage (features derived partly after outcome), or handling of censoring/time-to-event aspects. It also does not clearly describe how patients lost to follow-up or missing events were managed. | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High risk High risk | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate concern Moderate concern |
| Setting | Clinical CKD care setting in China (single hospital). Retrospective EMR context. | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Unclear risk Unclear risk | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate concern Moderate concern |
| Intended Use of Predictive Model | To help clinicians identify CKD patients at high risk for cardiovascular disease, enabling early interventions or tailored monitoring. | - | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate concern Moderate concern |
| Statistical/Modeling Analysis | They evaluated performance using AUC, accuracy, sensitivity, specificity, F1-score on test set. The top model (XGBoost) had AUC ~ 0.89 in the test set. They also used SHAP to interpret feature importance. However, the analysis lacks detailed calibration metrics, external validation, detailed missing-data imputation strategies, robustness checks (e.g., sensitivity analyses), and explicit statements about prevention of overfitting or leakage. | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High risk High risk | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate concern Moderate concern |
| Overall Judgment | The study presents a promising approach with strong discrimination metrics and interpretability efforts, but methodological reporting gaps (temporal leakage risk, missing data handling, lack of external validation) undermine reliability. | ![Biomedinformatics 05 00067 i003 Biomedinformatics 05 00067 i003]() High RoB High RoB | ![Biomedinformatics 05 00067 i002 Biomedinformatics 05 00067 i002]() Moderate applicability concern Moderate applicability concern |
Table A51 is a PROBAST evaluation summary table that includes both methodological domains and key structures of the study, Zhu et al., 2024 [
40].
Zhu et al. (2024) [
40] developed an ML-based CVD risk prediction model in a cohort of 8894 CKD patients from a single Chinese center. The XGBoost model delivered strong discrimination (AUC ≈ 0.89 in test data) and was made interpretable via SHAP.
However, several common pitfalls in predictive modeling are apparent:
Leakage risk/sloppy timing windows: Without strong temporal separation (i.e., training on earlier data, testing on truly future data), there is a chance that features partly reflect future events or correlate with outcomes in unintended ways.
Undefined handling of missingness: The paper does not clearly explain how missing predictor values were handled (imputation, exclusion), which can bias model performance.
Outcome label drift across sites: The composite definition of CVD is broad and may not map cleanly to other cohorts; what qualifies as a CVD event may vary over time or by hospital coding practices.
Follow-up adequacy: Although the test sets were drawn from the same center, there is limited discussion of censoring, loss to follow-up, or how long predictive horizons are clinically meaningful.
Thus, while the “eye-catching number” of AUC ~ 0.89 suggests very high performance, it must be taken with caution. The lack of external validation and incomplete methodological transparency means the metric may not travel well to new patient populations or settings. In assessing models in this space, higher-quality evidence, i.e., multi-center external validations, clear reporting of missing-data strategies, rigorous temporal splitting, and calibration metrics, should carry greater weight than a single-center result, no matter how impressive the discrimination.
 ): Bienaimé et al., 2023 [26] and Schanstra et al., 2015 [29] demonstrate external validation, calibration slope, and multi-center reproducibility.
): Bienaimé et al., 2023 [26] and Schanstra et al., 2015 [29] demonstrate external validation, calibration slope, and multi-center reproducibility. ): Qin et al., 2019 [28] and Muiru et al., 2021 [30] demonstrate internal validation or repeated measures but lack calibration or independent replication.
): Qin et al., 2019 [28] and Muiru et al., 2021 [30] demonstrate internal validation or repeated measures but lack calibration or independent replication. ): Pizzini et al., 2017 [27] is a small pilot without validation or calibration, serving more as proof-of-concept than reliable evidence.
): Pizzini et al., 2017 [27] is a small pilot without validation or calibration, serving more as proof-of-concept than reliable evidence. ):
): ):
): ): Chan et al., 2021 [24], Ferguson et al., 2022 [31], and Tangri et al., 2024 [32], all performed external validation, demonstrated good calibration, and sometimes included DCA showing clinical utility.
): Chan et al., 2021 [24], Ferguson et al., 2022 [31], and Tangri et al., 2024 [32], all performed external validation, demonstrated good calibration, and sometimes included DCA showing clinical utility. ): Zou et al., 2022 [33] lacked external validation and calibration assessment, relying on internal data only.
): Zou et al., 2022 [33] lacked external validation and calibration assessment, relying on internal data only. ): Ma et al., 2023 [34], Chen et al., 2025 [35], Tran et al., 2024 [38], demonstrate sound calibration and/or external validation.
): Ma et al., 2023 [34], Chen et al., 2025 [35], Tran et al., 2024 [38], demonstrate sound calibration and/or external validation. ): Hung et al., 2022 [36], interpretable ML, but single-center only.
): Hung et al., 2022 [36], interpretable ML, but single-center only. ): Lin et al., 2023 [37], Kim et al., 2020 [39], Zhu et al., 2024 [40], rely on limited biomarker datasets, lack calibration or validation, and risk overfitting.
): Lin et al., 2023 [37], Kim et al., 2020 [39], Zhu et al., 2024 [40], rely on limited biomarker datasets, lack calibration or validation, and risk overfitting. High
High High, Participants are limited to a specific demographic (Cook County Hospital, Chicago), potentially limiting generalizability.
High, Participants are limited to a specific demographic (Cook County Hospital, Chicago), potentially limiting generalizability. /
/ Moderate to High
Moderate to High Moderate, Implementation in other settings could yield variable results due to equipment and imaging protocol differences.
Moderate, Implementation in other settings could yield variable results due to equipment and imaging protocol differences. Low
Low Low, The outcome is directly relevant to clinical practice.
Low, The outcome is directly relevant to clinical practice. Moderate
Moderate Low, Likely applicable since both tests relate to the same diagnostic event.
Low, Likely applicable since both tests relate to the same diagnostic event. Green = External validation (multi-site or independent cohort).
Green = External validation (multi-site or independent cohort).  Orange = limited or incomplete validation (small-sample temporal validation, narrow subgroup validation, or internal validation with methodological limitations).
Orange = limited or incomplete validation (small-sample temporal validation, narrow subgroup validation, or internal validation with methodological limitations).  Red = No external validation/preprint/exploratory approach.
Red = No external validation/preprint/exploratory approach. High
High High, The study population (transplant recipients in a tertiary center with available MRI) does not represent the full clinical spectrum of post-transplant patients.
High, The study population (transplant recipients in a tertiary center with available MRI) does not represent the full clinical spectrum of post-transplant patients. /
/ Moderate to High
Moderate to High Moderate, MRI protocols, scanners, and pre-processing steps are highly site-specific; generalization to other centers may be limited.
Moderate, MRI protocols, scanners, and pre-processing steps are highly site-specific; generalization to other centers may be limited. Low
Low Low, The biopsy grading directly answers the target condition and is appropriate for the study aim.
Low, The biopsy grading directly answers the target condition and is appropriate for the study aim. Moderate
Moderate Low to Moderate, The six-month interval could affect diagnostic consistency, but within chronic IFTA context, it’s partly acceptable.
Low to Moderate, The six-month interval could affect diagnostic consistency, but within chronic IFTA context, it’s partly acceptable. Green = External validation (multi-site or independent cohort).
Green = External validation (multi-site or independent cohort).  Yellow = Internal split or temporal validation only.
Yellow = Internal split or temporal validation only.  Orange = limited or incomplete validation (small-sample temporal validation, narrow subgroup validation, or internal validation with methodological limitations).
Orange = limited or incomplete validation (small-sample temporal validation, narrow subgroup validation, or internal validation with methodological limitations).  Red = No external validation/preprint/exploratory approach.
Red = No external validation/preprint/exploratory approach. Moderate
Moderate Low to Moderate, The sample (renal biopsies with chronic injury) represents the target clinical population, though external representativeness is uncertain.
Low to Moderate, The sample (renal biopsies with chronic injury) represents the target clinical population, though external representativeness is uncertain. Moderate
Moderate Moderate, Deep learning performance may vary with slide scanners, staining, and lab protocols, affecting real-world applicability.
Moderate, Deep learning performance may vary with slide scanners, staining, and lab protocols, affecting real-world applicability. Low
Low Low, Pathologist-based grading directly reflects the intended diagnostic construct.
Low, Pathologist-based grading directly reflects the intended diagnostic construct. Low
Low Low, The timing of analyses aligns with typical diagnostic workflows.
Low, The timing of analyses aligns with typical diagnostic workflows. Green = External validation (multi-site or independent cohort).
Green = External validation (multi-site or independent cohort).  Yellow = Internal split or temporal validation only.
Yellow = Internal split or temporal validation only.  Orange = limited or incomplete validation (small-sample temporal validation, narrow subgroup validation, or internal validation with methodological limitations).
Orange = limited or incomplete validation (small-sample temporal validation, narrow subgroup validation, or internal validation with methodological limitations). Low-Moderate
Low-Moderate Low, The population represents patients undergoing renal biopsy, which matches the intended clinical target population for IFTA grading.
Low, The population represents patients undergoing renal biopsy, which matches the intended clinical target population for IFTA grading. Low
Low Low, The model directly addresses automated IFTA grading on digitized WSIs, the intended purpose of the index test.
Low, The model directly addresses automated IFTA grading on digitized WSIs, the intended purpose of the index test. Moderate
Moderate Low, Reference standard is clinically appropriate, using expert nephropathologists’ evaluation of renal biopsy WSIs.
Low, Reference standard is clinically appropriate, using expert nephropathologists’ evaluation of renal biopsy WSIs. Low-Moderate
Low-Moderate Low, Data handling, timing, and grading criteria are consistent, reflecting intended workflow for WSI analysis.
Low, Data handling, timing, and grading criteria are consistent, reflecting intended workflow for WSI analysis. Green = External validation (multi-site or independent cohort).
Green = External validation (multi-site or independent cohort).  Yellow = Internal split or temporal validation only.
Yellow = Internal split or temporal validation only.  Orange = limited or incomplete validation (small-sample temporal validation, narrow subgroup validation, or internal validation with methodological limitations).
Orange = limited or incomplete validation (small-sample temporal validation, narrow subgroup validation, or internal validation with methodological limitations). Moderate
Moderate Moderate, Population reflects patients undergoing biopsy but may not represent broader populations or those without biopsy, limiting generalizability to all kidney disease patients.
Moderate, Population reflects patients undergoing biopsy but may not represent broader populations or those without biopsy, limiting generalizability to all kidney disease patients. Low
Low Low, The index test is directly relevant to the clinical task of non-invasive IFTA assessment.
Low, The index test is directly relevant to the clinical task of non-invasive IFTA assessment. Low-Moderate
Low-Moderate Low, The reference standard is clinically appropriate and directly measures the target condition.
Low, The reference standard is clinically appropriate and directly measures the target condition. Low
Low Low, Flow and timing are appropriate for evaluating the DL model against the biopsy reference standard.
Low, Flow and timing are appropriate for evaluating the DL model against the biopsy reference standard. Green = External validation (multi-site or independent cohort).
Green = External validation (multi-site or independent cohort).  Yellow = Internal split or temporal validation only.
Yellow = Internal split or temporal validation only.  Orange = limited or incomplete validation (small-sample temporal validation, narrow subgroup validation, or internal validation with methodological limitations).
Orange = limited or incomplete validation (small-sample temporal validation, narrow subgroup validation, or internal validation with methodological limitations). Low-Moderate
Low-Moderate Low, Study samples are representative of patients undergoing renal biopsy for glomerulosclerosis and IFTA assessment, the intended target population.
Low, Study samples are representative of patients undergoing renal biopsy for glomerulosclerosis and IFTA assessment, the intended target population. Low
Low Low, The index test directly addresses the clinical task of automated segmentation and quantitation of renal histologic injury.
Low, The index test directly addresses the clinical task of automated segmentation and quantitation of renal histologic injury. Moderate
Moderate Low, Expert pathologist annotations are clinically relevant and appropriate for training and validating the model.
Low, Expert pathologist annotations are clinically relevant and appropriate for training and validating the model. Low
Low Low, Flow and timing reflect intended use, with proper separation of training and test datasets.
Low, Flow and timing reflect intended use, with proper separation of training and test datasets. Green = External validation (multi-site or independent cohort).
Green = External validation (multi-site or independent cohort).  Yellow = Internal split or temporal validation only.
Yellow = Internal split or temporal validation only.  Orange = limited or incomplete validation (small-sample temporal validation, narrow subgroup validation, or internal validation with methodological limitations).
Orange = limited or incomplete validation (small-sample temporal validation, narrow subgroup validation, or internal validation with methodological limitations). Moderate, selection bias possible; not fully representative of all transplant patients
Moderate, selection bias possible; not fully representative of all transplant patients High, multiple model testing, risk of overfitting, small validation sets relative to training
High, multiple model testing, risk of overfitting, small validation sets relative to training Low, clinically accepted standard; reference outcome is relevant and reliable
Low, clinically accepted standard; reference outcome is relevant and reliable Moderate, grading differences across cohorts and batch effects may introduce misclassification bias
Moderate, grading differences across cohorts and batch effects may introduce misclassification bias Moderate, timing differences and cross-sectional data may limit prediction consistency
Moderate, timing differences and cross-sectional data may limit prediction consistency Moderate, datasets may not represent real-world clinical populations
Moderate, datasets may not represent real-world clinical populations Moderate, potential clinical use, but external validation and clinical implementation pending
Moderate, potential clinical use, but external validation and clinical implementation pending Green = External validation (multi-site or independent cohort).
Green = External validation (multi-site or independent cohort).  Orange = limited or incomplete validation (small-sample temporal validation, narrow subgroup validation, or internal validation with methodological limitations).
Orange = limited or incomplete validation (small-sample temporal validation, narrow subgroup validation, or internal validation with methodological limitations).  Red = No external validation/preprint/exploratory approach.
Red = No external validation/preprint/exploratory approach. High RoB, with
High RoB, with  Moderate Concerns for Applicability.
Moderate Concerns for Applicability. Low-Moderate, Prospective and clinically relevant, but limited sample and single cohort reduce representativeness.
Low-Moderate, Prospective and clinically relevant, but limited sample and single cohort reduce representativeness. Moderate, LASSO penalization reduces overfitting risk, but internal-only validation and data-driven selection of biomarkers may inflate performance estimates.
Moderate, LASSO penalization reduces overfitting risk, but internal-only validation and data-driven selection of biomarkers may inflate performance estimates. Low, Comparator is appropriate, widely accepted, and clinically meaningful.
Low, Comparator is appropriate, widely accepted, and clinically meaningful. Low, Objective and precise measurement of kidney function minimizes outcome misclassification.
Low, Objective and precise measurement of kidney function minimizes outcome misclassification. Low, Clear temporal sequence supports valid prediction; prospective data collection minimizes bias.
Low, Clear temporal sequence supports valid prediction; prospective data collection minimizes bias. Low, Well-controlled research setting; consistent sample handling and assay validation.
Low, Well-controlled research setting; consistent sample handling and assay validation. Low, Intended use is clinically relevant and aligned with current CKD management goals.
Low, Intended use is clinically relevant and aligned with current CKD management goals. Green = External validation (multi-site or independent cohort).
Green = External validation (multi-site or independent cohort).  Yellow = Internal split or temporal validation only.
Yellow = Internal split or temporal validation only.  Orange = limited or incomplete validation (small-sample temporal validation, narrow subgroup validation, or internal validation with methodological limitations).
Orange = limited or incomplete validation (small-sample temporal validation, narrow subgroup validation, or internal validation with methodological limitations). Moderate Overall RoB, with
Moderate Overall RoB, with  Low Applicability Concern
Low Applicability Concern Moderate, Clinically relevant CKD population but small, single-center, and possibly non-representative sample.
Moderate, Clinically relevant CKD population but small, single-center, and possibly non-representative sample. Moderate, Simple derivation method, but internal-only validation, small sample, and data-driven thresholding raise overfitting risk.
Moderate, Simple derivation method, but internal-only validation, small sample, and data-driven thresholding raise overfitting risk. Low, eGFR is a gold-standard clinical reference for kidney function.
Low, eGFR is a gold-standard clinical reference for kidney function. Low, Outcome is standardized, measurable, and clinically meaningful.
Low, Outcome is standardized, measurable, and clinically meaningful. Low, Appropriate temporal relationship between predictors and outcome.
Low, Appropriate temporal relationship between predictors and outcome. Low, Controlled clinical and analytical environment ensures reliable data quality.
Low, Controlled clinical and analytical environment ensures reliable data quality. Low, Intended use aligns with nephrology practice and unmet clinical need.
Low, Intended use aligns with nephrology practice and unmet clinical need. Green = External validation (multi-site or independent cohort).
Green = External validation (multi-site or independent cohort).  Orange = limited or incomplete validation (small-sample temporal validation, narrow subgroup validation, or internal validation with methodological limitations).
Orange = limited or incomplete validation (small-sample temporal validation, narrow subgroup validation, or internal validation with methodological limitations). Moderate Overall RoB with
Moderate Overall RoB with  Low Applicability Concern
Low Applicability Concern Moderate
Moderate High, inpatient sample limits generalizability to screening or primary-care settings.
High, inpatient sample limits generalizability to screening or primary-care settings. High, simple univariable analysis; no validation or adjustment for overfitting.
High, simple univariable analysis; no validation or adjustment for overfitting. Moderate, biomarkers clinically measurable but not yet standardized for DKD diagnosis.
Moderate, biomarkers clinically measurable but not yet standardized for DKD diagnosis. High, predictor-outcome dependency likely inflates AUCs.
High, predictor-outcome dependency likely inflates AUCs. Moderate, UAE widely accepted but not ideal for early-stage DKD reference.
Moderate, UAE widely accepted but not ideal for early-stage DKD reference. Moderate, objective lab-based outcome but lacks temporal dimension.
Moderate, objective lab-based outcome but lacks temporal dimension. Moderate, relevant to early DKD diagnosis but not progression prediction.
Moderate, relevant to early DKD diagnosis but not progression prediction. High
High High, not predictive; diagnostic only.
High, not predictive; diagnostic only. Moderate
Moderate High, single-center; potential institutional bias.
High, single-center; potential institutional bias. Moderate, appropriate for hypothesis generation only.
Moderate, appropriate for hypothesis generation only. High, limited use beyond internal diagnostic context.
High, limited use beyond internal diagnostic context. High overall RoB
High overall RoB High applicability concern
High applicability concern Yellow = Internal split or temporal validation only.
Yellow = Internal split or temporal validation only.  Red = No external validation/preprint/exploratory approach.
Red = No external validation/preprint/exploratory approach. Low-Moderate
Low-Moderate Low, representative CKD and at-risk populations suitable for diagnostic and prognostic use.
Low, representative CKD and at-risk populations suitable for diagnostic and prognostic use. Low, standardized proteomic measurement, objective analysis.
Low, standardized proteomic measurement, objective analysis. Low, proteomic test applicable to intended CKD risk stratification setting.
Low, proteomic test applicable to intended CKD risk stratification setting. Moderate, potential incorporation bias with overlapping filtration markers.
Moderate, potential incorporation bias with overlapping filtration markers. Low, consistent with clinical standards for CKD staging and progression.
Low, consistent with clinical standards for CKD staging and progression. Low, flow appropriate and timing consistent.
Low, flow appropriate and timing consistent. Low, progression analysis based on follow-up data aligns with intended use.
Low, progression analysis based on follow-up data aligns with intended use. Low overall
Low overall Low overall
Low overall Green = External validation (multi-site or independent cohort).
Green = External validation (multi-site or independent cohort).  Yellow = Internal split or temporal validation only.
Yellow = Internal split or temporal validation only.  Orange = limited or incomplete validation (small-sample temporal validation, narrow subgroup validation, or internal validation with methodological limitations).
Orange = limited or incomplete validation (small-sample temporal validation, narrow subgroup validation, or internal validation with methodological limitations). Low-Moderate, selection limited to relatively healthy women, potentially introducing bias.
Low-Moderate, selection limited to relatively healthy women, potentially introducing bias. Moderate, mostly middle-aged Black women with HIV; may not represent general CKD or HIV-positive male populations.
Moderate, mostly middle-aged Black women with HIV; may not represent general CKD or HIV-positive male populations. Low-Moderate, laboratory methods robust, but no pre-specified diagnostic cut-offs.
Low-Moderate, laboratory methods robust, but no pre-specified diagnostic cut-offs. Some concern, biomarkers used as exploratory indicators, not validated diagnostic tests.
Some concern, biomarkers used as exploratory indicators, not validated diagnostic tests. High, absence of a defined reference standard limits diagnostic accuracy assessment.
High, absence of a defined reference standard limits diagnostic accuracy assessment. High, reference variables do not constitute a diagnostic criterion for CKD.
High, reference variables do not constitute a diagnostic criterion for CKD. Low, clear temporal structure and uniform application of tests.
Low, clear temporal structure and uniform application of tests. Low, appropriate interval and consistent follow-up across participants.
Low, appropriate interval and consistent follow-up across participants. Green = External validation (multi-site or independent cohort).
Green = External validation (multi-site or independent cohort).  Yellow = Internal split or temporal validation only.
Yellow = Internal split or temporal validation only.  Red = No external validation/preprint/exploratory approach.
Red = No external validation/preprint/exploratory approach. Low risk
Low risk Low risk
Low risk Some concern
Some concern Moderate
Moderate Green = External validation (multi-site or independent cohort).
Green = External validation (multi-site or independent cohort).  Yellow = Internal split or temporal validation only.
Yellow = Internal split or temporal validation only. 
 Low risk
Low risk Unclear
Unclear Low risk
Low risk Some concern
Some concern Green = External validation (multi-site or independent cohort).
Green = External validation (multi-site or independent cohort).  Yellow = Internal split or temporal validation only.
Yellow = Internal split or temporal validation only. 
 High risk
High risk Low risk
Low risk High
High Green = External validation (multi-site or independent cohort).
Green = External validation (multi-site or independent cohort).  Yellow = Internal split or temporal validation only.
Yellow = Internal split or temporal validation only.  (not applicable as a true diagnostic accuracy study)
(not applicable as a true diagnostic accuracy study) Low risk
Low risk Low risk
Low risk Low risk
Low risk Low
Low Green = External validation (multi-site or independent cohort).
Green = External validation (multi-site or independent cohort).  Some concern, selection limited to women with preserved renal function; may not represent patients with advanced CKD or male populations.
Some concern, selection limited to women with preserved renal function; may not represent patients with advanced CKD or male populations. Moderate, findings apply mainly to women with HIV and may not generalize to all HIV or CKD populations.
Moderate, findings apply mainly to women with HIV and may not generalize to all HIV or CKD populations. Some concern, robust internal modeling, but no external or temporal validation; unclear internal validation (e.g., bootstrapping).
Some concern, robust internal modeling, but no external or temporal validation; unclear internal validation (e.g., bootstrapping). Moderate, model exploratory, not clinically implemented; predictive performance not reported.
Moderate, model exploratory, not clinically implemented; predictive performance not reported. High, absence of comparator limits interpretation of predictive improvement or incremental value.
High, absence of comparator limits interpretation of predictive improvement or incremental value. Some concern, not designed for model comparison or validation.
Some concern, not designed for model comparison or validation. Some concern, surrogate outcomes, not direct measures of kidney disease progression or patient-level endpoints.
Some concern, surrogate outcomes, not direct measures of kidney disease progression or patient-level endpoints. Moderate, outcome biologically meaningful but limited for clinical prediction utility.
Moderate, outcome biologically meaningful but limited for clinical prediction utility. Low, consistent and appropriate timing for longitudinal biomarker change evaluation.
Low, consistent and appropriate timing for longitudinal biomarker change evaluation. Low, timing consistent with biological plausibility for kidney biomarker change.
Low, timing consistent with biological plausibility for kidney biomarker change. Low, standardized data collection and laboratory protocols reduce bias.
Low, standardized data collection and laboratory protocols reduce bias. Some concern, research setting may differ from clinical practice environments.
Some concern, research setting may differ from clinical practice environments. Some concern, model not yet developed for clinical prediction; exploratory by design.
Some concern, model not yet developed for clinical prediction; exploratory by design. Moderate, informative for biomarker research, but not directly applicable to clinical prediction or screening.
Moderate, informative for biomarker research, but not directly applicable to clinical prediction or screening. Green = External validation (multi-site or independent cohort).
Green = External validation (multi-site or independent cohort).  Yellow = Internal split or temporal validation only.
Yellow = Internal split or temporal validation only.  Red = No external validation/preprint/exploratory approach.
Red = No external validation/preprint/exploratory approach. Some concern, single-center dataset, potential selection bias from missing data exclusion, unclear external representativeness.
Some concern, single-center dataset, potential selection bias from missing data exclusion, unclear external representativeness. Moderate concern, findings may not generalize beyond a Chinese tertiary-care population.
Moderate concern, findings may not generalize beyond a Chinese tertiary-care population. Some concern, appropriate internal validation but possible overfitting; no external or temporal validation.
Some concern, appropriate internal validation but possible overfitting; no external or temporal validation. Low concern, clearly defined model, interpretable, replicable in similar data contexts.
Low concern, clearly defined model, interpretable, replicable in similar data contexts. Some concern, comparison limited to internal ML models; lack of benchmark limits context for clinical improvement.
Some concern, comparison limited to internal ML models; lack of benchmark limits context for clinical improvement. Moderate concern, comparators appropriate for model development, but not for clinical performance comparison.
Moderate concern, comparators appropriate for model development, but not for clinical performance comparison. Low risk, objective, clinically established outcome, standardized data source.
Low risk, objective, clinically established outcome, standardized data source. Low concern, outcome relevant for DN screening in diabetes populations.
Low concern, outcome relevant for DN screening in diabetes populations. Some concern, unclear predictor-outcome chronology; possible temporal bias.
Some concern, unclear predictor-outcome chronology; possible temporal bias. Moderate concern, timing acceptable for diagnostic screening, but limits prognostic inference.
Moderate concern, timing acceptable for diagnostic screening, but limits prognostic inference. Low risk, consistent data acquisition, uniform diagnostic methods.
Low risk, consistent data acquisition, uniform diagnostic methods. Some concern, applicability limited to hospital settings with similar laboratory infrastructure.
Some concern, applicability limited to hospital settings with similar laboratory infrastructure. Some concern, exploratory model; not externally validated or integrated into clinical workflow.
Some concern, exploratory model; not externally validated or integrated into clinical workflow. Moderate concern, potential clinical value but requires further validation for real-world implementation.
Moderate concern, potential clinical value but requires further validation for real-world implementation. Overall RoB: Moderate
Overall RoB: Moderate Overall Applicability: Moderate
Overall Applicability: Moderate Green = External validation (multi-site or independent cohort).
Green = External validation (multi-site or independent cohort).  Yellow = Internal split or temporal validation only.
Yellow = Internal split or temporal validation only. Some concern, retrospective, secondary dataset selection; unclear if all consecutive DN and NC samples were included; possible spectrum bias due to dataset curation.
Some concern, retrospective, secondary dataset selection; unclear if all consecutive DN and NC samples were included; possible spectrum bias due to dataset curation. Some concern, data derived from public gene-expression repositories, not real-world clinical diagnostic populations.
Some concern, data derived from public gene-expression repositories, not real-world clinical diagnostic populations. Some concern, model training and testing based on retrospective datasets; no blinding; potential overfitting; no pre-specified diagnostic threshold reported.
Some concern, model training and testing based on retrospective datasets; no blinding; potential overfitting; no pre-specified diagnostic threshold reported. Low concern, gene-expression-based model aligns with molecular diagnostic purpose; methodologically appropriate.
Low concern, gene-expression-based model aligns with molecular diagnostic purpose; methodologically appropriate. Low risk, standard diagnostic definitions are likely applied in source datasets.
Low risk, standard diagnostic definitions are likely applied in source datasets. Low concern, reference standard appropriate for DN diagnosis.
Low concern, reference standard appropriate for DN diagnosis. Some concern, heterogeneous datasets with varying collection protocols and unknown blinding; no consistent sample flow reporting.
Some concern, heterogeneous datasets with varying collection protocols and unknown blinding; no consistent sample flow reporting. Moderate concern, applicability limited by cross-dataset integration and differences in tissue vs. blood validation samples.
Moderate concern, applicability limited by cross-dataset integration and differences in tissue vs. blood validation samples. Overall RoB: Moderate
Overall RoB: Moderate Overall Applicability: Moderate
Overall Applicability: Moderate Green = External validation (multi-site or independent cohort).
Green = External validation (multi-site or independent cohort).  Yellow = Internal split or temporal validation only.
Yellow = Internal split or temporal validation only. Low,
Low,  Some concern,
Some concern,  High).
High). Some concern, modest sample size; potential selection bias due to dropouts (10% loss); limited diversity (single-center Japanese cohort).
Some concern, modest sample size; potential selection bias due to dropouts (10% loss); limited diversity (single-center Japanese cohort). Moderate, population specific to Japanese DKD patients with mid-stage disease; external generalizability limited.
Moderate, population specific to Japanese DKD patients with mid-stage disease; external generalizability limited. Some concern, internal validation robust, but external validation absent; unclear model transparency or reproducibility; potential overfitting given small sample and complex model.
Some concern, internal validation robust, but external validation absent; unclear model transparency or reproducibility; potential overfitting given small sample and complex model. Moderate, explainable ML enhances interpretability, but implementation may vary by analytic infrastructure.
Moderate, explainable ML enhances interpretability, but implementation may vary by analytic infrastructure. Low, inclusion of conventional and ML comparators strengthens credibility.
Low, inclusion of conventional and ML comparators strengthens credibility. Low, comparator choice appropriate for methodologic benchmarking.
Low, comparator choice appropriate for methodologic benchmarking. Low, clearly defined, objective, clinically meaningful outcome.
Low, clearly defined, objective, clinically meaningful outcome. Low, outcome directly relevant to DKD progression prediction.
Low, outcome directly relevant to DKD progression prediction. Low, appropriate timing between predictors and outcome; prospective design minimizes bias.
Low, appropriate timing between predictors and outcome; prospective design minimizes bias. Low, suitable for longitudinal prediction of decline.
Low, suitable for longitudinal prediction of decline. Low, consistent laboratory and analytic procedures minimize bias.
Low, consistent laboratory and analytic procedures minimize bias. Some concern, single-center design limits generalizability to other healthcare settings or ethnic populations.
Some concern, single-center design limits generalizability to other healthcare settings or ethnic populations. Some concern, not yet validated for clinical application or regulatory standards.
Some concern, not yet validated for clinical application or regulatory standards. Moderate, promising as a biomarker discovery tool, but not ready for deployment in patient management.
Moderate, promising as a biomarker discovery tool, but not ready for deployment in patient management. Overall RoB: Moderate
Overall RoB: Moderate Overall Applicability: Moderate
Overall Applicability: Moderate Green = External validation (multi-site or independent cohort).
Green = External validation (multi-site or independent cohort).  Yellow = Internal split or temporal validation only.
Yellow = Internal split or temporal validation only. Low,
Low,  Some concern,
Some concern,  High).
High). Low, representative, well-described cohort; random selection minimizes selection bias.
Low, representative, well-described cohort; random selection minimizes selection bias. Low, broad CKD spectrum and racial diversity enhance generalizability to DKD populations.
Low, broad CKD spectrum and racial diversity enhance generalizability to DKD populations. Low, transparent model development; overfitting minimized via cross-validation; clear reporting of variable selection and penalization.
Low, transparent model development; overfitting minimized via cross-validation; clear reporting of variable selection and penalization. Low, approach and predictors feasible in similar metabolomics datasets; clinically interpretable outputs.
Low, approach and predictors feasible in similar metabolomics datasets; clinically interpretable outputs. Low, appropriate comparison and internal replication improve credibility.
Low, appropriate comparison and internal replication improve credibility. Low, methods align with modern standards for omics model evaluation.
Low, methods align with modern standards for omics model evaluation. Low, clearly defined, objective, clinically relevant outcomes with validated measurement methods.
Low, clearly defined, objective, clinically relevant outcomes with validated measurement methods. Low, directly relevant to DKD progression prediction.
Low, directly relevant to DKD progression prediction. Low, appropriate prospective design ensures temporal relationship between predictors and outcomes.
Low, appropriate prospective design ensures temporal relationship between predictors and outcomes. Low, suitable duration for modeling DKD progression.
Low, suitable duration for modeling DKD progression. Low, strong methodological rigor; consistent analytical standards.
Low, strong methodological rigor; consistent analytical standards. Low, high applicability to U.S. clinical research and biobank settings.
Low, high applicability to U.S. clinical research and biobank settings. Some concern, currently exploratory; not validated externally or clinically implemented.
Some concern, currently exploratory; not validated externally or clinically implemented. Some concern, clinical translation pending replication and assay standardization.
Some concern, clinical translation pending replication and assay standardization. Overall RoB: Low
Overall RoB: Low Overall Applicability: Some Concern (requires external validation)
Overall Applicability: Some Concern (requires external validation) Green = External validation (multi-site or independent cohort).
Green = External validation (multi-site or independent cohort).  Yellow = Internal split or temporal validation only.
Yellow = Internal split or temporal validation only.  Some concern, multi-center, but selection may favor patients with biobank consent and available plasma; some missing baseline data imputed using ±1 year window.
Some concern, multi-center, but selection may favor patients with biobank consent and available plasma; some missing baseline data imputed using ±1 year window. Some concern, mostly U.S. urban centers; may not generalize to rural or non-U.S. populations.
Some concern, mostly U.S. urban centers; may not generalize to rural or non-U.S. populations. Some concern, RF is complex; model performance may be overestimated in derivation set, though cross-validation was performed.
Some concern, RF is complex; model performance may be overestimated in derivation set, though cross-validation was performed. Some concern, use of proprietary biomarkers limits immediate clinical applicability; EHR integration feasible only where high-quality longitudinal data exist.
Some concern, use of proprietary biomarkers limits immediate clinical applicability; EHR integration feasible only where high-quality longitudinal data exist. Low, appropriate standard-of-care comparisons included.
Low, appropriate standard-of-care comparisons included. Low, KDIGO widely used; clinical model generalizable.
Low, KDIGO widely used; clinical model generalizable. Low, clear, validated, and meaningful outcome.
Low, clear, validated, and meaningful outcome. Low, aligns with intended clinical use for DKD progression.
Low, aligns with intended clinical use for DKD progression. Low, longitudinal follow-up sufficient to capture outcome events.
Low, longitudinal follow-up sufficient to capture outcome events. Low, follow-up duration clinically relevant for DKD progression.
Low, follow-up duration clinically relevant for DKD progression. Low, standardized assay reduces measurement bias.
Low, standardized assay reduces measurement bias. Some concern, biobank-based, urban population; may not generalize to broader outpatient or international settings.
Some concern, biobank-based, urban population; may not generalize to broader outpatient or international settings. Some concern, proprietary tests; intended use is clinical, but real-world implementation not yet evaluated.
Some concern, proprietary tests; intended use is clinical, but real-world implementation not yet evaluated. Some concern, risk score relies on biomarker availability and EHR integration.
Some concern, risk score relies on biomarker availability and EHR integration. Some concern, low bias for outcome and predictor measurement; moderate bias in participant selection and model complexity.
Some concern, low bias for outcome and predictor measurement; moderate bias in participant selection and model complexity. Some concern, applicability mainly for U.S. biobank populations; broader implementation pending validation.
Some concern, applicability mainly for U.S. biobank populations; broader implementation pending validation. Green = External validation (multi-site or independent cohort).
Green = External validation (multi-site or independent cohort).  Yellow = Internal split or temporal validation only.
Yellow = Internal split or temporal validation only. Low, Comprehensive provincial databases minimize selection bias; inclusion criteria clear and reproducible.
Low, Comprehensive provincial databases minimize selection bias; inclusion criteria clear and reproducible. Low, Broad CKD population across all G1-G5 stages; representative of routine clinical care.
Low, Broad CKD population across all G1-G5 stages; representative of routine clinical care. Some concern, Although advanced imputation used, model complexity (RF) may obscure predictor interpretability and calibration across subgroups.
Some concern, Although advanced imputation used, model complexity (RF) may obscure predictor interpretability and calibration across subgroups. Low, Uses routinely collected lab data; readily available in most health systems.
Low, Uses routinely collected lab data; readily available in most health systems. Low, Appropriate and well-established comparators included.
Low, Appropriate and well-established comparators included. Low, Comparators are clinically relevant benchmarks.
Low, Comparators are clinically relevant benchmarks. Low, Hard clinical outcomes derived from standardized lab and administrative data.
Low, Hard clinical outcomes derived from standardized lab and administrative data. Low, Outcome clinically meaningful and applicable across CKD stages.
Low, Outcome clinically meaningful and applicable across CKD stages. Low, Adequate follow-up duration for CKD progression.
Low, Adequate follow-up duration for CKD progression. Low, Consistent time windows and clinical relevance.
Low, Consistent time windows and clinical relevance. Low, Real-world data minimizes bias from selective recruitment; centralized labs ensure measurement consistency.
Low, Real-world data minimizes bias from selective recruitment; centralized labs ensure measurement consistency. Some concern, Applicability may vary in countries lacking centralized lab data or universal healthcare systems.
Some concern, Applicability may vary in countries lacking centralized lab data or universal healthcare systems. Low, Clear intended use with feasible data inputs; internal and external validation performed.
Low, Clear intended use with feasible data inputs; internal and external validation performed. Low, Widely applicable to clinical practice given use of standard labs.
Low, Widely applicable to clinical practice given use of standard labs. Low RoB
Low RoB Some concern, Highly applicable to systems with similar data infrastructure; may need calibration elsewhere.
Some concern, Highly applicable to systems with similar data infrastructure; may need calibration elsewhere. Green = External validation (multi-site or independent cohort).
Green = External validation (multi-site or independent cohort).  Yellow = Internal split or temporal validation only.
Yellow = Internal split or temporal validation only. Low, Randomized, well-characterized cohorts minimize selection bias.
Low, Randomized, well-characterized cohorts minimize selection bias. Some concern, Population restricted to type 2 diabetes in clinical trial settings, possibly healthier and more adherent than routine practice.
Some concern, Population restricted to type 2 diabetes in clinical trial settings, possibly healthier and more adherent than routine practice. Low, Predictors objectively measured; minimal missingness (<3%, except glucose). Transparent pre-specified variable list from prior study.
Low, Predictors objectively measured; minimal missingness (<3%, except glucose). Transparent pre-specified variable list from prior study. Low, Uses standard clinical lab parameters routinely collected in care.
Low, Uses standard clinical lab parameters routinely collected in care. Low, Comparators appropriate, established, and relevant to current clinical practice.
Low, Comparators appropriate, established, and relevant to current clinical practice. Low, Directly comparable to clinical risk tools used worldwide.
Low, Directly comparable to clinical risk tools used worldwide. Low, Objective, reproducible, and adjudicated outcome definitions.
Low, Objective, reproducible, and adjudicated outcome definitions. Low, Outcomes align with KDIGO and FDA-recommended endpoints for CKD trials.
Low, Outcomes align with KDIGO and FDA-recommended endpoints for CKD trials. Low, Consistent follow-up and timing across studies; clear baseline and outcome assessment windows.
Low, Consistent follow-up and timing across studies; clear baseline and outcome assessment windows. Low, Time frame appropriate for CKD progression prediction.
Low, Time frame appropriate for CKD progression prediction. Low, Data completeness, quality control, and outcome adjudication robust.
Low, Data completeness, quality control, and outcome adjudication robust. Some concern, Controlled trial environment may limit generalizability to routine care or non-trial populations.
Some concern, Controlled trial environment may limit generalizability to routine care or non-trial populations. Low, Clearly defined purpose; model feasible for integration.
Low, Clearly defined purpose; model feasible for integration. Low, Broadly applicable in clinical decision support if similar data infrastructure exists.
Low, Broadly applicable in clinical decision support if similar data infrastructure exists. Low RoB
Low RoB Some concern, Excellent validation but restricted to SGLT2i trial population with type 2 diabetes; may need recalibration for general CKD populations.
Some concern, Excellent validation but restricted to SGLT2i trial population with type 2 diabetes; may need recalibration for general CKD populations. Green = External validation (multi-site or independent cohort).
Green = External validation (multi-site or independent cohort).  Yellow = Internal split or temporal validation only.
Yellow = Internal split or temporal validation only. Low, Well-defined, consecutive, pathologically confirmed cohort minimizes misclassification and selection bias.
Low, Well-defined, consecutive, pathologically confirmed cohort minimizes misclassification and selection bias. High, Restricted to biopsy-confirmed DKD in hospital setting; not representative of broader T2DM populations with clinically diagnosed DKD.
High, Restricted to biopsy-confirmed DKD in hospital setting; not representative of broader T2DM populations with clinically diagnosed DKD. Low, Predictors measured objectively; consistent definitions; appropriate handling of missingness.
Low, Predictors measured objectively; consistent definitions; appropriate handling of missingness. Some concern, Reliance on biopsy-specific and pathology variables limits applicability in routine, non-biopsy DKD care.
Some concern, Reliance on biopsy-specific and pathology variables limits applicability in routine, non-biopsy DKD care. Low, Comparator models appropriate; fair model comparison across consistent datasets.
Low, Comparator models appropriate; fair model comparison across consistent datasets. Low, Comparisons relevant to model-selection purpose.
Low, Comparisons relevant to model-selection purpose. Low, Objective, standard ESRD definition; ascertainment likely accurate.
Low, Objective, standard ESRD definition; ascertainment likely accurate. Low, Outcome clinically relevant to DKD prognosis; matches intended use.
Low, Outcome clinically relevant to DKD prognosis; matches intended use. Low, Prospective follow-up adequate for ESRD prediction; outcome timing clearly defined.
Low, Prospective follow-up adequate for ESRD prediction; outcome timing clearly defined. Low, Appropriate prediction horizon for ESRD in advanced DKD.
Low, Appropriate prediction horizon for ESRD in advanced DKD. Some concern, High-quality data but limited generalizability to community or primary-care settings.
Some concern, High-quality data but limited generalizability to community or primary-care settings. High, Specialized hospital cohort; may not represent broader DKD care environments.
High, Specialized hospital cohort; may not represent broader DKD care environments. Low, Clearly stated clinical purpose; model interpretable and feasible for use where lab data available.
Low, Clearly stated clinical purpose; model interpretable and feasible for use where lab data available. Some concern, Model designed for advanced DKD at tertiary level; limited for early-stage DKD or general diabetic populations.
Some concern, Model designed for advanced DKD at tertiary level; limited for early-stage DKD or general diabetic populations. Some concern/Moderate risk, Internal validation only, modest sample size.
Some concern/Moderate risk, Internal validation only, modest sample size. High concern, Limited to biopsy-proven DKD; may not generalize to broader T2DM populations or other ethnicities.
High concern, Limited to biopsy-proven DKD; may not generalize to broader T2DM populations or other ethnicities. Green = External validation (multi-site or independent cohort).
Green = External validation (multi-site or independent cohort).  Yellow = Internal split or temporal validation only.
Yellow = Internal split or temporal validation only.  Red = No external validation/preprint/exploratory approach.
Red = No external validation/preprint/exploratory approach. Some concern, Restriction to English and specific databases may have introduced selection bias and publication bias.
Some concern, Restriction to English and specific databases may have introduced selection bias and publication bias. Low concern, The included patient populations (T2D with/without DKD) match the review’s stated target population for AI-based DKD prediction.
Low concern, The included patient populations (T2D with/without DKD) match the review’s stated target population for AI-based DKD prediction. Some concern, Variable reporting of model development and validation among included studies; unclear thresholding and calibration metrics in some.
Some concern, Variable reporting of model development and validation among included studies; unclear thresholding and calibration metrics in some. Low concern, The AI models were evaluated for DKD prediction as intended; no mismatch between review scope and test purpose.
Low concern, The AI models were evaluated for DKD prediction as intended; no mismatch between review scope and test purpose. Some concern, Heterogeneity in DKD definitions across studies (clinical vs. pathological confirmation).
Some concern, Heterogeneity in DKD definitions across studies (clinical vs. pathological confirmation). Low concern, All definitions align with real-world diagnostic criteria for DKD or CKD progression.
Low concern, All definitions align with real-world diagnostic criteria for DKD or CKD progression. Some concern, Inconsistent reporting of temporal relationships between predictor measurement and DKD outcomes across included studies.
Some concern, Inconsistent reporting of temporal relationships between predictor measurement and DKD outcomes across included studies. Low concern, The studies collectively address the intended question of early prediction of DKD, regardless of specific follow-up intervals.
Low concern, The studies collectively address the intended question of early prediction of DKD, regardless of specific follow-up intervals. Some concern, Lack of quantitative synthesis and limited quality appraisal details for each included study.
Some concern, Lack of quantitative synthesis and limited quality appraisal details for each included study. Low concern, The review’s conclusions appropriately reflected the included evidence without overgeneralizing applicability.
Low concern, The review’s conclusions appropriately reflected the included evidence without overgeneralizing applicability. Moderate RoB
Moderate RoB Low applicability concern
Low applicability concern Green = External validation (multi-site or independent cohort).
Green = External validation (multi-site or independent cohort).  Yellow = Internal split or temporal validation only.
Yellow = Internal split or temporal validation only. Some concern, Retrospective design, unclear whether all consecutive DN biopsies were included; potential for selection bias toward good-quality slides.
Some concern, Retrospective design, unclear whether all consecutive DN biopsies were included; potential for selection bias toward good-quality slides. Low concern, Study population (biopsy-confirmed DN) reflects the intended target population for histopathologic classification.
Low concern, Study population (biopsy-confirmed DN) reflects the intended target population for histopathologic classification. Some concern, Model trained and validated on retrospective labeled data; unclear if testing was blinded to reference standard; possible overfitting and lack of external dataset validation.
Some concern, Model trained and validated on retrospective labeled data; unclear if testing was blinded to reference standard; possible overfitting and lack of external dataset validation. Low concern, The index test (CNN model) aligns directly with the intended clinical use (automated morphological classification).
Low concern, The index test (CNN model) aligns directly with the intended clinical use (automated morphological classification). Low risk, Reference standard based on validated, widely accepted histopathologic criteria (RPS 2010). Consensus reading reduces misclassification risk.
Low risk, Reference standard based on validated, widely accepted histopathologic criteria (RPS 2010). Consensus reading reduces misclassification risk. Low concern, Standard reflects current diagnostic practice; appropriate for evaluating CNN-based methods.
Low concern, Standard reflects current diagnostic practice; appropriate for evaluating CNN-based methods. Some concern, Same dataset source used for both training and testing; unclear independence of test set; lack of prospective validation; no time interval between index and reference tests.
Some concern, Same dataset source used for both training and testing; unclear independence of test set; lack of prospective validation; no time interval between index and reference tests. Some concern, Applicability limited by single-center design and lack of external dataset; performance may vary across staining protocols or scanners.
Some concern, Applicability limited by single-center design and lack of external dataset; performance may vary across staining protocols or scanners. Overall RoB: Moderate
Overall RoB: Moderate Overall Applicability: Moderate
Overall Applicability: Moderate Green = External validation (multi-site or independent cohort).
Green = External validation (multi-site or independent cohort).  Yellow = Internal split or temporal validation only.
Yellow = Internal split or temporal validation only. Some concern, Potential selection bias due to retrospective data extraction and unclear inclusion/exclusion handling.
Some concern, Potential selection bias due to retrospective data extraction and unclear inclusion/exclusion handling. Low concern, Reflects typical diabetic population encountered in clinical settings.
Low concern, Reflects typical diabetic population encountered in clinical settings. High risk, Limited transparency in feature selection, internal validation only, and unclear handling of missing or correlated data.
High risk, Limited transparency in feature selection, internal validation only, and unclear handling of missing or correlated data. Some concern, Model dependent on EMR data structure; reproducibility across systems uncertain.
Some concern, Model dependent on EMR data structure; reproducibility across systems uncertain. High risk, Absence of comparator limits evaluation of incremental value and clinical benefit.
High risk, Absence of comparator limits evaluation of incremental value and clinical benefit. Some concern, Lacks benchmark comparison for interpretability in clinical context.
Some concern, Lacks benchmark comparison for interpretability in clinical context. Low risk, Outcome clinically relevant, objective, and based on standard renal indicators.
Low risk, Outcome clinically relevant, objective, and based on standard renal indicators. Low concern, Outcome definition aligns with nephrology practice.
Low concern, Outcome definition aligns with nephrology practice. Some concern, Retrospective follow-up with unclear frequency and consistency of measurements across patients.
Some concern, Retrospective follow-up with unclear frequency and consistency of measurements across patients. Low concern, Clinically realistic prediction horizon for monitoring DKD progression.
Low concern, Clinically realistic prediction horizon for monitoring DKD progression. Some concern, Variability in data collection across institutions and EMR systems may affect model generalizability.
Some concern, Variability in data collection across institutions and EMR systems may affect model generalizability. Low concern, EMR-based design applicable to modern healthcare environments.
Low concern, EMR-based design applicable to modern healthcare environments. Low risk, Intended use clinically appropriate and clearly stated.
Low risk, Intended use clinically appropriate and clearly stated. Low concern, Applicable for clinical decision support once validated.
Low concern, Applicable for clinical decision support once validated. Overall RoB: Moderate-High
Overall RoB: Moderate-High Overall Applicability: Moderate
Overall Applicability: Moderate Green = External validation (multi-site or independent cohort).
Green = External validation (multi-site or independent cohort).  Yellow = Internal split or temporal validation only.
Yellow = Internal split or temporal validation only.  Red = No external validation/preprint/exploratory approach.
Red = No external validation/preprint/exploratory approach. Some concern, Moderate sample size; single-center data may not capture population heterogeneity; retrospective selection may introduce bias.
Some concern, Moderate sample size; single-center data may not capture population heterogeneity; retrospective selection may introduce bias. Some concern, South Indian hospital population; external generalizability to other regions or healthcare systems uncertain.
Some concern, South Indian hospital population; external generalizability to other regions or healthcare systems uncertain. Some concern, Appropriate algorithms and interpretability tools used, but potential overfitting due to small dataset; internal validation only.
Some concern, Appropriate algorithms and interpretability tools used, but potential overfitting due to small dataset; internal validation only. Low concern, Predictors based on routinely collected lab and clinical parameters; feasible for clinical adoption.
Low concern, Predictors based on routinely collected lab and clinical parameters; feasible for clinical adoption. High risk, Lack of comparator prevents evaluation of incremental value beyond existing clinical criteria.
High risk, Lack of comparator prevents evaluation of incremental value beyond existing clinical criteria. Some concern, Limits clinical interpretability relative to standard DKD staging frameworks.
Some concern, Limits clinical interpretability relative to standard DKD staging frameworks. Low risk, Outcomes based on standard KDIGO staging and eGFR; reliable and objective.
Low risk, Outcomes based on standard KDIGO staging and eGFR; reliable and objective. Low concern, Consistent with accepted nephrology definitions and clinical relevance.
Low concern, Consistent with accepted nephrology definitions and clinical relevance. Some concern, Retrospective timing may affect consistency of follow-up and missing data patterns.
Some concern, Retrospective timing may affect consistency of follow-up and missing data patterns. Low concern, Reflects clinically realistic patient follow-up intervals for DKD.
Low concern, Reflects clinically realistic patient follow-up intervals for DKD. Some concern, Institutional data quality and recording practices could vary; limits external reproducibility.
Some concern, Institutional data quality and recording practices could vary; limits external reproducibility. Some concern, Single-center scope may affect generalizability across diverse healthcare infrastructures.
Some concern, Single-center scope may affect generalizability across diverse healthcare infrastructures. Low risk, Purpose is well-defined, clinically relevant, and ethically appropriate.
Low risk, Purpose is well-defined, clinically relevant, and ethically appropriate. Low concern, Intended use aligns with clinical decision support in nephrology.
Low concern, Intended use aligns with clinical decision support in nephrology. Overall RoB: Moderate
Overall RoB: Moderate Overall Applicability: Moderate
Overall Applicability: Moderate Green = External validation (multi-site or independent cohort).
Green = External validation (multi-site or independent cohort).  Yellow = Internal split or temporal validation only.
Yellow = Internal split or temporal validation only.  Red = No external validation/preprint/exploratory approach.
Red = No external validation/preprint/exploratory approach. Low
Low Moderate
Moderate Moderate
Moderate Low
Low Low
Low Low
Low Low
Low Low
Low Moderate
Moderate Moderate
Moderate Moderate
Moderate Moderate
Moderate Low
Low Low
Low Moderate
Moderate Moderate
Moderate Green = External validation (multi-site or independent cohort).
Green = External validation (multi-site or independent cohort).  Yellow = Internal split or temporal validation only.
Yellow = Internal split or temporal validation only.  Low
Low Low
Low Moderate
Moderate Low
Low Low
Low Low
Low Low
Low Low
Low Low
Low Low
Low Moderate
Moderate Moderate
Moderate Moderate
Moderate Low
Low Low
Low Low
Low Moderate
Moderate Moderate
Moderate Green = External validation (multi-site or independent cohort).
Green = External validation (multi-site or independent cohort).  Yellow = Internal split or temporal validation only.
Yellow = Internal split or temporal validation only. Moderate
Moderate Low
Low Low
Low Low
Low Moderate
Moderate Low
Low Low
Low Low
Low Low
Low Low
Low Low-to-Moderate
Low-to-Moderate Low
Low Green = External validation (multi-site or independent cohort).
Green = External validation (multi-site or independent cohort).  Yellow = Internal split or temporal validation only.
Yellow = Internal split or temporal validation only. High/Unclear, Selective recruitment (urine proteomics + pathology cohort) with limited reporting on inclusion/exclusion; possible spectrum bias.
High/Unclear, Selective recruitment (urine proteomics + pathology cohort) with limited reporting on inclusion/exclusion; possible spectrum bias. Moderate, Participants unlikely to reflect general T2DM population; community-clinic applicability uncertain.
Moderate, Participants unlikely to reflect general T2DM population; community-clinic applicability uncertain. Moderate/Unclear, Unclear blinding; thresholds not pre-specified; same dataset used for discovery and model training → overfitting risk.
Moderate/Unclear, Unclear blinding; thresholds not pre-specified; same dataset used for discovery and model training → overfitting risk. Moderate, Uses specialized proteomics pipeline; external platforms may differ → limited generalizability.
Moderate, Uses specialized proteomics pipeline; external platforms may differ → limited generalizability. Low → Moderate, Pathology-based reference appropriate but possibly heterogeneous (biopsy vs. clinical classification); unclear if consistent across subjects.
Low → Moderate, Pathology-based reference appropriate but possibly heterogeneous (biopsy vs. clinical classification); unclear if consistent across subjects. Moderate, Standard relevant to DKD but not necessarily identical to routine clinical endpoints.
Moderate, Standard relevant to DKD but not necessarily identical to routine clinical endpoints. High, Incomplete information on timing between index and reference tests; unclear follow-up; potential verification bias.
High, Incomplete information on timing between index and reference tests; unclear follow-up; potential verification bias. Moderate, Specialized research workflow limits applicability to routine timelines and sampling logistics.
Moderate, Specialized research workflow limits applicability to routine timelines and sampling logistics. Overall RoB: High, Unclear patient selection + limited reporting on timing increase risk.
Overall RoB: High, Unclear patient selection + limited reporting on timing increase risk. Overall Applicability: Moderate, Study setting and assay platform differ from standard clinical practice; external validation needed.
Overall Applicability: Moderate, Study setting and assay platform differ from standard clinical practice; external validation needed. Yellow = Internal split or temporal validation only.
Yellow = Internal split or temporal validation only.  Orange = limited or incomplete validation (small-sample temporal validation, narrow subgroup validation, or internal validation with methodological limitations).
Orange = limited or incomplete validation (small-sample temporal validation, narrow subgroup validation, or internal validation with methodological limitations).  Red = No external validation/preprint/exploratory approach.
Red = No external validation/preprint/exploratory approach. Low → Moderate, Participants included clear diagnostic categories (T2DM with/without DKD), but sampling method and exclusion criteria were not fully described; possible selection bias if matched retrospectively.
Low → Moderate, Participants included clear diagnostic categories (T2DM with/without DKD), but sampling method and exclusion criteria were not fully described; possible selection bias if matched retrospectively. Low, DKD and control populations clinically relevant; findings likely applicable to standard T2DM cohorts with albuminuria-based classification.
Low, DKD and control populations clinically relevant; findings likely applicable to standard T2DM cohorts with albuminuria-based classification. Moderate, ML approach (e.g., SVM, RF) applied without fully independent test set; unclear blinding to reference standard; internal validation only.
Moderate, ML approach (e.g., SVM, RF) applied without fully independent test set; unclear blinding to reference standard; internal validation only. Moderate, Omics workflows and normalization pipelines may not generalize across assay platforms; batch effects could limit external use.
Moderate, Omics workflows and normalization pipelines may not generalize across assay platforms; batch effects could limit external use. Low, Diagnostic definitions followed established DKD criteria (albuminuria, eGFR decline); reference standard appropriate and consistently applied.
Low, Diagnostic definitions followed established DKD criteria (albuminuria, eGFR decline); reference standard appropriate and consistently applied. Low, Reference outcomes align well with clinical practice and guidelines.
Low, Reference outcomes align well with clinical practice and guidelines. Moderate, Timing between urine collection and DKD classification not explicitly stated; unclear if temporal gaps could bias associations.
Moderate, Timing between urine collection and DKD classification not explicitly stated; unclear if temporal gaps could bias associations. Low → Moderate, Acceptable for cross-sectional biomarker screening but limited for longitudinal prediction.
Low → Moderate, Acceptable for cross-sectional biomarker screening but limited for longitudinal prediction. Overall RoB: Moderate, Limited external validation and potential for overfitting; otherwise methodologically reasonable.
Overall RoB: Moderate, Limited external validation and potential for overfitting; otherwise methodologically reasonable. Overall Applicability: Low Concern, Results relevant to typical DKD diagnostic settings, but omics reproducibility remains a constraint.
Overall Applicability: Low Concern, Results relevant to typical DKD diagnostic settings, but omics reproducibility remains a constraint. Green = External validation (multi-site or independent cohort).
Green = External validation (multi-site or independent cohort).  Yellow = Internal split or temporal validation only.
Yellow = Internal split or temporal validation only.  Orange = limited or incomplete validation (small-sample temporal validation, narrow subgroup validation, or internal validation with methodological limitations).
Orange = limited or incomplete validation (small-sample temporal validation, narrow subgroup validation, or internal validation with methodological limitations). Low
Low Low
Low Moderate
Moderate Low
Low Low
Low Low
Low Low
Low Low
Low Moderate
Moderate Moderate
Moderate High
High Moderate
Moderate Low
Low Low
Low High
High Moderate
Moderate Moderate-High
Moderate-High Low
Low Green = External validation (multi-site or independent cohort).
Green = External validation (multi-site or independent cohort).  Yellow = Internal split or temporal validation only.
Yellow = Internal split or temporal validation only.  Orange = limited or incomplete validation (small-sample temporal validation, narrow subgroup validation, or internal validation with methodological limitations).
Orange = limited or incomplete validation (small-sample temporal validation, narrow subgroup validation, or internal validation with methodological limitations).  Red = No external validation/preprint/exploratory approach.
Red = No external validation/preprint/exploratory approach. Low
Low Low
Low Low
Low Low
Low Low
Low Low
Low Low
Low Low
Low Moderate
Moderate Moderate
Moderate High
High Moderate
Moderate Low
Low Low
Low Moderate
Moderate Moderate
Moderate Moderate
Moderate Low
Low Green = External validation (multi-site or independent cohort).
Green = External validation (multi-site or independent cohort).  Yellow = Internal split or temporal validation only.
Yellow = Internal split or temporal validation only.  Red = No external validation/preprint/exploratory approach.
Red = No external validation/preprint/exploratory approach. Moderate risk, retrospective design may introduce selection bias and missingness in clinical-genetic linkage.
Moderate risk, retrospective design may introduce selection bias and missingness in clinical-genetic linkage. Low concern, representative of tertiary diabetic populations.
Low concern, representative of tertiary diabetic populations. Moderate risk, details of model tuning and cross-validation split not fully reported; unclear if feature leakage was avoided.
Moderate risk, details of model tuning and cross-validation split not fully reported; unclear if feature leakage was avoided. Moderate concern, algorithm may overfit tertiary-center EMR data, limiting community translation.
Moderate concern, algorithm may overfit tertiary-center EMR data, limiting community translation. High risk, absence of external biological validation may weaken subtype credibility.
High risk, absence of external biological validation may weaken subtype credibility. Moderate concern, relies on EMR and genetic proxies rather than clinical pathology confirmation.
Moderate concern, relies on EMR and genetic proxies rather than clinical pathology confirmation. Moderate risk, potential temporal mismatch between clinical and genomic data points.
Moderate risk, potential temporal mismatch between clinical and genomic data points. Low concern, overall timing reasonable for computational phenotyping.
Low concern, overall timing reasonable for computational phenotyping. High risk, limited transparency and missing calibration or generalizability metrics.
High risk, limited transparency and missing calibration or generalizability metrics. High concern, lack of independent validation and reproducibility assessment.
High concern, lack of independent validation and reproducibility assessment. High overall RoB
High overall RoB Moderate applicability concern
Moderate applicability concern Green = External validation (multi-site or independent cohort).
Green = External validation (multi-site or independent cohort).  Yellow = Internal split or temporal validation only.
Yellow = Internal split or temporal validation only.  Red = No external validation/preprint/exploratory approach.
Red = No external validation/preprint/exploratory approach. High
High High
High Unclear
Unclear Unclear
Unclear Unclear
Unclear Unclear
Unclear Unclear
Unclear Unclear
Unclear High
High Moderate
Moderate Unclear
Unclear Unclear
Unclear Moderate
Moderate Unclear
Unclear Moderate
Moderate High
High High
High High
High High
High Unclear
Unclear Moderate
Moderate Unclear
Unclear Moderate
Moderate High
High High
High High
High Moderate
Moderate High
High Moderate
Moderate Yellow = Internal split or temporal validation only.
Yellow = Internal split or temporal validation only.  Red = No external validation/preprint/exploratory approach.
Red = No external validation/preprint/exploratory approach. Unclear risk, The review included studies of diabetic patients with and without nephropathy, but inclusion criteria across studies varied widely (different stages of DN, different diabetes types, and demographic variability). No explicit description of how participants were selected in included studies.
Unclear risk, The review included studies of diabetic patients with and without nephropathy, but inclusion criteria across studies varied widely (different stages of DN, different diabetes types, and demographic variability). No explicit description of how participants were selected in included studies. Unclear concern, Applicability limited by heterogeneity in populations (T1DM vs. T2DM, ethnic differences, sample sources).
Unclear concern, Applicability limited by heterogeneity in populations (T1DM vs. T2DM, ethnic differences, sample sources). High risk, The review summarized various ML approaches (e.g., RF, SVM, LASSO), but many primary studies lacked validation procedures or transparent performance metrics. The review did not critically appraise algorithm robustness or validation quality.
High risk, The review summarized various ML approaches (e.g., RF, SVM, LASSO), but many primary studies lacked validation procedures or transparent performance metrics. The review did not critically appraise algorithm robustness or validation quality. Moderate concern, Some ML models aimed to identify candidate biomarkers (not diagnostic tools), so their direct clinical applicability is limited.
Moderate concern, Some ML models aimed to identify candidate biomarkers (not diagnostic tools), so their direct clinical applicability is limited. Unclear risk, DN definitions varied (e.g., based on eGFR, albuminuria, biopsy, or clinical diagnosis). The review did not standardize or stratify based on diagnostic criteria.
Unclear risk, DN definitions varied (e.g., based on eGFR, albuminuria, biopsy, or clinical diagnosis). The review did not standardize or stratify based on diagnostic criteria. Unclear concern, Variability in diagnostic definitions affects generalizability of biomarker findings.
Unclear concern, Variability in diagnostic definitions affects generalizability of biomarker findings. High risk, The review did not evaluate timing consistency or completeness of datasets in primary studies. Missing data handling and participant flow were not discussed.
High risk, The review did not evaluate timing consistency or completeness of datasets in primary studies. Missing data handling and participant flow were not discussed. Moderate concern, Potential temporal mismatch between biomarker sampling and DN diagnosis may bias interpretation.
Moderate concern, Potential temporal mismatch between biomarker sampling and DN diagnosis may bias interpretation. High RoB, No formal quality or bias assessment (e.g., QUADAS-2, PROBAST) applied; heterogeneous populations, methods, and validation standards.
High RoB, No formal quality or bias assessment (e.g., QUADAS-2, PROBAST) applied; heterogeneous populations, methods, and validation standards. Moderate applicability concern, Review useful for identifying research trends but limited clinical translation due to poor standardization and unclear diagnostic validity.
Moderate applicability concern, Review useful for identifying research trends but limited clinical translation due to poor standardization and unclear diagnostic validity. Yellow = Internal split or temporal validation only.
Yellow = Internal split or temporal validation only.  Red = No external validation/preprint/exploratory approach.
Red = No external validation/preprint/exploratory approach. Unclear risk, Sampling may not reflect broader DKD populations (mostly moderate to severe stages).
Unclear risk, Sampling may not reflect broader DKD populations (mostly moderate to severe stages). Unclear concern, Applicability may be limited to similar hospital-based populations.
Unclear concern, Applicability may be limited to similar hospital-based populations. High risk, Possible overfitting and unclear missing data handling.
High risk, Possible overfitting and unclear missing data handling. Moderate concern, KAI may not generalize to settings with different patient characteristics or lab standards.
Moderate concern, KAI may not generalize to settings with different patient characteristics or lab standards. Unclear risk, Reference measures acceptable but not standardized across datasets.
Unclear risk, Reference measures acceptable but not standardized across datasets. Low concern, Consistent with clinical definitions of kidney function.
Low concern, Consistent with clinical definitions of kidney function. High risk, Potential bias due to inconsistent timing and incomplete follow-up data.
High risk, Potential bias due to inconsistent timing and incomplete follow-up data. Moderate concern, Limited longitudinal data reduce applicability for real-world progression prediction.
Moderate concern, Limited longitudinal data reduce applicability for real-world progression prediction. High overall RoB
High overall RoB Moderate applicability concern
Moderate applicability concern Green = External validation (multi-site or independent cohort).
Green = External validation (multi-site or independent cohort).  Yellow = Internal split or temporal validation only.
Yellow = Internal split or temporal validation only.  Red = No external validation/preprint/exploratory approach.
Red = No external validation/preprint/exploratory approach. Unclear
Unclear Unclear
Unclear High
High Moderate
Moderate Unclear
Unclear Moderate
Moderate Unclear
Unclear Low
Low High
High Moderate
Moderate Unclear
Unclear Moderate
Moderate Moderate
Moderate High
High Moderate
Moderate High
High Moderate
Moderate Green = External validation (multi-site or independent cohort).
Green = External validation (multi-site or independent cohort).  Yellow = Internal split or temporal validation only.
Yellow = Internal split or temporal validation only.  Red = No external validation/preprint/exploratory approach.
Red = No external validation/preprint/exploratory approach. Unclear
Unclear Moderate
Moderate High
High Moderate
Moderate Unclear
Unclear Moderate
Moderate Unclear
Unclear Low
Low High
High Moderate
Moderate Unclear
Unclear Moderate
Moderate Moderate
Moderate High
High Moderate
Moderate High
High Moderate
Moderate Green = External validation (multi-site or independent cohort).
Green = External validation (multi-site or independent cohort).  Yellow = Internal split or temporal validation only.
Yellow = Internal split or temporal validation only.  Red = No external validation/preprint/exploratory approach.
Red = No external validation/preprint/exploratory approach. Unclear risk
Unclear risk Moderate concern
Moderate concern High risk
High risk Moderate concern
Moderate concern Unclear risk
Unclear risk Moderate concern
Moderate concern Unclear risk
Unclear risk Low concern
Low concern High risk
High risk Moderate concern
Moderate concern Unclear risk
Unclear risk Moderate concern
Moderate concern Moderate concern
Moderate concern High risk
High risk Moderate concern
Moderate concern High risk
High risk Moderate concern
Moderate concern Green = External validation (multi-site or independent cohort).
Green = External validation (multi-site or independent cohort).  Yellow = Internal split or temporal validation only.
Yellow = Internal split or temporal validation only.  Red = No external validation/preprint/exploratory approach.
Red = No external validation/preprint/exploratory approach. Unclear
Unclear Moderate
Moderate Unclear
Unclear Moderate
Moderate Unclear
Unclear Moderate
Moderate Low
Low Low
Low High
High Moderate
Moderate Unclear
Unclear Moderate
Moderate Moderate
Moderate High
High Moderate
Moderate High
High Moderate
Moderate Green = External validation (multi-site or independent cohort).
Green = External validation (multi-site or independent cohort).  Yellow = Internal split or temporal validation only.
Yellow = Internal split or temporal validation only.  Red = No external validation/preprint/exploratory approach.
Red = No external validation/preprint/exploratory approach. Unclear, while external, differences in cohort characteristics suggest possible selection or spectrum bias.
Unclear, while external, differences in cohort characteristics suggest possible selection or spectrum bias. Unclear, setting (French outpatient CKD stage 4–5) may limit generalizability to other countries, dialysis cohorts, or more heterogeneous CKD populations.
Unclear, setting (French outpatient CKD stage 4–5) may limit generalizability to other countries, dialysis cohorts, or more heterogeneous CKD populations. Unclear, The validation uses an existing model, but details of model adaptation, calibration and predictor measurement in the new cohort are limited.
Unclear, The validation uses an existing model, but details of model adaptation, calibration and predictor measurement in the new cohort are limited. Unclear, Model developed in one dataset may perform differently in new populations with different baseline risks and features.
Unclear, Model developed in one dataset may perform differently in new populations with different baseline risks and features. Unclear, absence of comparator limits assessment of relative performance.
Unclear, absence of comparator limits assessment of relative performance. Unclear, Without benchmark models, it’s hard to evaluate added value in this setting.
Unclear, Without benchmark models, it’s hard to evaluate added value in this setting. Low, Outcome is well-defined and appropriate.
Low, Outcome is well-defined and appropriate. Low, Applicability of outcome is good for clinical mortality prediction in CKD stage 4–5.
Low, Applicability of outcome is good for clinical mortality prediction in CKD stage 4–5. High, The potential for bias is elevated because of insufficient reporting of timing, missing follow-up handling, and potential changes in care over time.
High, The potential for bias is elevated because of insufficient reporting of timing, missing follow-up handling, and potential changes in care over time. Moderate concern, The 2-year horizon is clinically relevant, but differences in cohorts and treatment eras may affect transportability.
Moderate concern, The 2-year horizon is clinically relevant, but differences in cohorts and treatment eras may affect transportability. Unclear, Single region may limit heterogeneity; applicability to broader settings uncertain.
Unclear, Single region may limit heterogeneity; applicability to broader settings uncertain. Moderate concern, The setting is relevant for outpatient CKD patients, but generalization to other geographies, healthcare systems or dialysis populations is uncertain.
Moderate concern, The setting is relevant for outpatient CKD patients, but generalization to other geographies, healthcare systems or dialysis populations is uncertain. Moderate concern, Intended use is compatible with the validation setting, but the lack of broad transportability reduces practical applicability.
Moderate concern, Intended use is compatible with the validation setting, but the lack of broad transportability reduces practical applicability. High, Key modeling aspects (calibration, missing data, model updating) are inadequately reported, increasing bias risk.
High, Key modeling aspects (calibration, missing data, model updating) are inadequately reported, increasing bias risk. Moderate concern, The model shows reasonable discrimination, but limited detail and moderate specificity raise concerns about real-world performance.
Moderate concern, The model shows reasonable discrimination, but limited detail and moderate specificity raise concerns about real-world performance. High RoB
High RoB Moderate applicability concern
Moderate applicability concern Green = External validation (multi-site or independent cohort).
Green = External validation (multi-site or independent cohort).  Yellow = Internal split or temporal validation only.
Yellow = Internal split or temporal validation only.  Red = No external validation/preprint/exploratory approach.
Red = No external validation/preprint/exploratory approach. Unclear
Unclear Moderate
Moderate Unclear
Unclear Moderate
Moderate Unclear
Unclear Moderate
Moderate Low
Low Low
Low High
High Moderate
Moderate Unclear
Unclear Moderate
Moderate High
High Moderate
Moderate High RoB
High RoB Moderate applicability concern
Moderate applicability concern Green = External validation (multi-site or independent cohort).
Green = External validation (multi-site or independent cohort).  Yellow = Internal split or temporal validation only.
Yellow = Internal split or temporal validation only.  Red = No external validation/preprint/exploratory approach.
Red = No external validation/preprint/exploratory approach. Unclear risk
Unclear risk Moderate concern, single-center data may not generalize to other populations or health systems
Moderate concern, single-center data may not generalize to other populations or health systems High risk
High risk Moderate concern, model may be overfit to center-specific patterns
Moderate concern, model may be overfit to center-specific patterns Unclear risk
Unclear risk Moderate concern
Moderate concern Unclear risk
Unclear risk Moderate concern
Moderate concern High risk
High risk Moderate concern
Moderate concern Unclear risk
Unclear risk Moderate concern
Moderate concern Moderate concern
Moderate concern High risk
High risk Moderate concern
Moderate concern High RoB
High RoB Moderate applicability concern
Moderate applicability concern Yellow = Internal split or temporal validation only.
Yellow = Internal split or temporal validation only.  Red = No external validation/preprint/exploratory approach.
Red = No external validation/preprint/exploratory approach.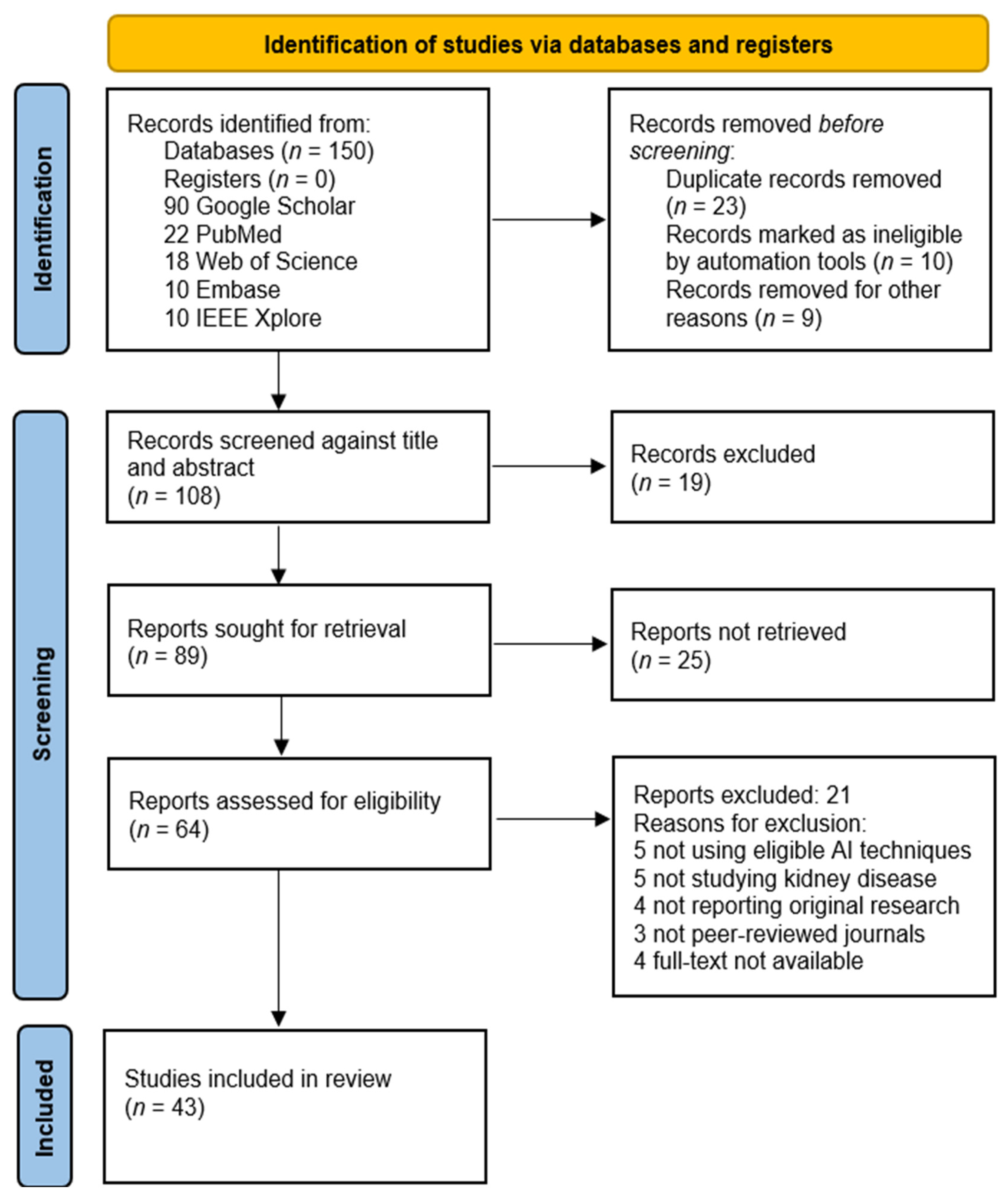
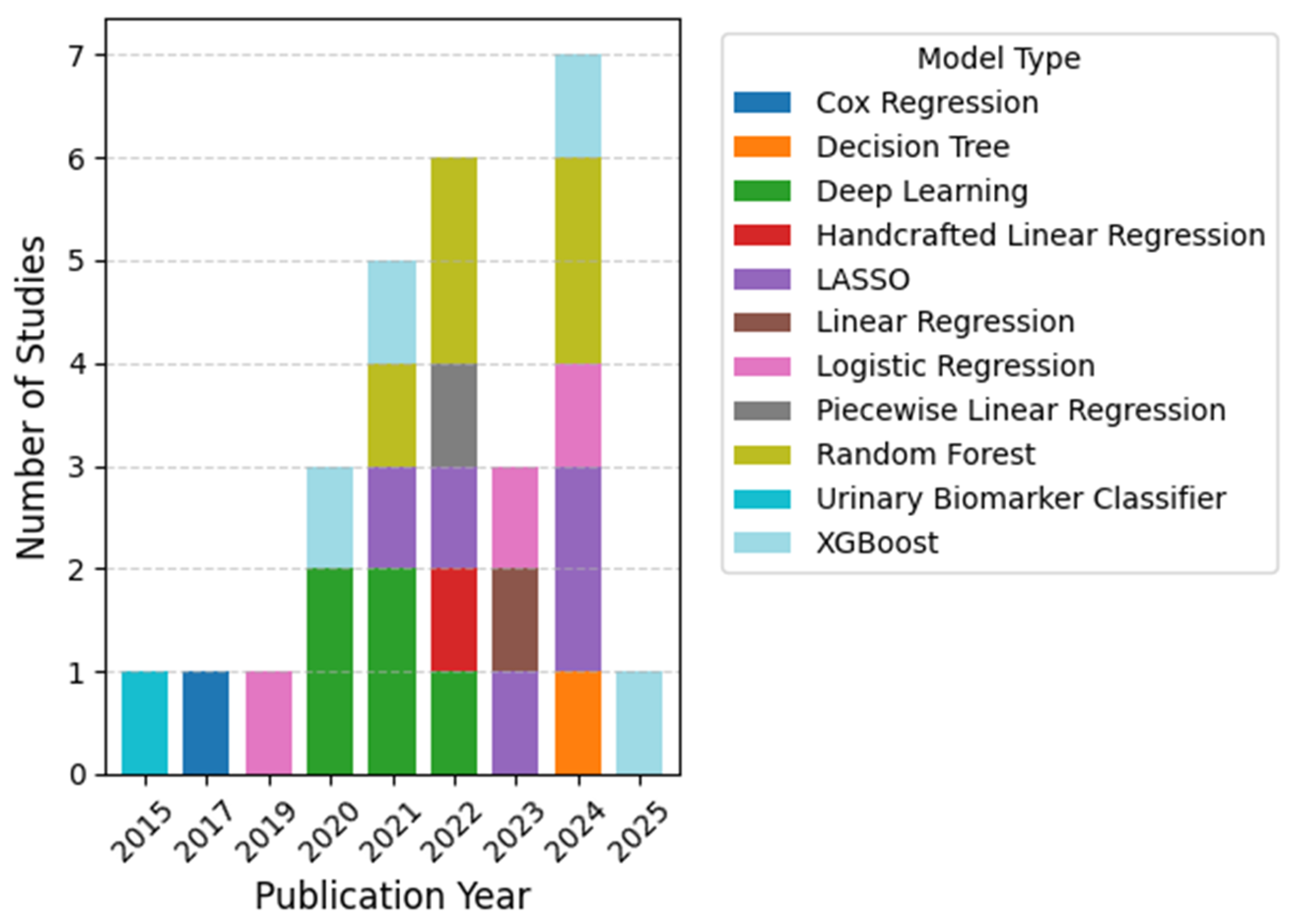
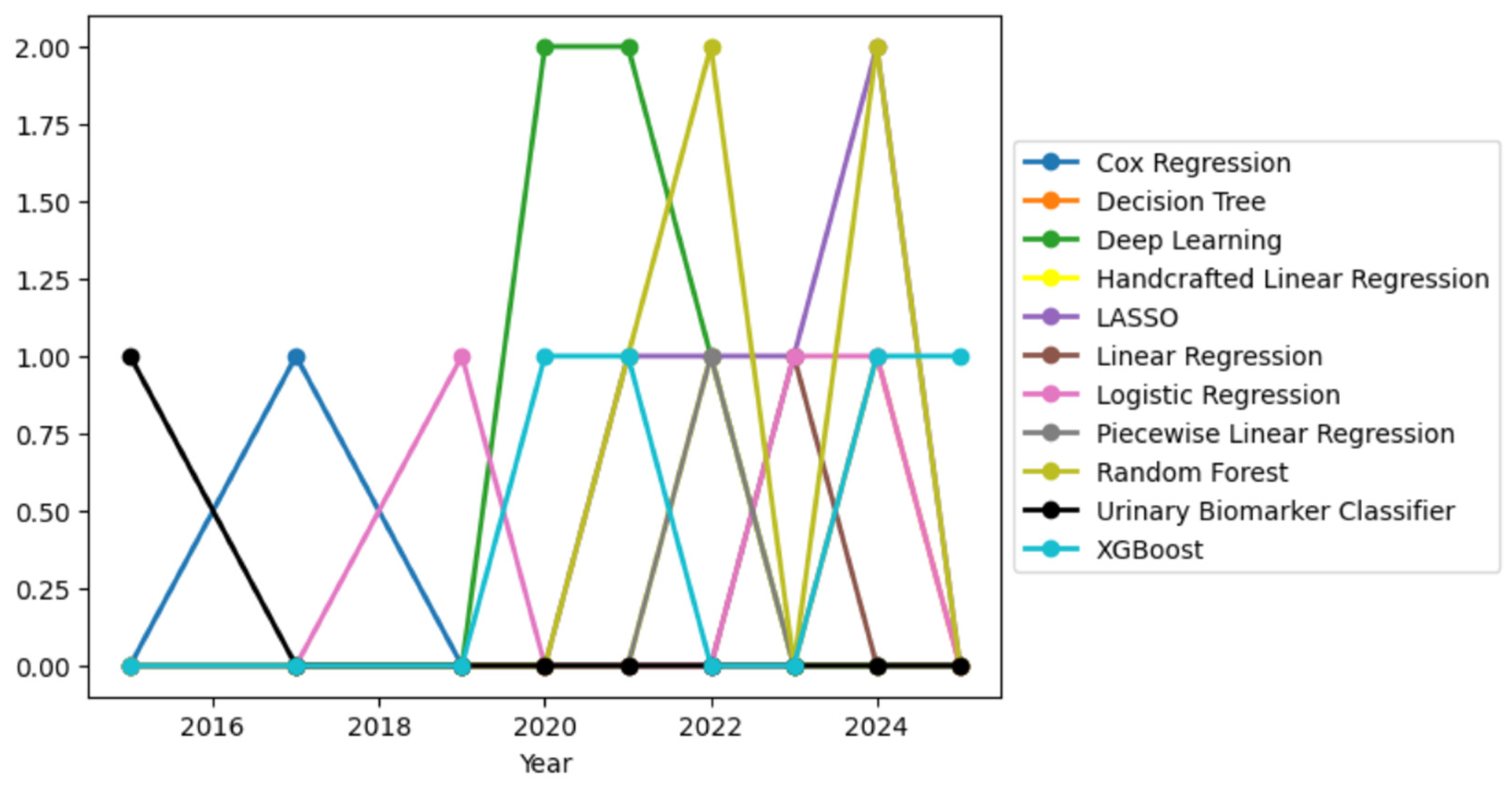
 High, External or Temporal Validation
High, External or Temporal Validation Moderate, Internal Validation only
Moderate, Internal Validation only  High, External or Temporal Validation
High, External or Temporal Validation Moderate, Internal Validation only
Moderate, Internal Validation only Low, No Validation
Low, No Validation Low, No Validation
Low, No Validation High, External or Temporal Validation
High, External or Temporal Validation Green = External validation (multi-site or independent cohort).
Green = External validation (multi-site or independent cohort).  Yellow = Internal split or temporal validation only.
Yellow = Internal split or temporal validation only.  Red = No external validation/preprint/exploratory approach.
Red = No external validation/preprint/exploratory approach. High, External Validation
High, External Validation Low (small sample, no validation)
Low (small sample, no validation) Moderate (internal split validation only)
Moderate (internal split validation only) High (multi-cohort validation)
High (multi-cohort validation) Moderate (observational diagnostic study; internal validation only)
Moderate (observational diagnostic study; internal validation only) Green = External validation (multi-site or independent cohort).
Green = External validation (multi-site or independent cohort).  Yellow = Internal split or temporal validation only.
Yellow = Internal split or temporal validation only.  Red = No external validation/preprint/exploratory approach.
Red = No external validation/preprint/exploratory approach. Moderate (internal validation only)
Moderate (internal validation only) Moderate (internal cross-validation only)
Moderate (internal cross-validation only) High (external validation, robust calibration)
High (external validation, robust calibration) High (independent test dataset; good reproducibility)
High (independent test dataset; good reproducibility) Green = External validation (multi-site or independent cohort).
Green = External validation (multi-site or independent cohort).  Yellow = Internal split or temporal validation only.
Yellow = Internal split or temporal validation only.  High, External Validation
High, External Validation High, External Validation
High, External Validation High, External Validation
High, External Validation  Moderate (internal split only)
Moderate (internal split only) Green = External validation (multi-site or independent cohort).
Green = External validation (multi-site or independent cohort).  Yellow = Internal split or temporal validation only.
Yellow = Internal split or temporal validation only.  Systematic Review (Narrative)
Systematic Review (Narrative) External Cohort
External Cohort Temporal Split (Internal)
Temporal Split (Internal) Single-Center Retrospective
Single-Center Retrospective Meta-Analysis (Pooled)
Meta-Analysis (Pooled) External Cohort
External Cohort Multi-Cohort Validation
Multi-Cohort Validation Green = External validation (multi-site or independent cohort).
Green = External validation (multi-site or independent cohort).  Yellow = Internal split or temporal validation only.
Yellow = Internal split or temporal validation only.  Red = No external validation/preprint/exploratory approach.
Red = No external validation/preprint/exploratory approach. External Validation (multi-site pathology + omics).
External Validation (multi-site pathology + omics). Internal Validation (5-fold CV).
Internal Validation (5-fold CV). External Validation (multi-hospital cohort).
External Validation (multi-hospital cohort). Internal Temporal Split.
Internal Temporal Split. No External Validation (medRxiv preprint).
No External Validation (medRxiv preprint). Reviewed Externally Validated Models.
Reviewed Externally Validated Models. Review of Validated Models.
Review of Validated Models. External Validation (European cohort).
External Validation (European cohort). Green = External validation (multi-site or independent cohort).
Green = External validation (multi-site or independent cohort).  Yellow = Internal split or temporal validation only.
Yellow = Internal split or temporal validation only.  Red = No external validation/preprint/exploratory approach.
Red = No external validation/preprint/exploratory approach. High
High High
High Moderate
Moderate Low
Low High
High Low
Low Low
Low Green = External validation (multi-site or independent cohort).
Green = External validation (multi-site or independent cohort).  Yellow = Internal split or temporal validation only.
Yellow = Internal split or temporal validation only.  Red = No external validation/preprint/exploratory approach.
Red = No external validation/preprint/exploratory approach. (Some concern)
(Some concern) (Some concern)
(Some concern) (Low concern)
(Low concern) (Some concern)
(Some concern) (Some concern)
(Some concern) (Some concern)
(Some concern) (Low concern)
(Low concern) (Low concern)
(Low concern) (Some concern)
(Some concern) (Some concern)
(Some concern) (Low concern)
(Low concern) (Some concern)
(Some concern) (Some concern)
(Some concern) (Some concern)
(Some concern) (Low concern)
(Low concern) (Some concern)
(Some concern) (Some concern)
(Some concern) (Low concern)
(Low concern) (Low concern)
(Low concern) (Some concern)
(Some concern) (Low concern)
(Low concern) (Some concern)
(Some concern) (Some concern)
(Some concern) (High concern)
(High concern) (Low concern)
(Low concern) (Low concern)
(Low concern) (Low concern)
(Low concern) (Low concern)
(Low concern) (Some concern)
(Some concern) (Low concern)
(Low concern) (Some concern)
(Some concern) (Low concern)
(Low concern) (Some concern)
(Some concern) (Low concern)
(Low concern) (Some concern)
(Some concern) (Low concern)
(Low concern) (Some concern)
(Some concern) (Some concern)
(Some concern) (Some concern)
(Some concern) (Low concern)
(Low concern) (Some concern)
(Some concern) (Some concern)
(Some concern) (Some concern)
(Some concern) (Some concern)
(Some concern) (Low concern)
(Low concern) Green = External validation (multi-site or independent cohort).
Green = External validation (multi-site or independent cohort).  Yellow = Internal split or temporal validation only.
Yellow = Internal split or temporal validation only.  Red = No external validation/preprint/exploratory approach.
Red = No external validation/preprint/exploratory approach. External temporal validation
External temporal validation Internal Cross-Validation only
Internal Cross-Validation only External dataset
External dataset Single-center Cross validation
Single-center Cross validation Development only
Development only Internal Cross Validation
Internal Cross Validation Internal validation
Internal validation Internal Cross Validation
Internal Cross Validation External multi-cohort
External multi-cohort Internal Cross Validation
Internal Cross Validation Internal Cross Validation
Internal Cross Validation Discovery only
Discovery only Internal Cross Validation only
Internal Cross Validation only Preprint; no validation
Preprint; no validation Internal Cross Validation
Internal Cross Validation Internal Cross Validation
Internal Cross Validation External validation
External validation Preprint
Preprint Internal Cross Validation
Internal Cross Validation External validation
External validation External (Singapore)
External (Singapore) External (NephroTest)
External (NephroTest) Small cohort
Small cohort Internal validation
Internal validation External (Proteomic data)
External (Proteomic data) Internal
Internal External cohort
External cohort External (CANVAS, CREDENCE)
External (CANVAS, CREDENCE) Internal
Internal External
External Internal Cross Validation
Internal Cross Validation Internal
Internal Internal
Internal External validation
External validation Internal
Internal Internal
Internal No external validation
No external validation Internal
Internal External cohort
External cohort Green = External validation (multi-site or independent cohort).
Green = External validation (multi-site or independent cohort).  Yellow = Internal split or temporal validation only.
Yellow = Internal split or temporal validation only.  Red = No external validation/preprint/exploratory approach.
Red = No external validation/preprint/exploratory approach.






