Uncovering the Diagnostic Power of Radiomic Feature Significance in Automated Lung Cancer Detection: An Integrative Analysis of Texture, Shape, and Intensity Contributions
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Acquisition and Preprocessing
2.2. Radiomic Feature Extraction
2.2.1. First-Order Statistics
- Mean Intensity: The mean value of the voxels comprised within the ROI, reflecting to a great extent the average density of the tissue.
- Skewness: Characterizes the asymmetry of the voxel intensity distribution and, hence, the heterogeneity of the tumor.
- Kurtosis: Describing the property ‘peakedness’; in other words, whether a distribution of voxel intensities is bunched around or spread out from the mean.
- Energy: This is the sum of the square of the voxel values, which may be related to the aggressiveness of the tumor or its metabolic activity.
2.2.2. Shape-Based Features
- Sphericity: A measure of how spherical (round) the tumor is, where values approaching 1 depict structures near perfectly spherical. Lower sphericity values indicate that highly aggressive and invasive tumors are more characteristic.
- Compactness: This shows how much the shape of a tumor is spherical or elongated and can thus be an indication of its invasive power.
- Surface-area-to-volume Ratio: This measure compares the surface complexity of the tumor to its volume. The higher the ratio, the greater the chance for irregular growth patterns—often associated with malignancy.
- Elongation: Measures the deviation in the tumor shape from a perfect sphere and may give clues of its infiltration into surrounding tissues.
2.2.3. Texture-Based Features
- GLCM: GLCM features describe the frequency of pixel intensity pairs for a predefined spatial relationship. GLCM Entropy may serve as an example and denote the complexity of the variation in voxel intensities. The greater the values, the greater the heterogeneity, which is usually associated with malignancies. Other significant GLCM features include contrast, which describes the difference between high and low intensities, and correlation, which is the measure of linear dependencies between the intensity of voxels.
- GLRLM: This matrix provides the length of consecutively sharing voxels with the same intensity value in a given direction. The related features to this are the GLRLM (Gray-level Run Length Matrix) Short Run Emphasis, which manifests the presence of small homogeneous regions inside the tumor, while the GLRLM Long Run Emphasis gives information about the boundless homogeneous regions. These will be relevant for identifying the fibrotic regions or regions bearing necrosis inside the tumor.
- Grayscale-level Size Zone Matrix (GLSZM): GLSZM is very similar to the GLRLM in that it quantifies regions of identical intensities; no directional information is taken into account, however. Important features rely on this matrix: Zone Size Non-Uniformity and Large Zone Emphasis may be useful to detect the presence of large homogeneous areas, which are generally indicative of late disease stages.
- Wavelet Features: Refer to those signal features extracted through the application of wavelet filters on images for capturing texture at multiple resolutions. This multi-scale analysis is critical in order to detect subtle patterns both in fine and coarse details, thus offering a more nuanced understanding of tumor heterogeneity.
2.2.4. Multi-Scale and Multi-Dimensional Feature Extraction
2.2.5. Innovative Aspects of Feature Extraction
- Manual and Semi-Automated Segmentation: The tumor ROIs were segmented using a great deal of care by manual delineation of expert radiologists combined with semi-automated algorithms to achieve the best precision together with reducing observer variability.
- Standardization and Reproducibility: All extraction was carried out by strictly adhering to guidelines provided by the IBSI, with the aim of having all features reproducible across different imaging systems and acquisition protocols.
- High-throughput Feature Extraction: PyRadiomics thus enables the extraction of hundreds of features in one image effectively. High-throughput feature extraction in imaging data is of high importance to machine learning applications, especially in cases where the volume of data is immense.
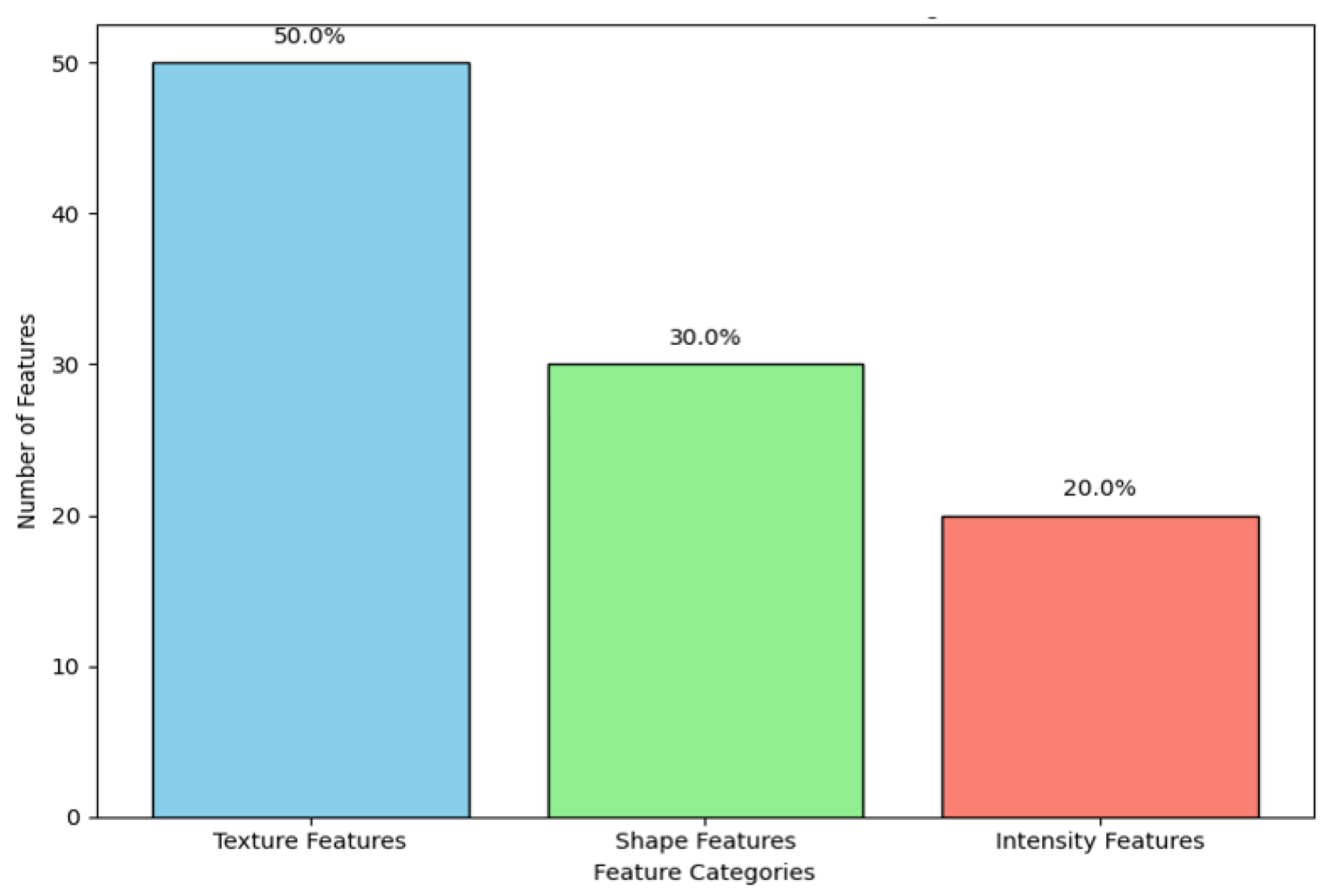
- Finding the Best and Most Useful Features: Out of the 350 features that were collected, tests using forward stepwise correlation analysis and tests for test–retest variability were used to get rid of features that were duplicated or not relevant. We were able to minimize overfitting without reducing model accuracy by focusing the analysis on the most stable and informative features.
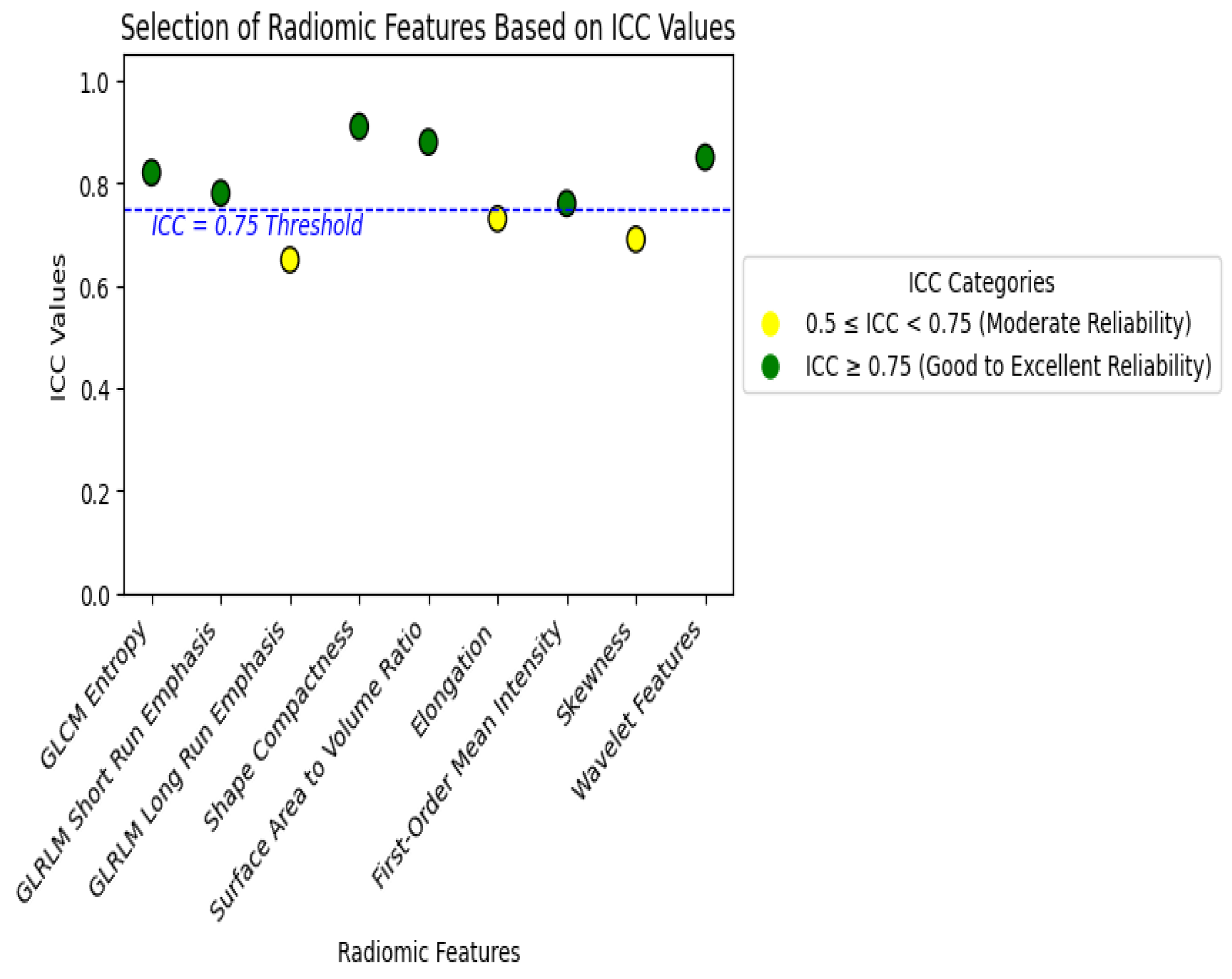
2.3. Feature Selection and Stability Analysis
2.3.1. Feature Stability and Reproducibility
2.3.2. Correlation and Dimensionality Reduction
2.3.3. Feature Selection Techniques
- LASSO Regression: LASSO introduced regularization into the feature selection process by applying a penalty to the magnitude of the coefficients associated with each feature. This is a powerful method in high-dimensional datasets, as it shrinks the coefficients of less important features to zero, effectively removing them from the model.
- Recursive Feature Elimination (RFE): This second feature selection method provided a wrapper method, working in conjunction with a machine learning model. RFE recursively removed the least important features based on their contribution to model performance until only the optimal set of features remained. This method ensures that the final feature set is tailored to the specific model being used, maximizing accuracy and interpretability [20].
2.4. Machine Learning Model Development
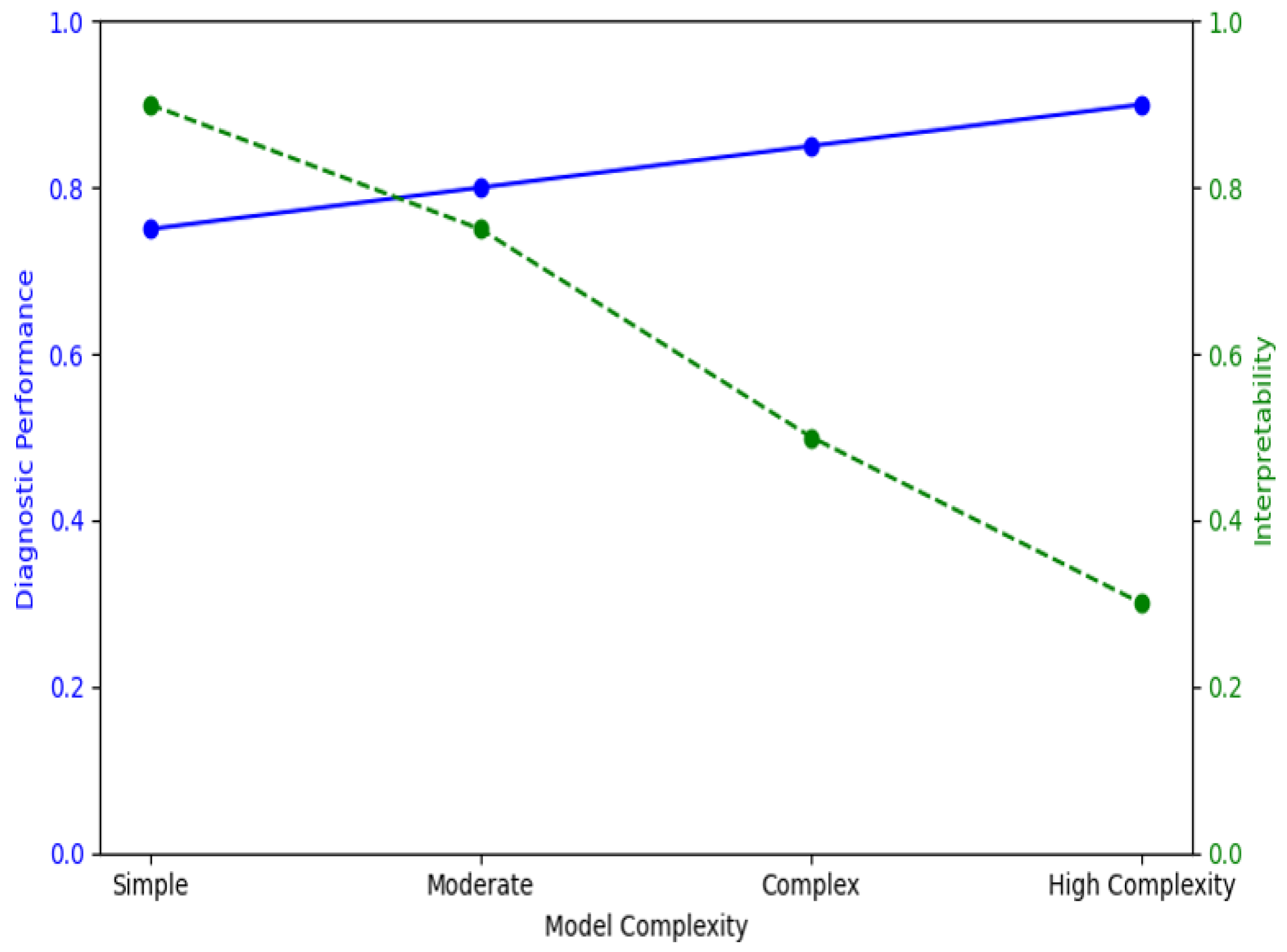
2.4.1. Model Architecture and Choice
- DenseNet-201 (CNN Model): DenseNet-201 is a popular CNN architecture that effectively performs image feature extraction through dense connections between layers. This dense connectivity mitigates the vanishing gradient problem by helping in reusing features across layers to learn more complex imaging tasks like detecting cancer. To this end, in our study, we trained the DenseNet model separately on both raw CT images and feature maps to enable the network to learn from spatial patterns (raw images) as well as quantitative tumor characteristics (radiomics-driven features) [23].
- XGBoost (Gradient-boosted Decision Tree Model): XGBoost is a powerful machine learning algorithm that is known to perform well on tabular data and has become very popular in the world of Kaggle competitions, where it generally reaches SOTA (State-of-the-Art) scores. Because of its feature importance ranking and model interpretability, CNNs are not very good at being mean for image data. Training the XGBoost model on extracted features allowed us to test the importance that individual features hold in diagnostic tasks and thus infer biological interpretability [24]. The model was then trained on the entire set of extracted features, and feature importance was evaluated using SHAP values and permutation importance to rank these race predictors (explained in Section 2.3).
2.4.2. Training Process and Cross-Validation
- Five-fold Cross-validation: The dataset was divided into five equal-sized subsets, and in each fold of the cross-validation, one subset played the role of a validation set, while the other four subsets played the role of training sets. This was repeated five times in such a way that each subset acted as the validation set once. Averaging over five folds provides a robust estimate of accuracy, sensitivity, specificity, and AUC-ROC for model performance.
- Data Augmentation: For the training of the CNN to enhance generalization, data augmentation techniques were used on the CT images. Then, several augmentation techniques—like random rotations, flipping, cropping, and scaling—were used to introduce patient positioning and variability in scanner settings, which will increase distortions and the diversity of the training.
- Hyperparameter Tuning: These activities also involve the tuning of the learning rate, batch size, and dropout rate by a grid search strategy. The early stopping in this CNN model stops the training when it observes that there is no more improvement in validation loss within five successive epochs to avoid overfitting and saving computational resources. Similarly, XGBoost hyperparameters, such as, but not limited to, those involving the maximum depth of trees, learning rate, and regularization parameters, have been tuned in a manner that predictive performance is maximized at the possible cost of overfitting.
2.4.3. Model Optimization and Loss Functions
2.4.4. Performance Metrics
- Accuracy: It refers to the measure of the portion of properly classified images, thus giving the general performance of the model.
- Sensitivity (recall): It is the capability of the model to correctly identify malignant cases. High sensitivity is rather important in cancer diagnosis because it will reduce false negatives.
- Specificity: The percentage of healthy cases correct that this model should identify in order to avoid the misclassification of a benign case as malignant.
- AUC-ROC (Area Under the Receiver Operating Characteristic Curve): A metric that will provide the model performance for distinguishing malignant from benign cases across all possible classification thresholds. The high value of AUC-ROC means it would be unproblematic for this classification model to make good distinctions between the classes, even if there is a class imbalance in this dataset.
2.5. Feature Importance and Interpretability
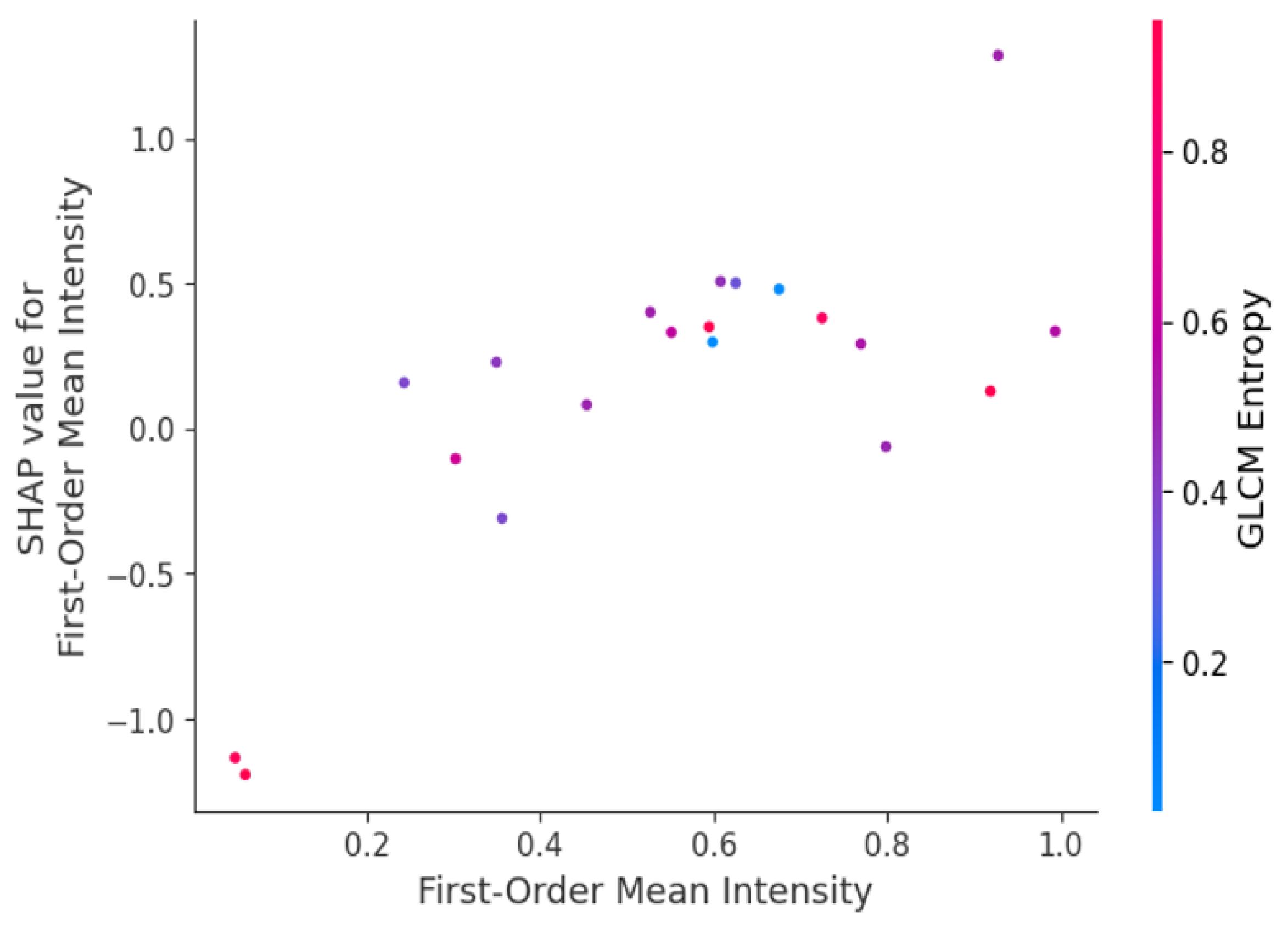
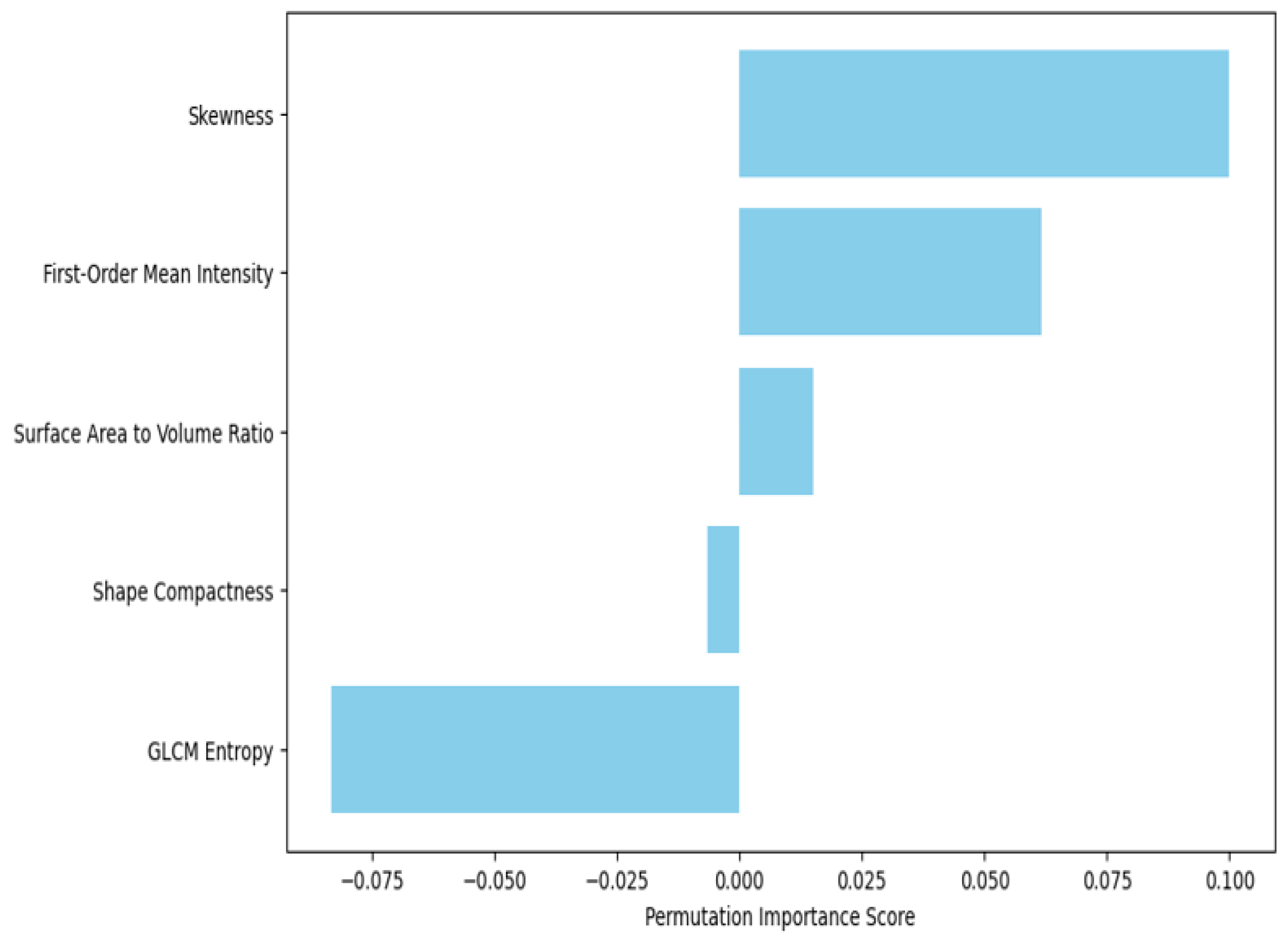
- Global SHAP Analysis: We averaged the SHAP values of every feature on the dataset and ranked the features by their general importance. Indeed, for every test we ran in this work, the features that ranked in the top places included GLCM Entropy and GLRLM Run Length Non-Uniformity; thus, our intuition that tumor heterogeneity was one of the key factors in malignant/benign tumor classification was legitimate.
- Local SHAP Analysis: SHAP also allowed us to drill into the contribution of features on a case-by-case basis. For example, in the specific cases of early-stage lung cancer, features related to first-order skewness, which reflects the asymmetry of voxel intensity distribution, have a high SHAP value and, hence, are critical for the right classification of small irregular tumors. That is very important for local interpretability in clinical applications, as it would tell the radiologists which features drove the model’s decision for each given patient.
2.6. Tumor-Specific Features Relevance
- Small Tumors (<2 cm): Earlier-stage tumors, which, due to their small size, are often very hard to detect because they do not cause much morphological disruption. For these cases, we expected that texture-based features would convey the most information since these features capture the subtle heterogeneity in the tumor that might not be obvious to the naked eye from the raw images. This was indeed the case, as all texture features ranked higher in importance for smaller tumors, in keeping with our hypothesis that early malignancies are more micro-heterogeneous.
- Medium-sized Tumors (2–4 cm): In tumors of intermediate size, it was expected that both the texture- and shape-based features had a balanced contribution. For such tumors, shape elongation and GLCM correlation were important features, for they extracted information on the change in geometry of the tumor during growth and more structured texture patterns within the tumor mass.
- Large Tumors (>4 cm): We expected that with the larger size and more advanced development of tumors, shape-based features such as surface-area-to-volume ratio and compactness would turn out to be the most informative measures in distinguishing between malignant and benign status.
- Early-stage Tumors (Stage I–II): The critical contribution of texture-based features became more pronounced in low-stage tumors, which present with more subtle imaging characteristics. For example, GLCM contrast and GLRLM long-run emphasis constituted the most informative features for early-stage tumors, underscoring the importance of capturing those fine-grained textural variations arising from early intratumoral heterogeneity. These findings hint that texture features may be used as an early sign of malignancy, therefore opening up perspectives that allow for earlier and more accurate detection.
- Late-stage Tumors (Stage III–IV): While advancing to the more advanced stages, tumor shapes become increasingly irregular due to invasive growth patterns. In late-stage tumors, shape-based features such as elongation, surface irregularity, and sphericity were most important. These features captured the complex structural deformations occurring with invasion into adjacent tissues and more pronounced morphological characteristics of these tumors. More than that, the texture features related to necrosis and heterogeneity remained salient at these stages and were probably related to the increased inner complexity of the advanced tumor.
- Central tumors: Tumors around the central bronchi and mediastinum may, for instance, have more complex relationships with vessels and airways. For them, shape-based features such as convexity and elongation had greater relevance since the feature set was biased toward distinguishing irregular growth patterns that result from confinement imposed by surrounding anatomical structures. In addition, GLCM correlation performed really well in detecting tumor heterogeneity of central lung tumors that may show complex interaction with the surrounding tissues and thus exhibit variant intensity patterns in CT images.
- Peripheral tumors: Tumors that originate in peripheral lung tissues, away from the bronchi and mediastinum, grow more freely in most cases and adopt more rounded or nodular shapes. In such cases, texture-based features were of higher importance, reflecting heterogeneity and an irregular internal texture of the tumor. Moreover, the surface-area-to-volume ratio played an important role in the detection of an irregular tumor shape since peripheral tumors are less confined by structure compared with central tumors.
- Small early-stage peripheral tumors: These are best described by features of texture—for example, GLCM Entropy, which captured the subtle heterogeneity within the tumor. Shape features are less important for this subgroup since these tumors have not adopted those irregular morphologies typical of more advanced cancers.
- Large late-stage central tumors: On the contrary, these larger tumors, located near the central part of the lungs, were more characterized by shape-based features, such as surface irregularity and elongation, describing the invasive and irregular growth of the tumor in an advanced stage of cancer.
3. Results
3.1. Model Performance
3.2. Radiomic Feature Importance and Model Insights
3.3. Tumor Size and Feature Relevance
3.3.1. Radiomic Feature Relevance for Small Tumors (<2 cm)
3.3.2. Radiomic Feature Relevance for Medium-Sized Tumors (2–4 cm)
3.3.3. Radiomic Feature Relevance for Large Tumors (>4 cm)
| Radiomic Feature | Tumor Size | Importance Score |
|---|---|---|
| GLCM Entropy | Small | 0.72 |
| GLCM Entropy | Medium | 0.81 |
| GLCM Entropy | Large | 0.88 |
| Shape Compactness | Small | 0.65 |
| Shape Compactness | Medium | 0.75 |
| Shape Compactness | Large | 0.84 |
| Surface-Area-to-Volume Ratio | Small | 0.68 |
| Surface-Area-to-Volume Ratio | Medium | 0.78 |
| Surface-Area-to-Volume Ratio | Large | 0.83 |
| First-Order Mean Intensity | Small | 0.55 |
| First-Order Mean Intensity | Medium | 0.63 |
| First-Order Mean Intensity | Large | 0.71 |
| Skewness | Small | 0.49 |
| Skewness | Medium | 0.56 |
3.3.4. Comparative Analysis of Feature Importance by Tumor Size
3.4. Comparative Analysis Between Models
4. Discussion
4.1. Comparison of Model Performance with and Without Texture Features
4.2. Shape-Based Features and Tumor Progression
4.2.1. Comparative Analysis of Shape Features in Different Tumor Stages
4.2.2. Clinical Implications of Shape-Based Features in Tumor Progression
4.2.3. Stability of Selected Features
- Cross-Validation Stability: K-fold cross-validation was implemented to make sure that the features selected were generalizing and robust. It provided an estimation of how consistent the top features’ contributions were to the modeling performance across different splits of the training and validation sets. The top features, such as GLCM Entropy and shape compactness found in Table 8, are more or less similar in their ranking across all folds.
- Bootstrap Aggregation (Bagging): To further assess feature stability, we conducted bootstrap aggregation, where multiple models had been trained on different subsets of the data. This technique allowed us to assess how each feature’s importance varied across the different samples and ensure that the selected features in these samples were constantly ranked as important. The obtained results are presented in Table 9 and give evidence of the high stability of the selected features among different bootstrapping iterations.
4.2.4. Impact of Feature Selection on Model Performance
4.3. Clinical Implications of Radiomic Feature Selection
- Early detection and diagnosis: Features like GLCM Entropy enable the models to capture subtle intratumoral heterogeneity for early detection of malignant tissues with high sensitivity, thereby reducing the likelihood of false negatives. This is very critical for early intervention and treatment.
- Assessment of tumor aggressiveness: The shape-related features quantify, for example, the surface-area-to-volume ratio, which bears information on identifying tumors with irregular and invasive growth patterns, hence reflecting aggressiveness. These features provide assistance in grading the cancer and help deduce a suitable treatment strategy, including metastasis potential.
- Personalized diagnostic approach: Integration of stable, interpretable features in the diagnosis models enables personalized insights in each case. Clinicians will understand the underlying reasons for every prediction using SHAP values and permutation importance to make more informed, personalized decisions in patient care.
4.4. Limitations and Future Directions
4.4.1. Limitations
Data Diversity and Generalizability
- Variability in Imaging Protocols: This would further lead to inconsistency in feature extraction due to differences in resolution, slice thickness, and different reconstruction algorithms; thus, the models may predict variably. These limitations could have been mitigated if standardization of imaging protocols across all institutions was considered, or, even further still, with a simulation of variability in training data by data augmentation.
Feature Robustness and Reproducibility
Model Interpretability and Clinical Integration
4.4.2. Future Directions
Standardization of Imaging Protocols
Feature Robustness Across Diverse Cohorts
Development of Explainable AI (XAI) Models
Integration with Genomic and Clinical Data
Reproducibility and FAIR Principles
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. Available online: https://www.who.int/news-room/fact-sheets/detail/cancer (accessed on 2 October 2024).
- Scapicchio, C.; Gabelloni, M.; Barucci, A.; Cioni, D.; Saba, L.; Neri, E. A deep look into radiomics. Radiol. Med. 2021, 126, 1296–1311. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Lou, X.; Kong, N.; Xu, M.; Gao, C. Can quantitative peritumoral CT radiomics features predict the prognosis of patients with non-small cell lung cancer? A systematic review. Eur. Radiol. 2022, 33, 2105–2117. [Google Scholar] [CrossRef] [PubMed]
- Raptis, S.; Ilioudis, C.; Theodorou, K. From pixels to prognosis: Unveiling radiomics models with SHAP and LIME for enhanced interpretability. Biomed. Phys. Eng. Express 2024, 10, 035016. [Google Scholar] [CrossRef] [PubMed]
- Marcilio, W.E.; Eler, D.M. From explanations to feature selection: Assessing SHAP values as feature selection mechanism. In Proceedings of the 2020 33rd SIBGRAPI Conference on Graphics, Patterns and Images (SIBGRAPI), Porto de Galinhas, Brazil, 7–10 November 2020; pp. 340–347. [Google Scholar] [CrossRef]
- Liu, L.; Chen, X.; Petinrin, O.O.; Zhang, W.; Rahaman, S.; Tang, Z.-R.; Wong, K.-C. Machine Learning Protocols in Early Cancer Detection Based on Liquid Biopsy: A Survey. Life 2021, 11, 638. [Google Scholar] [CrossRef]
- Neri, E.; Del Re, M.; Paiar, F.; Erba, P.; Cocuzza, P.; Regge, D.; Danesi, R. Radiomics and liquid biopsy in oncology: The holons of systems medicine. Insights Imaging 2018, 9, 915–924. [Google Scholar] [CrossRef]
- Taşcı, E.; Uğur, A. Shape and Texture Based Novel Features for Automated Juxtapleural Nodule Detection in Lung CTs. J. Med. Syst. 2015, 39, 46. [Google Scholar] [CrossRef]
- Kalendralis, P.; Shi, Z.; Traverso, A.; Choudhury, A.; Sloep, M.; Zhovannik, I.; Starmans, M.P.; Grittner, D.; Feltens, P.; Monshouwer, R.; et al. FAIR-compliant clinical, radiomics and DICOM metadata of RIDER, interobserver, Lung1 and head-Neck1 TCIA collections. Med. Phys. 2020, 47, 5931–5940. [Google Scholar] [CrossRef]
- Wee, L.; Aerts, H.J.; Kalendralis, P.; Dekker, A. Data from NSCLC-Radiomics-Interobserver1. Cancer Imaging Arch. 2019. [Google Scholar] [CrossRef]
- ISBI. Available online: https://theibsi.github.io/ (accessed on 2 October 2024).
- van Griethuysen, J.J.M.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.H.; Fillion-Robin, J.-C.; Pieper, S.; Aerts, H.J.W.L. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef]
- Dhawan, A.P. Medical Image Analysis, 2nd ed.; IEEE Press series on biomedical engineering, no. 17; Wiley-IEEE Press: Hoboken, NJ, USA, 2011. [Google Scholar]
- Mall, P.K.; Singh, P.K.; Yadav, D. GLCM Based Feature Extraction and Medical X-RAY Image Classification using Machine Learning Techniques. In Proceedings of the 2019 IEEE Conference on Information and Communication Technology, Allahabad, India, 6–8 December 2019; pp. 1–6. [Google Scholar] [CrossRef]
- Koo, T.K.; Li, M.Y. A Guideline of Selecting and Reporting Intraclass Correlation Coefficients for Reliability Research. J. Chiropr. Med. 2016, 15, 155–163. [Google Scholar] [CrossRef]
- Lambin, P.; Leijenaar, R.T.H.; Deist, T.M.; Peerlings, J.; de Jong, E.E.C.; van Timmeren, J.; Sanduleanu, S.; Larue, R.T.H.M.; Even, A.J.G.; Jochems, A.; et al. Radiomics: The bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 2017, 14, 749–762. [Google Scholar] [CrossRef] [PubMed]
- Parmar, C.; Leijenaar, R.T.H.; Grossmann, P.; Velazquez, E.R.; Bussink, J.; Rietveld, D.; Rietbergen, M.M.; Haibe-Kains, B.; Lambin, P.; Aerts, H.J. Radiomic feature clusters and Prognostic Signatures specific for Lung and Head & Neck cancer. Sci. Rep. 2015, 5, 11044. [Google Scholar] [CrossRef]
- Benesty, J.; Chen, J.; Huang, Y.; Cohen, I. Pearson Correlation Coefficient. In Noise Reduction in Speech Processing; Springer Topics in Signal Processing, 2; Springer: Berlin/Heidelberg, Germany, 2009; pp. 1–4. [Google Scholar] [CrossRef]
- Vasquez, M.M.; Hu, C.; Roe, D.J.; Chen, Z.; Halonen, M.; Guerra, S. Least absolute shrinkage and selection operator type methods for the identification of serum biomarkers of overweight and obesity: Simulation and application. BMC Med. Res. Methodol. 2016, 16, 154. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Jeong, J.C. Enhanced recursive feature elimination. In Proceedings of the Sixth International Conference on Machine Learning and Applications (ICMLA 2007), Cincinnati, OH, USA, 13–15 December 2007; pp. 429–435. [Google Scholar] [CrossRef]
- Nohara, Y.; Matsumoto, K.; Soejima, H.; Nakashima, N. Explanation of machine learning models using shapley additive explanation and application for real data in hospital. Comput. Methods Programs Biomed. 2022, 214, 106584. [Google Scholar] [CrossRef] [PubMed]
- Raptis, S.; Softa, V.; Angelidis, G.; Ilioudis, C.; Theodorou, K. Automation Radiomics in Predicting Radiation Pneumonitis (RP). Automation 2023, 4, 191–209. [Google Scholar] [CrossRef]
- Guo, W.; Xu, Z.; Zhang, H. Interstitial lung disease classification using improved DenseNet. Multimed. Tools Appl. 2019, 78, 30615–30626. [Google Scholar] [CrossRef]
- Iranzad, R.; Liu, X.; Chaovalitwongse, W.A.; Hippe, D.; Wang, S.; Han, J.; Thammasorn, P.; Duan, C.; Zeng, J.; Bowen, S. Gradient boosted trees for spatial data and its application to medical imaging data. IISE Trans. Healthc. Syst. Eng. 2022, 12, 165–179. [Google Scholar] [CrossRef]
- Raptis, S.; Tsougos, I.; Theodorou, K.; Ilioudis, C. Harmonizing Radiomics and Interpretable AI: Precision and Transparency in Oncological Prognostication. In Proceedings of the 2024 IEEE International Symposium on Biomedical Imaging (ISBI), Athens, Greece, 27–30 May 2024; pp. 1–4. [Google Scholar] [CrossRef]
- Lim, W.; Ridge, C.A.; Nicholson, A.G.; Mirsadraee, S. The 8th lung cancer TNM classification and clinical staging system: Review of the changes and clinical implications. Quant. Imaging Med. Surg. 2018, 8, 709–718. [Google Scholar] [CrossRef]
- Demirjian, N.L.; Varghese, B.A.; Cen, S.Y.; Hwang, D.H.; Aron, M.; Siddiqui, I.; Fields, B.K.K.; Lei, X.; Yap, F.Y.; Rivas, M.; et al. CT-based radiomics stratification of tumor grade and TNM stage of clear cell renal cell carcinoma. Eur. Radiol. 2022, 32, 2552–2563. [Google Scholar] [CrossRef]
- Dwivedi, R.; Dave, D.; Naik, H.; Singhal, S.; Omer, R.; Patel, P.; Qian, B.; Wen, Z.; Shah, T.; Morgan, G.; et al. Explainable AI (XAI): Core Ideas, Techniques, and Solutions. ACM Comput. Surv. 2023, 55, 1–33. [Google Scholar] [CrossRef]
- Marvin, G.; Jjingo, D.; Nakatumba-Nabende, J.; Alam, M.G.R. Local Interpretable Model-Agnostic Explanations for Online Maternal Healthcare. In Proceedings of the 2023 2nd International Conference on Smart Technologies and Systems for Next Generation Computing (ICSTSN), Villupuram, India, 21–22 April 2023; pp. 1–6. [Google Scholar] [CrossRef]
- Suara, S.; Jha, A.; Sinha, P.; Sekh, A.A. Is Grad-CAM Explainable in Medical Images? In Computer Vision and Image Processing; Kaur, H., Jakhetiya, V., Goyal, P., Khanna, P., Raman, B., Kumar, S., Eds.; Communications in Computer and Information Science; Springer Nature Switzerland: Cham, Switzerland, 2024; Volume 2009, pp. 124–135. [Google Scholar] [CrossRef]
- Kierner, S.; Kucharski, J.; Kierner, Z. Taxonomy of hybrid architectures involving rule-based reasoning and machine learning in clinical decision systems: A scoping review. J. Biomed. Inform. 2023, 144, 104428. [Google Scholar] [CrossRef]
- Saxena, S.; Jena, B.; Gupta, N.; Das, S.; Sarmah, D.; Bhattacharya, P.; Nath, T.; Paul, S.; Fouda, M.M.; Kalra, M.; et al. Role of Artificial Intelligence in Radiogenomics for Cancers in the Era of Precision Medicine. Cancers 2022, 14, 2860. [Google Scholar] [CrossRef]



| Model | Accuracy | Sensitivity | Specificity | AUC-ROC | Key Findings |
|---|---|---|---|---|---|
| DenseNet-201 (CNN) | 92.4% | 91.6% | 93.2% | 0.94 | Combined radiomic feature maps and CT images improved performance. Captured tumor heterogeneity and morphology effectively. |
| XGBoost (Radiomics) | 89.7% | 88.4% | 90.5% | 0.90 | Relied on radiomic features like GLCM Entropy and shape compactness. Strong performance without raw image data. |
| Tumor Size | Dominant Features | Key Insights |
|---|---|---|
| Small (<2 cm) | Texture (GLCM Entropy, GLRLM Short Run Emphasis) | Texture features capture subtle heterogeneity, critical for early-stage cancer detection. Shape features are less relevant due to uniform shape. |
| Medium (2–4 cm) | Balanced (Shape Compactness, GLCM Correlation) | Both texture and shape features contribute equally. Shape irregularities begin to appear, while texture heterogeneity remains significant. |
| Large (>4 cm) | Shape (Surface-area-to-volume Ratio, Shape Elongation) | Shape features dominate, capturing the irregular, invasive morphology of advanced tumors. Texture features still provide insights into heterogeneity. |
| Radiomic Feature | Mean SHAP Value (DenseNet-201) | Mean SHAP Value (XGBoost) | Permutation Importance (DenseNet-201) | Permutation Importance (XGBoost) |
|---|---|---|---|---|
| GLCM Entropy | 0.47 | 0.55 | 0.63 | 0.66 |
| Shape Compactness | 0.35 | 0.41 | 0.52 | 0.58 |
| Surface-Area-to-Volume Ratio | 0.29 | 0.49 | 0.48 | 0.60 |
| GLRLM Run Length Non-Uniformity | 0.42 | 0.37 | 0.58 | 0.54 |
| Tumor Stage | Dominant Shape Features | Key Insights |
|---|---|---|
| Early Stage (I–II) | Shape Compactness, Elongation | Tumors exhibit more regular shapes; shape features less relevant but still important in certain cases. |
| Late Stage (III–IV) | Surface-Area-to-Volume Ratio, Shape Compactness, Elongation | Tumors show significant morphological irregularities; shape features critical for detecting invasiveness. |
| Model | Accuracy (Before Feature Selection) | Accuracy (After Feature Selection) | AUC-ROC (Before Feature Selection) | AUC-ROC (After Feature Selection) |
|---|---|---|---|---|
| DenseNet-201 (CNN) | 85.7% | 92.4% | 0.88 | 0.94 |
| XGBoost | 83.2% | 89.7% | 0.84 | 0.90 |
| Model | Accuracy | Sensitivity (Recall) | Specificity | AUC-ROC |
|---|---|---|---|---|
| DenseNet-201 (with texture) | 92.4% | 91.6% | 93.2% | 0.94 |
| DenseNet-201 (without texture) | 85.7% | 82.3% | 87.5% | 0.88 |
| XGBoost (with texture) | 89.7% | 88.4% | 90.5% | 0.90 |
| XGBoost (without texture) | 83.2% | 80.6% | 85.0% | 0.84 |
| Feature | Average Rank (Fold 1) | Average Rank (Fold 2) | Average Rank (Fold 3) | Stability |
| GLCM Entropy | 1 | 1 | 1 | High |
| Shape Compactness | 2 | 2 | 2 | High |
| Surface-Area-to-Volume Ratio | 3 | 3 | 3 | High |
| GLRLM Run Length Non-Uniformity | 4 | 4 | 4 | High |
| First-Order Mean Intensity | 5 | 5 | 5 | Medium |
| Feature | Bootstrap Iteration 1 | Bootstrap Iteration 2 | Bootstrap Iteration 3 | Stability |
| GLCM Entropy | 1 | 1 | 1 | High |
| Shape Compactness | 2 | 2 | 2 | High |
| Surface-Area-to-Volume Ratio | 3 | 3 | 3 | High |
| GLRLM Run Length Non-Uniformity | 4 | 4 | 4 | High |
| First-Order Mean Intensity | 5 | 5 | 5 | Medium |
| Imaging Parameter | Impact on Feature Extraction | Impact on Model Performance |
|---|---|---|
| Slice Thickness | Affects texture-based feature consistency | Reduces model generalizability |
| Reconstruction Algorithm | Alters intensity and shape features | Increases risk of overfitting |
| Scanner Type | Introduces variability in intensity values | Decreases reproducibility |
| XAI Technique | Description | Applicability |
|---|---|---|
| SHAP Analysis | Explains feature importance for individual predictions | Useful for feature-level interpretation |
| Local Interpretable Model-agnostic Explanations (LIME) [29] | Explains model predictions in a localized context | Helps in model transparency for clinicians |
| Grad-CAM [30] | Visualizes areas of the image that influence model predictions | Suitable for deep learning interpretability |
| Decision Trees/Rule-based Models [31] | Models that produce rules or trees, offering straightforward interpretability | May be used as baseline models for radiomic features, offering clear, interpretable rules, though potentially less accuracy than complex models |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Raptis, S.; Ilioudis, C.; Theodorou, K. Uncovering the Diagnostic Power of Radiomic Feature Significance in Automated Lung Cancer Detection: An Integrative Analysis of Texture, Shape, and Intensity Contributions. BioMedInformatics 2024, 4, 2400-2425. https://doi.org/10.3390/biomedinformatics4040129
Raptis S, Ilioudis C, Theodorou K. Uncovering the Diagnostic Power of Radiomic Feature Significance in Automated Lung Cancer Detection: An Integrative Analysis of Texture, Shape, and Intensity Contributions. BioMedInformatics. 2024; 4(4):2400-2425. https://doi.org/10.3390/biomedinformatics4040129
Chicago/Turabian StyleRaptis, Sotiris, Christos Ilioudis, and Kiki Theodorou. 2024. "Uncovering the Diagnostic Power of Radiomic Feature Significance in Automated Lung Cancer Detection: An Integrative Analysis of Texture, Shape, and Intensity Contributions" BioMedInformatics 4, no. 4: 2400-2425. https://doi.org/10.3390/biomedinformatics4040129
APA StyleRaptis, S., Ilioudis, C., & Theodorou, K. (2024). Uncovering the Diagnostic Power of Radiomic Feature Significance in Automated Lung Cancer Detection: An Integrative Analysis of Texture, Shape, and Intensity Contributions. BioMedInformatics, 4(4), 2400-2425. https://doi.org/10.3390/biomedinformatics4040129