Assessment of ChatGPT in Recommending Immunohistochemistry Panels for Salivary Gland Tumors
Abstract
1. Introduction
2. Materials and Methods
2.1. Tumor Types and Selection Criteria
2.2. Pathologist Consensus and Scoring Criteria
- Literature Review and Evidence-Based Marker Selection:
- ○
- Each pathologist independently reviewed current clinical guidelines, consensus reports, and peer-reviewed studies relevant to IHC marker selection for salivary gland tumors.
- ○
- A comprehensive list of essential and ancillary markers was compiled based on documented diagnostic utility.
- Delphi Method for Consensus:
- ○
- A modified Delphi approach was used to refine the reference IHC panels.
- ○
- Pathologists participated in three rounds of blinded reviews where they rated the importance of each marker for specific tumor types. Discrepancies and disagreements identified between reviewers were discussed collectively, and markers were re-evaluated in subsequent rounds until consensus was achieved. Final agreement required at least 85% concordance among reviewers for each tumor type.
- Final Validation and Benchmarking:
- ○
- The finalized IHC panels were cross-validated against real-world pathology case datasets.
- ○
- Benchmarking was conducted by comparing the finalized panels to existing guideline-based recommendations from WHO, the Armed Forces Institute of Pathology (AFIP), and other pathology authorities.
- ○
- The validated reference panels served as the gold standard for evaluating ChatGPT’s IHC marker recommendations.
2.3. Chatbot Evaluation Framework
2.4. Data Collection
- Chatbot Response (IHC Markers): Markers recommended for each tumor type or pair.
- Pathologist Reference Panel: The finalized benchmark IHC panel was established by consensus among the participating pathologists.
- Scoring System: Responses were evaluated using a three-component scoring system:
- Accuracy Score: Inclusion of essential primary markers (1–3 scale);
- Completeness Score: Inclusion of secondary markers (1–3 scale);
- Relevance Score: Exclusion of unnecessary markers (1–3 scale);
- Composite Score: Sum of the three scores (range 3–9);
- Consistency Score: Assessed across repetitions (High, Moderate, Low variability).
2.5. Scoring Criteria
- Accuracy: Evaluates whether the chatbot included essential primary markers.
- ○
- 3 (High Accuracy): All primary markers are present.
- ○
- 2 (Moderate Accuracy): Most primary markers are present, but one essential marker is missing.
- ○
- 1 (Low Accuracy): Two or more essential markers are missing.
- Completeness: Checks if the chatbot included necessary secondary markers.
- ○
- 3 (High Completeness): Both primary and secondary markers are included.
- ○
- 2 (Moderate Completeness): Only primary markers are included.
- ○
- 1 (Low Completeness): Secondary markers are missing.
- Relevance: Assesses whether unnecessary markers are recommended.
- ○
- 3 (High Relevance): Only essential markers are recommended.
- ○
- 2 (Moderate Relevance): One irrelevant marker is included.
- ○
- 1 (Low Relevance): Two or more irrelevant markers are included.
- Composite Score:
- ○
- Calculated as the sum of accuracy, completeness, and relevance scores, yielding a possible range from 3 to 9 for each repetition.
2.6. Definition of a Rule-Based System and ChatGPT Prompt Framework
2.7. Statistical Analyses
- ▪
- Sensitivity: Measures correct identification of essential markers.
- ▪
- Specificity: Measures the correct exclusion of non-essential markers.
- ▪
- Accuracy: Overall correctness of recommendations.
3. Results
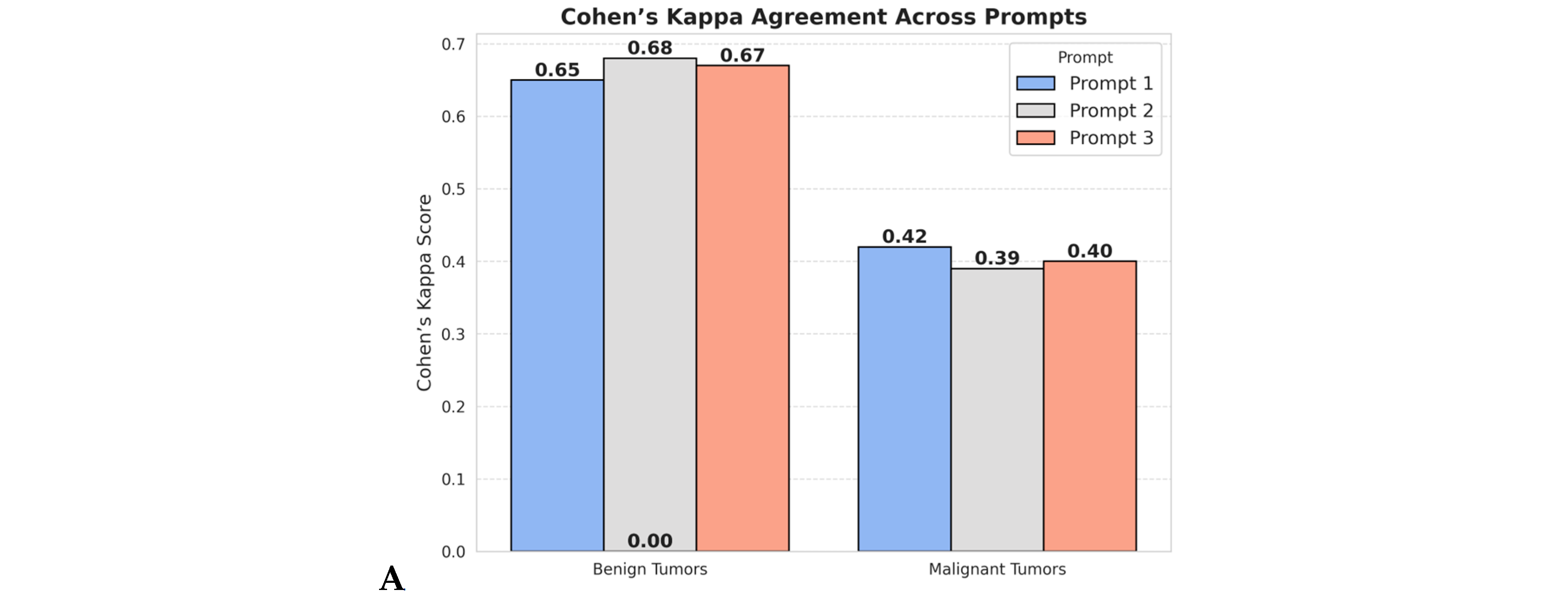
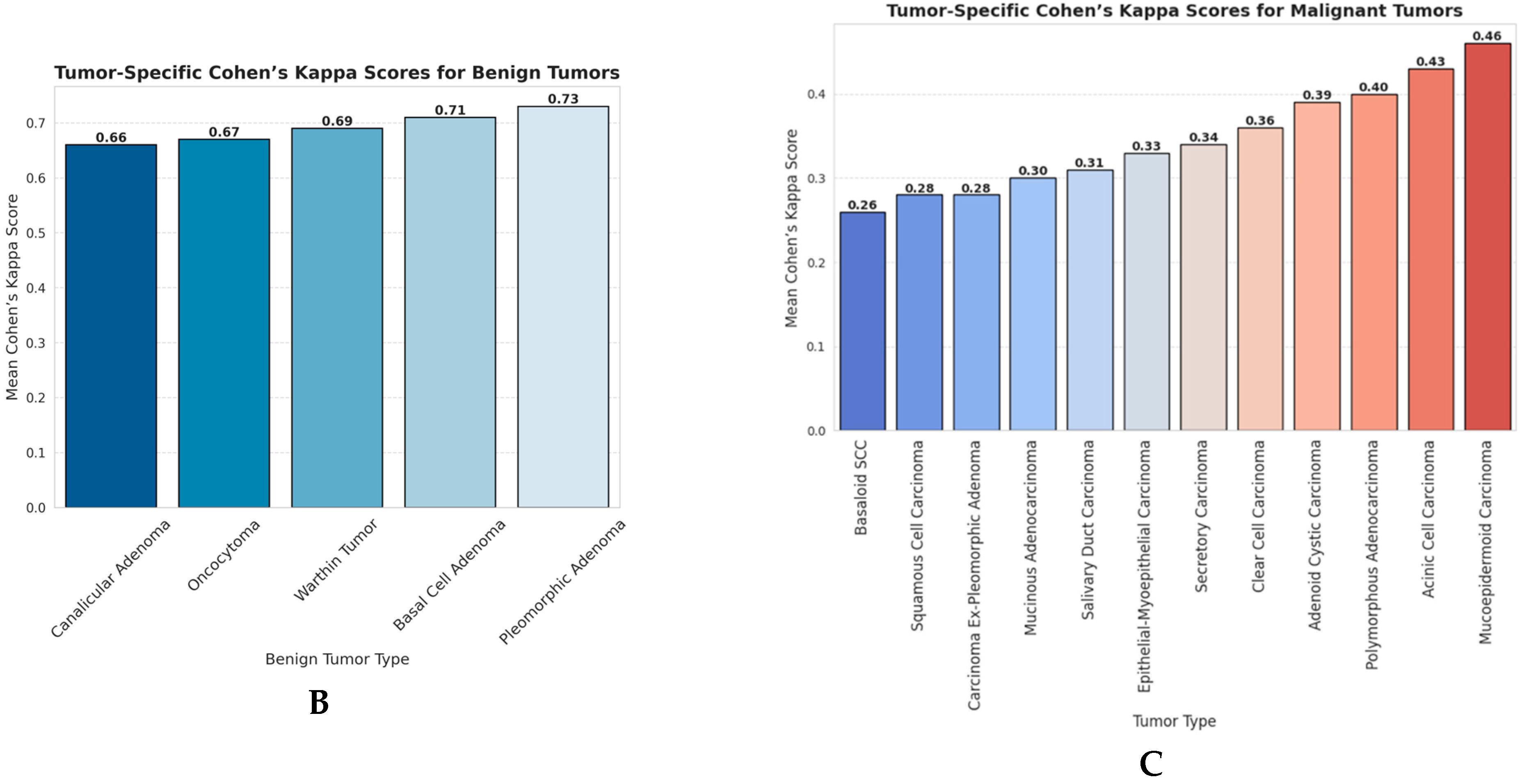
3.1. Agreement Between ChatGPT and Pathologist Panel (Cohen’s Kappa Analysis)
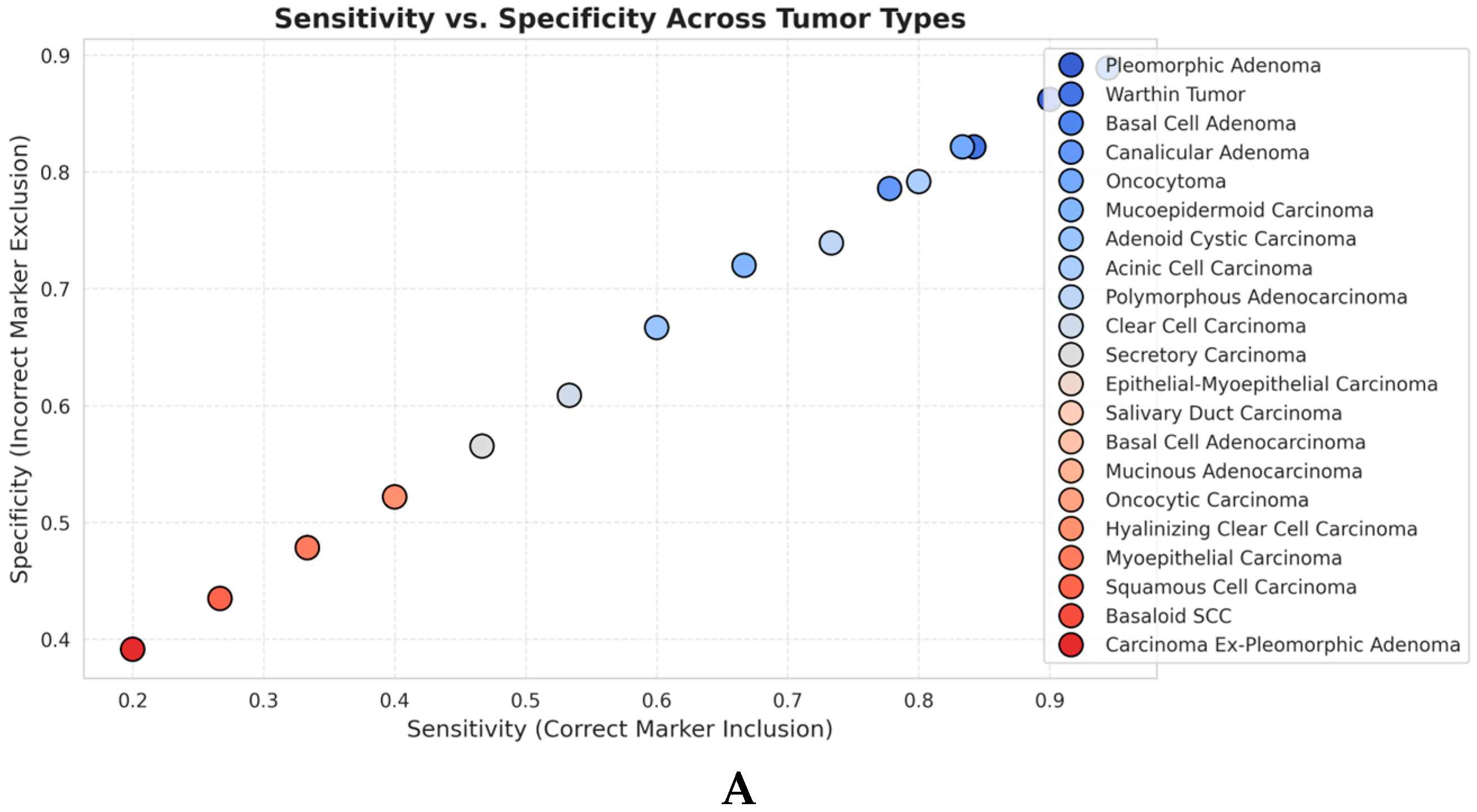
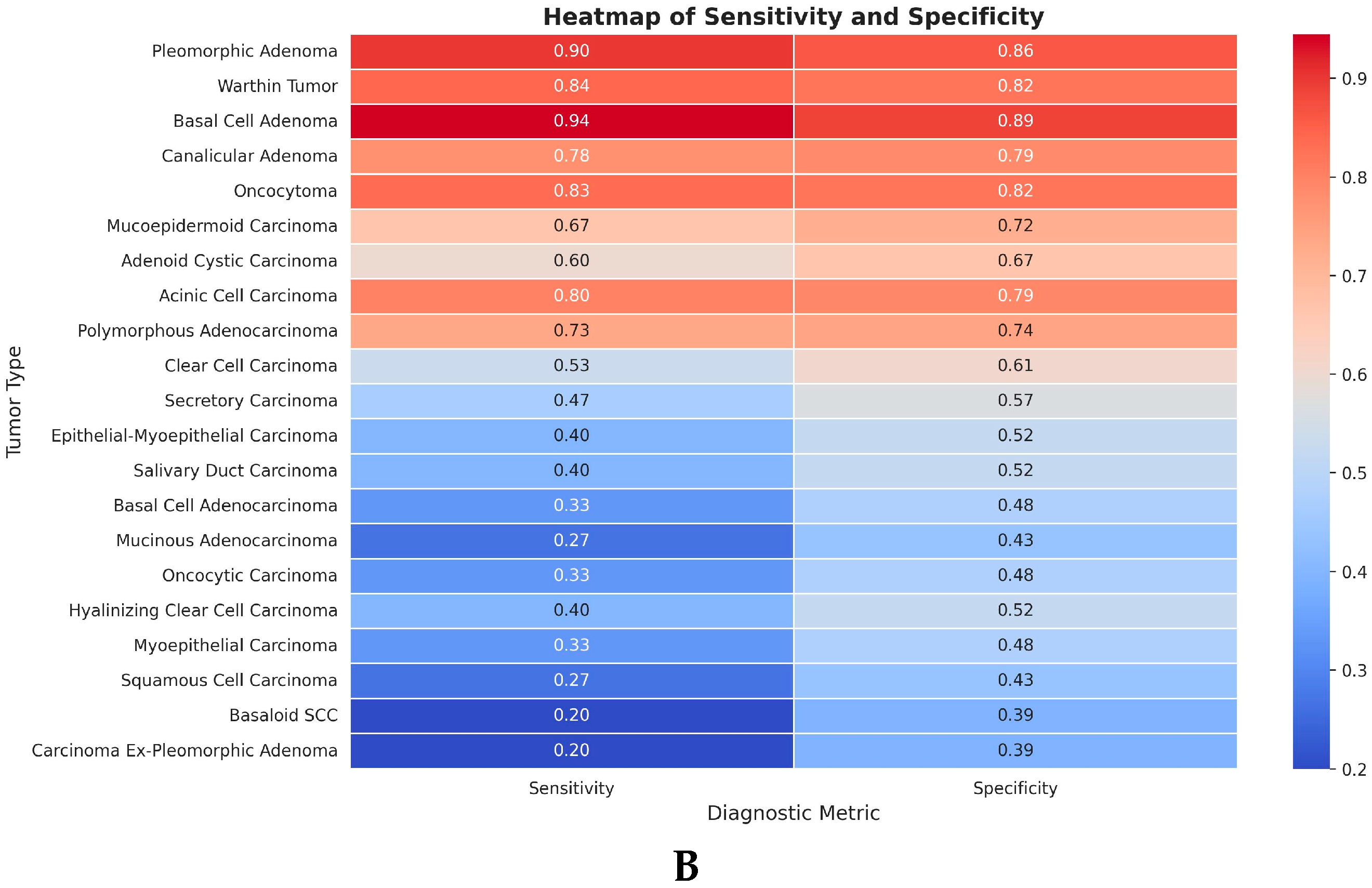
3.2. Sensitivity and Specificity Analyses
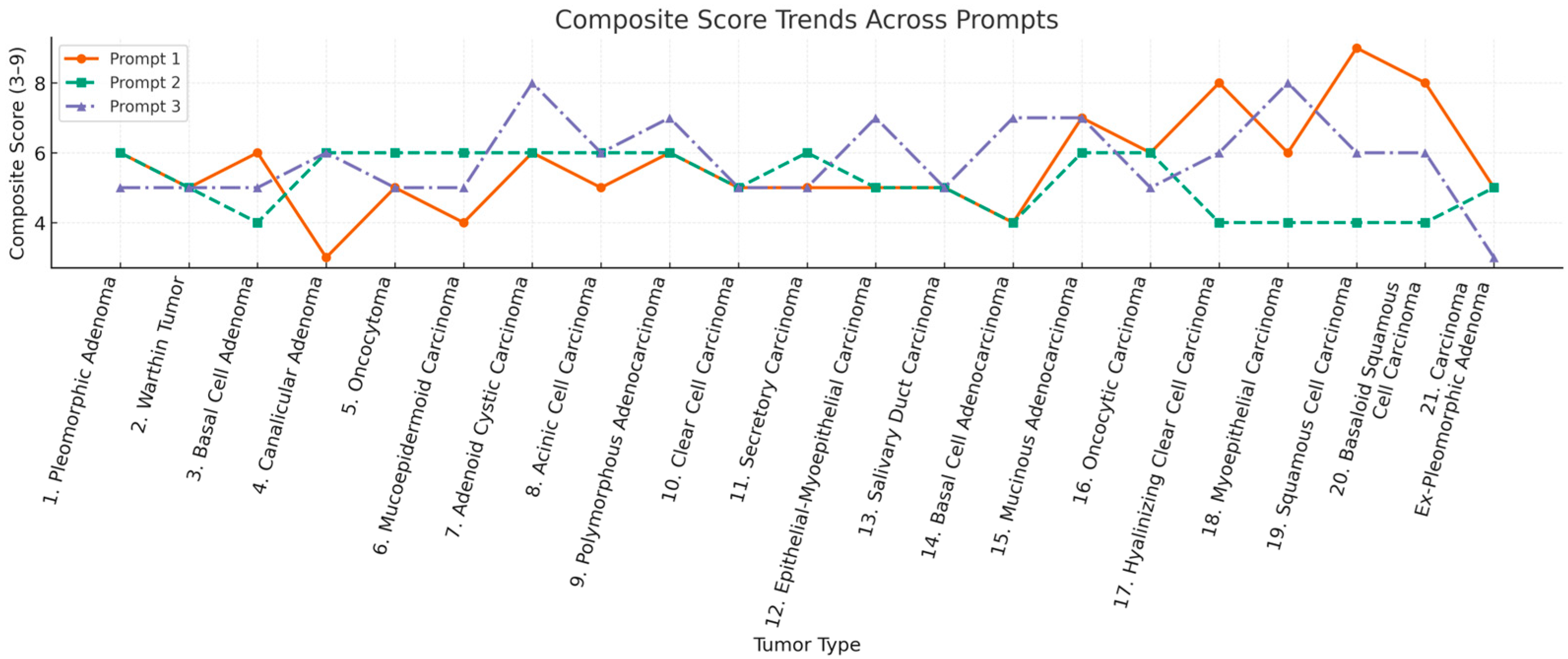
3.3. Consistency and Stability of ChatGPT’s Recommendations
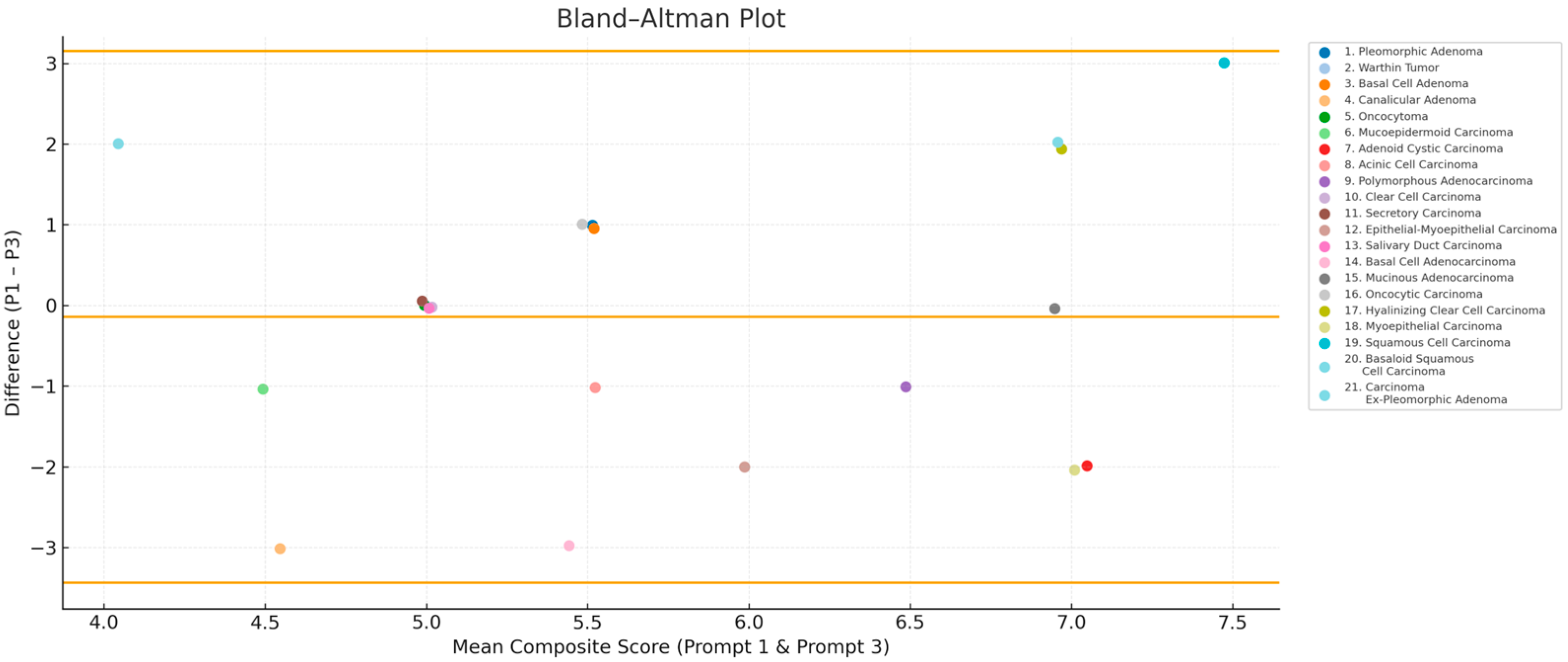
3.4. Bland–Altman Analysis: ChatGPT Score Variability
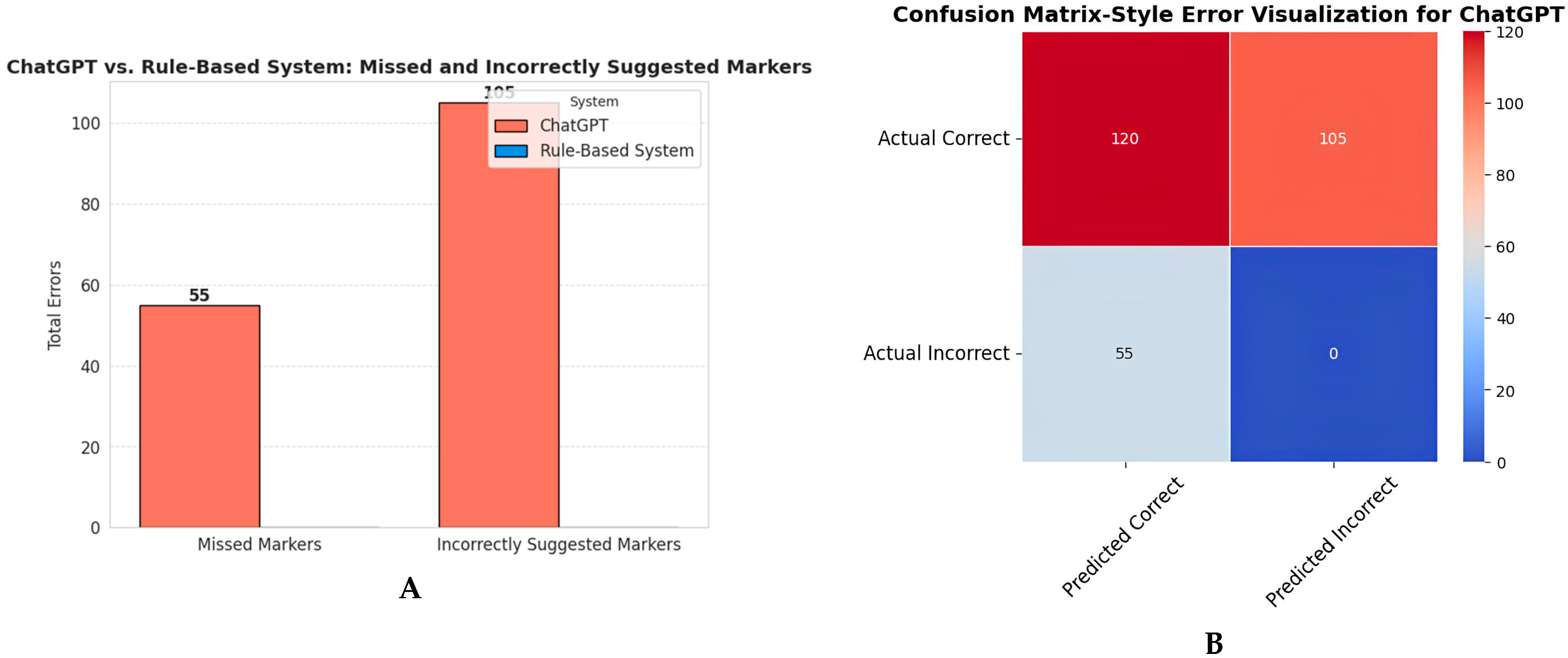
3.5. Chatbot vs. Rule-Based System Comparison
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
References
- Stenman, G.; Persson, F.; Andersson, M.K. Diagnostic and Therapeutic Implications of New Molecular Biomarkers in Salivary Gland Cancers. Oral Oncol. 2014, 50, 683–690. [Google Scholar] [CrossRef]
- Skalova, A.; Vanecek, T.; Simpson, R.H.W.; Michal, M. Molecular Advances in Salivary Gland Pathology and Their Practical Application. Diagn. Histopathol. 2012, 18, 388–396. [Google Scholar] [CrossRef]
- Speight, P.M.; Barrett, A.W. Salivary Gland Tumours: Diagnostic Challenges and an Update on the Latest WHO Classification. Diagn. Histopathol. 2020, 26, 147–158. [Google Scholar] [CrossRef]
- Iyer, J.; Hariharan, A.; Cao, U.M.N.; Mai, C.T.T.; Wang, A.; Khayambashi, P.; Nguyen, B.H.; Safi, L.; Tran, S.D. An Overview on the Histogenesis and Morphogenesis of Salivary Gland Neoplasms and Evolving Diagnostic Approaches. Cancers 2021, 13, 3910. [Google Scholar] [CrossRef]
- Kohale, M.G.; Dhobale, A.V.; Bankar, N.J.; Noman, O.; Hatgaonkar, K.; Mishra, V. Immunohistochemistry in Pathology: A Review. J. Cell Biotechnol. 2023, 9, 131–138. [Google Scholar] [CrossRef]
- Fang, R.; Wang, X.T.; Xia, Q.Y.; Zhou, X.J.; Rao, Q. Precision in Diagnostic Molecular Pathology Based on Immunohistochemistry. Crit. Rev. Oncog. 2017, 22, 451–469. [Google Scholar] [CrossRef]
- McCrary, M.R.; Galambus, J.; Chen, W. Evaluating the Diagnostic Performance of a Large Language Model-powered Chatbot for Providing Immunohistochemistry Recommendations in Dermatopathology. J. Cutan. Pathol. 2024, 51, 689–695. [Google Scholar] [CrossRef]
- Kumar, M.; Fatima, Z.H.; Goyal, P.; Qayyumi, B. Looking through the Same Lens—Immunohistochemistry for Salivary Gland Tumors: A Narrative Review on Testing and Management Strategies. Cancer Res. Stat. Treat. 2024, 7, 62–71. [Google Scholar] [CrossRef]
- Sultan, A.S.; Elgharib, M.A.; Tavares, T.; Jessri, M.; Basile, J.R. The Use of Artificial Intelligence, Machine Learning and Deep Learning in Oncologic Histopathology. J. Oral Pathol. Med. 2020, 49, 849–856. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Sun, K.; Gao, Y.; Wang, K.; Yu, G. Preparing Data for Artificial Intelligence in Pathology with Clinical-Grade Performance. Diagnostics 2023, 13, 3115. [Google Scholar] [CrossRef]
- Acs, B.; Rantalainen, M.; Hartman, J. Artificial Intelligence as the next Step towards Precision Pathology. J. Intern. Med. 2020, 288, 62–81. [Google Scholar] [CrossRef]
- Choi, S.; Kim, S. Artificial Intelligence in the Pathology of Gastric Cancer. J. Gastric Cancer 2023, 23, 410–427. [Google Scholar] [CrossRef] [PubMed]
- Frosolini, A.; Catarzi, L.; Benedetti, S.; Latini, L.; Chisci, G.; Franz, L.; Gennaro, P.; Gabriele, G. The Role of Large Language Models (LLMs) in Providing Triage for Maxillofacial Trauma Cases: A Preliminary Study. Diagnostics 2024, 14, 839. [Google Scholar] [CrossRef] [PubMed]
- Abdelsamea, M.M.; Zidan, U.; Senousy, Z.; Gaber, M.M.; Rakha, E.; Ilyas, M. A Survey on Artificial Intelligence in Histopathology Image Analysis. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 2022, 12, e1474. [Google Scholar] [CrossRef]
- Kalra, S.; Tizhoosh, H.R.; Shah, S.; Choi, C.; Damaskinos, S.; Safarpoor, A.; Shafiei, S.; Babaie, M.; Diamandis, P.; Campbell, C.J.V.; et al. Pan-Cancer Diagnostic Consensus through Searching Archival Histopathology Images Using Artificial Intelligence. NPJ Digit. Med. 2020, 3, 31. [Google Scholar] [CrossRef]
- Stenzinger, A.; Alber, M.; Allgäuer, M.; Jurmeister, P.; Bockmayr, M.; Budczies, J.; Lennerz, J.; Eschrich, J.; Kazdal, D.; Schirmacher, P.; et al. Artificial Intelligence and Pathology: From Principles to Practice and Future Applications in Histomorphology and Molecular Profiling. Semin. Cancer Biol. 2022, 84, 129–143. [Google Scholar] [CrossRef] [PubMed]
- Homeyer, A.; Lotz, J.; Schwen, L.O.; Weiss, N.; Romberg, D.; Höfener, H.; Zerbe, N.; Hufnagl, P. Artificial Intelligence in Pathology: From Prototype to Product. J. Pathol. Inform. 2021, 12, 13. [Google Scholar] [CrossRef]
- McGenity, C.; Clarke, E.L.; Jennings, C.; Matthews, G.; Cartlidge, C.; Freduah-Agyemang, H.; Stocken, D.D.; Treanor, D. Artificial Intelligence in Digital Pathology: A Systematic Review and Meta-Analysis of Diagnostic Test Accuracy. NPJ Digit. Med. 2024, 7, 114. [Google Scholar] [CrossRef]
- Colling, R.; Pitman, H.; Oien, K.; Rajpoot, N.; Macklin, P.; Bachtiar, V.; Booth, R.; Bryant, A.; Bull, J.; Bury, J.; et al. Artificial Intelligence in Digital Pathology: A Roadmap to Routine Use in Clinical Practice. J. Pathol. 2019, 249, 143–150. [Google Scholar] [CrossRef]
- Schukow, C.; Smith, S.C.; Landgrebe, E.; Parasuraman, S.; Folaranmi, O.O.; Paner, G.P.; Amin, M.B. Application of ChatGPT in Routine Diagnostic Pathology: Promises, Pitfalls, and Potential Future Directions. Adv. Anat. Pathol. 2024, 31, 15–21. [Google Scholar] [CrossRef]
- WHO. Classification of Tumours Editorial Board. In Head and Neck Tumours, 5th ed.; International Agency for Research on Cancer: Lyon, France, 2022; Volume 9. [Google Scholar]
- Tschandl, P.; Rinner, C.; Apalla, Z.; Argenziano, G.; Codella, N.; Halpern, A.; Janda, M.; Lallas, A.; Longo, C.; Malvehy, J.; et al. Human–Computer Collaboration for Skin Cancer Recognition. Nat. Med. 2020, 26, 1229–1234. [Google Scholar] [CrossRef]
- Campanella, G.; Hanna, M.G.; Geneslaw, L.; Miraflor, A.; Werneck Krauss Silva, V.; Busam, K.J.; Brogi, E.; Reuter, V.E.; Klimstra, D.S.; Fuchs, T.J. Clinical-Grade Computational Pathology Using Weakly Supervised Deep Learning on Whole Slide Images. Nat. Med. 2019, 25, 1301–1309. [Google Scholar] [CrossRef]
- Korbar, B.; Olofson, A.; Miraflor, A.; Nicka, C.; Suriawinata, M.; Torresani, L.; Suriawinata, A.; Hassanpour, S. Deep Learning for Classification of Colorectal Polyps on Whole-Slide Images. J. Pathol. Inform. 2017, 8, 30. [Google Scholar] [CrossRef]
- Oon, M.L.; Syn, N.L.; Tan, C.L.; Tan, K.B.; Ng, S.B. Bridging Bytes and Biopsies: A Comparative Analysis of ChatGPT and Histopathologists in Pathology Diagnosis and Collaborative Potential. Histopathology 2024, 84, 601–613. [Google Scholar] [CrossRef]
- Ullah, E.; Parwani, A.; Baig, M.M.; Singh, R. Challenges and Barriers of Using Large Language Models (LLM) Such as ChatGPT for Diagnostic Medicine with a Focus on Digital Pathology—A Recent Scoping Review. Diagn. Pathol. 2024, 19, 43. [Google Scholar] [CrossRef] [PubMed]
- Reerds, S.T.H.; Uijen, M.J.M.; Van Engen-Van Grunsven, A.C.H.; Marres, H.A.M.; van Herpen, C.M.L.; Honings, J. Results of Histopathological Revisions of Majorsalivary Gland Neoplasms in Routine Clinical Practice. J. Clin. Pathol. 2022, 76, 374–378. [Google Scholar] [CrossRef]
- Wu, S.; Yue, M.; Zhang, J.; Li, X.; Li, Z.; Zhang, H.; Wang, X.; Han, X.; Cai, L.; Shang, J.; et al. The Role of Artificial Intelligence in Accurate Interpretation of HER2 Immunohistochemical Scores 0 and 1+ in Breast Cancer. Mod. Pathol. 2023, 36, 100054. [Google Scholar] [CrossRef] [PubMed]
- Waqas, A.; Bui, M.M.; Glassy, E.F.; El Naqa, I.; Borkowski, P.; Borkowski, A.A.; Rasool, G. Revolutionizing Digital Pathology with the Power of Generative Artificial Intelligence and Foundation Models. Lab. Investig. 2023, 103, 100255. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.K.; Lin, S.-Y.; Fick, D.M.; Shulman, R.W.; Lee, S.; Shrestha, P.; Santoso, K. Optimizing ChatGPT’s Interpretation and Reporting of Delirium Assessment Outcomes: Exploratory Study. JMIR Form. Res. 2024, 8, e51383. [Google Scholar] [CrossRef]
- Naved, B.A.; Luo, Y. Contrasting Rule and Machine Learning Based Digital Self Triage Systems in the USA. NPJ Digit. Med. 2024, 7, 381. [Google Scholar] [CrossRef]
- Hager, P.; Jungmann, F.; Holland, R.; Bhagat, K.; Hubrecht, I.; Knauer, M.; Vielhauer, J.; Makowski, M.; Braren, R.; Kaissis, G.; et al. Evaluation and Mitigation of the Limitations of Large Language Models in Clinical Decision-Making. Nat. Med. 2024, 30, 2613–2622. [Google Scholar] [CrossRef]
- Cuevas-Nunez, M.; Silberberg, V.I.A.; Arregui, M.; Jham, B.C.; Ballester-Victoria, R.; Koptseva, I.; de Tejada, M.J.B.G.; Posada-Caez, R.; Manich, V.G.; Bara-Casaus, J.; et al. Diagnostic Performance of ChatGPT-4.0 in Histopathological Description Analysis of Oral and Maxillofacial Lesions: A Comparative Study with Pathologists. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2025, 139, 453–461. [Google Scholar] [CrossRef]
- Doeleman, T.; Hondelink, L.M.; Vermeer, M.H.; van Dijk, M.R.; Schrader, A.M.R. Artificial Intelligence in Digital Pathology of Cutaneous Lymphomas: A Review of the Current State and Future Perspectives. Semin. Cancer Biol. 2023, 94, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Querzoli, G.; Veronesi, G.; Corti, B.; Nottegar, A.; Dika, E. Basic Elements of Artificial Intelligence Tools in the Diagnosis of Cutaneous Melanoma. Crit. Rev. Oncog. 2023, 28, 37–41. [Google Scholar] [CrossRef] [PubMed]
- Al Tibi, G.; Alexander, M.; Miller, S.; Chronos, N. A Retrospective Comparison of Medication Recommendations Between a Cardiologist and ChatGPT-4 for Hypertension Patients in a Rural Clinic. Cureus 2024, 16, e55789. [Google Scholar] [CrossRef]
- Moore, P.D.C.; Guinigundo, M.A.S. The Role of Biomarkers in Guiding Clinical Decision-Making in Oncology. J. Adv. Pract. Oncol. 2023, 14, 15–37. [Google Scholar] [CrossRef] [PubMed]
- Shin, D.; Arthur, G.; Caldwell, C.; Popescu, M.; Petruc, M.; Diaz-Arias, A.; Shyu, C.-R. A Pathologist-in-the-Loop IHC Antibody Test Selection Using the Entropy-Based Probabilistic Method. J. Pathol. Inform. 2012, 3, 1. [Google Scholar] [CrossRef]
| Tumor Classification | Tumor Type |
|---|---|
| Benign Salivary Gland Tumors | 1. Pleomorphic Adenoma (PA) |
| 2. Warthin Tumor (WT) | |
| 3. Basal Cell Adenoma (BCA) | |
| 4. Canalicular Adenoma (CA) | |
| 5. Oncocytoma (OC) | |
| Malignant Salivary Gland Tumors | Adenocarcinomas |
| 1. Mucoepidermoid Carcinoma (MEC) | |
| 2. Adenoid Cystic Carcinoma (AdCC) | |
| 3. Acinic Cell Carcinoma (AcCC) | |
| 4. Polymorphous Adenocarcinoma (PAC) | |
| 5. Clear Cell Carcinoma (CCC) | |
| 6. Secretory Carcinoma (MASC) | |
| 7. Epithelial–Myoepithelial Carcinoma(EMC) | |
| 8. Salivary Duct Carcinoma (SDC) | |
| 9. Basal Cell Adenocarcinoma (BCAC) | |
| 11. Mucinous Adenocarcinoma (MAC) | |
| 12. Oncocytic Carcinoma (Oca) | |
| 13. Hyalinizing Clear Cell Carcinoma (HCCC) | |
| 15. Myoepithelial Carcinoma (MC) | |
| 16. Squamous Cell Carcinoma (SCC) | |
| 17. Basaloid Squamous Cell Carcinoma (BSCC) | |
| 18. Carcinoma Ex-Pleomorphic Adenoma (Ca-exPA) |
| Tumor Type | Mean Composite Score |
|---|---|
| Pleomorphic Adenoma | 5.67 |
| Warthin Tumor | 5.00 |
| Basal Cell Adenoma | 5.00 |
| Canalicular Adenoma | 5.00 |
| Oncocytoma | 5.33 |
| Mucoepidermoid Carcinoma | 5.00 |
| Adenoid Cystic Carcinoma | 6.67 |
| Acinic Cell Carcinoma | 5.67 |
| Polymorphous Adenocarcinoma | 6.33 |
| Clear Cell Carcinoma | 5.00 |
| Secretory Carcinoma | 5.33 |
| Epithelial–Myoepithelial Carcinoma | 5.67 |
| Salivary Duct Carcinoma | 5.00 |
| Basal Cell Adenocarcinoma | 5.00 |
| Mucinous Adenocarcinoma | 6.67 |
| Oncocytic Carcinoma | 5.33 |
| Hyalinizing Clear Cell Carcinoma | 6.00 |
| Myoepithelial Carcinoma | 6.00 |
| Squamous Cell Carcinoma | 6.33 |
| Basaloid SCC | 6.00 |
| Carcinoma Ex-Pleomorphic Adenoma | 4.33 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cuevas-Nunez, M.; Galletti, C.; Fiorillo, L.; Meto, A.; Díaz-Castañeda, W.R.; Farahani, S.S.; Fadda, G.; Zuccalà, V.; Manich, V.G.; Bara-Casaus, J.; et al. Assessment of ChatGPT in Recommending Immunohistochemistry Panels for Salivary Gland Tumors. BioMedInformatics 2025, 5, 66. https://doi.org/10.3390/biomedinformatics5040066
Cuevas-Nunez M, Galletti C, Fiorillo L, Meto A, Díaz-Castañeda WR, Farahani SS, Fadda G, Zuccalà V, Manich VG, Bara-Casaus J, et al. Assessment of ChatGPT in Recommending Immunohistochemistry Panels for Salivary Gland Tumors. BioMedInformatics. 2025; 5(4):66. https://doi.org/10.3390/biomedinformatics5040066
Chicago/Turabian StyleCuevas-Nunez, Maria, Cosimo Galletti, Luca Fiorillo, Aida Meto, Wilmer Rodrigo Díaz-Castañeda, Shokoufeh Shahrabi Farahani, Guido Fadda, Valeria Zuccalà, Victor Gil Manich, Javier Bara-Casaus, and et al. 2025. "Assessment of ChatGPT in Recommending Immunohistochemistry Panels for Salivary Gland Tumors" BioMedInformatics 5, no. 4: 66. https://doi.org/10.3390/biomedinformatics5040066
APA StyleCuevas-Nunez, M., Galletti, C., Fiorillo, L., Meto, A., Díaz-Castañeda, W. R., Farahani, S. S., Fadda, G., Zuccalà, V., Manich, V. G., Bara-Casaus, J., & Fernández-Figueras, M.-T. (2025). Assessment of ChatGPT in Recommending Immunohistochemistry Panels for Salivary Gland Tumors. BioMedInformatics, 5(4), 66. https://doi.org/10.3390/biomedinformatics5040066












