Explainable Machine Learning-Based Approach to Identify People at Risk of Diabetes Using Physical Activity Monitoring
Abstract
1. Introduction
| Statement of Significance | |
| Problem | Undiagnosed type 2 diabetes mellitus (T2DM) remains a significant public health issue, often leading to delayed intervention and increased complications. |
| What is Already Known | Physical activity is closely linked to T2DM risk, and wearable devices enable continuous activity monitoring. Machine learning has shown potential in leveraging such data for early disease detection, but current models primarily rely on clinical or demographic data and often overlook physical activity patterns. |
| What This Paper Adds | This study introduces a novel approach by utilizing physical activity monitoring (PAM) data, combined with machine learning techniques, to identify individuals at risk of undiagnosed T2DM and prediabetes. By using features derived from PAM data in combination with age, the study demonstrates improved model performance and offers a noninvasive, low-cost method for diabetes risk screening at home. |
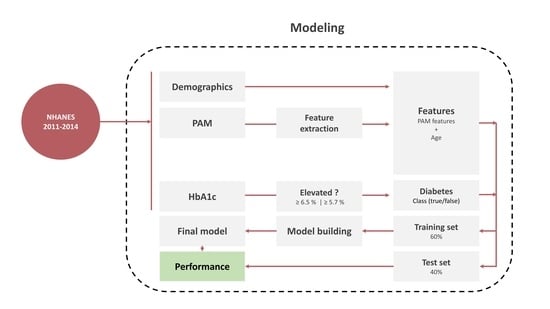
2. Methods
2.1. Data Sources
2.2. Physical Activity Monitoring (PAM)
2.3. Targets
2.4. Feature Engineering
2.5. Model Development
2.6. Model Assessment
2.7. Analyzing Feature Importance and Model Interpretability
3. Results
3.1. Model Performance
3.2. Feature Importance
4. Discussion
4.1. Comparison to Related Work
4.2. Strengths and Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Ong, K.L.; Stafford, L.K.; McLaughlin, S.A.; Boyko, E.J.; Vollset, S.E.; Smith, A.E.; Dalton, B.E.; Duprey, J.; Cruz, J.A.; Hagins, H.; et al. Global, regional, and national burden of diabetes from 1990 to 2021, with projections of prevalence to 2050: A systematic analysis for the Global Burden of Disease Study 2021. Lancet 2023, 402, 203–234. [Google Scholar] [CrossRef] [PubMed]
- Eppens, M.C.; Craig, M.E.; Cusumano, J.; Hing, S.; Chan, A.K.; Howard, N.J.; Silink, M.; Donaghue, K.C. Prevalence of Diabetes Complications in Adolescents with Type 2 Compared with Type 1 Diabetes. Diabetes Care 2006, 29, 1300–1306. [Google Scholar] [CrossRef]
- TODAY Study Group. Long-Term Complications in Youth-Onset Type 2 Diabetes. N. Engl. J. Med. 2021, 385, 416–426. [Google Scholar] [CrossRef]
- Tancredi, M.; Rosengren, A.; Svensson, A.M.; Kosiborod, M.; Pivodic, A.; Gudbjörnsdottir, S.; Wedel, H.; Clements, M.; Dahlqvist, S.; Lind, M. Excess Mortality among Persons with Type 2 Diabetes. N. Engl. J. Med. 2015, 373, 1720–1732. [Google Scholar] [CrossRef]
- Turner, R. Intensive blood-glucose control with sulphonylureas or insulin compared with conventional treatment and risk of complications in patients with type 2 diabetes (UKPDS 33). Lancet 1998, 352, 837–853. [Google Scholar] [CrossRef]
- Holman, R.R.; Paul, S.K.; Bethel, M.A.; Matthews, D.R.; Neil, H.A.W. 10-Year Follow-up of Intensive Glucose Control in Type 2 Diabetes. N. Engl. J. Med. 2008, 359, 1577–1589. [Google Scholar] [CrossRef]
- Selph, S.; Dana, T.; Blazina, I.; Bougatsos, C.; Patel, H.; Chou, R. Screening for Type 2 Diabetes Mellitus: A Systematic Review for the U.S. Preventive Services Task Force. Ann. Intern. Med. 2015, 162, 765–776. [Google Scholar] [CrossRef]
- Cichosz, S.L.; Fleischer, J.; Hoeyem, P.; Laugesen, E.; Poulsen, P.L.; Christiansen, J.S.; Ejskjær, N.; Hansen, T.K. Objective measurements of activity patterns in people with newly diagnosed Type 2 diabetes demonstrate a sedentary lifestyle. Diabet. Med. 2013, 30, 1063–1066. [Google Scholar] [CrossRef]
- Whelan, M.E.; Orme, M.W.; Kingsnorth, A.P.; Sherar, L.B.; Denton, F.L.; Esliger, D.W. Examining the use of glucose and physical activity self-monitoring technologies in individuals at moderate to high risk of developing type 2 diabetes: Randomized trial. JMIR mHealth uHealth 2019, 7, e14195. [Google Scholar] [CrossRef]
- Sidey-Gibbons, J.A.M.; Sidey-Gibbons, C.J. Machine learning in medicine: A practical introduction. BMC Med. Res. Methodol. 2019, 19, 64. [Google Scholar] [CrossRef]
- Bzdok, D.; Altman, N.; Krzywinski, M. Statistics versus machine learning. Nat. Methods 2018, 15, 233–234. [Google Scholar] [CrossRef] [PubMed]
- Demmer, R.T.; Zuk, A.M.; Rosenbaum, M.; Desvarieux, M. Prevalence of Diagnosed and Undiagnosed Type 2 Diabetes Mellitus Among US Adolescents: Results from the Continuous NHANES, 1999–2010. Am. J. Epidemiol. 2013, 178, 1106–1113. [Google Scholar] [CrossRef] [PubMed]
- Zoungas, S.; Woodward, M.; Li, Q.; Cooper, M.E.; Hamet, P.; Harrap, S.; Heller, S.; Marre, M.; Patel, A.; Poulter, N.; et al. Impact of age, age at diagnosis and duration of diabetes on the risk of macrovascular and microvascular complications and death in type 2 diabetes. Diabetologia 2014, 57, 2465–2474. [Google Scholar] [CrossRef] [PubMed]
- Herman, W.H.; Ye, W.; Griffin, S.J.; Simmons, R.K.; Davies, M.J.; Khunti, K.; Rutten, G.E.; Sandbaek, A.; Lauritzen, T.; Borch-Johnsen, K.; et al. Early detection and treatment of type 2 diabetes reduce cardiovascular morbidity and mortality: A simulation of the results of the Anglo-Danish-Dutch study of intensive treatment in people with screen-detected diabetes in primary care (ADDITION-Europe). Diabetes Care 2015, 38, 1449–1455. [Google Scholar] [CrossRef]
- Cichosz, S.L.; Jensen, M.H.; Olesen, S.S. Development and Validation of a Machine Learning Model to Predict Weekly Risk of Hypoglycemia in Patients with Type 1 Diabetes Based on Continuous Glucose Monitoring. Diabetes Technol. Ther. 2024, 26, 457–466. [Google Scholar] [CrossRef]
- Cichosz, S.L.; Hejlesen, O. Classification of Gastroparesis from Glycemic Variability in Type 1 Diabetes: A Proof-of-Concept Study. J. Diabetes Sci. Technol. 2022, 16, 1190–1195. [Google Scholar] [CrossRef]
- Cichosz, S.L.; Kronborg, T.; Jensen, M.H.; Hejlesen, O. Penalty weighted glucose prediction models could lead to better clinically usage. Comput. Biol. Med. 2021, 138, 104865. [Google Scholar] [CrossRef]
- Fregoso-Aparicio, L.; Noguez, J.; Montesinos, L.; García-García, J.A. Machine learning and deep learning predictive models for type 2 diabetes: A systematic review. Diabetol. Metab. Syndr. 2021, 13, 148. [Google Scholar] [CrossRef]
- Sambyal, N.; Saini, P.; Syal, R. A Review of Statistical and Machine Learning Techniques for Microvascular Complications in Type 2 Diabetes. Curr. Diabetes Rev. 2020, 17, 143–155. [Google Scholar] [CrossRef]
- Tigga, N.P.; Garg, S. Prediction of Type 2 Diabetes using Machine Learning Classification Methods. Procedia Comput. Sci. 2020, 167, 706–716. [Google Scholar] [CrossRef]
- Cichosz, S.L.; Lundby-Christensen, L.; Johansen, M.D.; Tarnow, L.; Almdal, T.P.; Hejlesen, O.K.; The, C.I.M.T. Trial Group. Prediction of excessive weight gain in insulin treated patients with type 2 diabetes. J. Diabetes 2016, 9, 325–331. [Google Scholar] [CrossRef] [PubMed]
- Bölen, M.C. From traditional wristwatch to smartwatch: Understanding the relationship between innovation attributes, switching costs and consumers’ switching intention. Technol. Soc. 2020, 63, 101439. [Google Scholar] [CrossRef]
- Weinstein, A.R.; Sesso, H.D.; Lee, I.M.; Cook, N.R.; Manson, J.E.; Buring, J.E.; Gaziano, J.M. Relationship of Physical Activity vs Body Mass Index with Type 2 Diabetes in Women. JAMA 2004, 292, 1188–1194. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.C.; Parker, J.D.; Clark, J.; Shin, H.C.; Rammon, J.R.; Burt, V.L. National Health and Nutrition Examination Survey: Estimation procedures, 2011–2014. Vital Health Stat 2018, 177, 1–26. [Google Scholar]
- Haffner, S.M. Epidemiology of Type 2 Diabetes: Risk Factors. Diabetes Care 1998, 21, C3–C6. [Google Scholar] [CrossRef]
- John, D.; Tang, Q.; Albinali, F.; Intille, S. An Open-Source Monitor-Independent Movement Summary for Accelerometer Data Processing. J. Meas. Phys. Behav. 2019, 2, 268–281. [Google Scholar] [CrossRef]
- American Diabetes Association Professional Practice Committee. 2. Classification and Diagnosis of Diabetes: Standards of Medical Care in Diabetes—2022. Diabetes Care 2022, 45, S17–S38. [Google Scholar] [CrossRef]
- Srinivasan, V.; Eswaran, C.; Sriraam, A.N. Artificial neural network based epileptic detection using time-domain and frequency-domain features. J. Med. Syst. 2005, 29, 647–660. [Google Scholar] [CrossRef]
- Hoffmann, R.G. Statistics in the Practice of Medicine. JAMA 1963, 185, 864–873. [Google Scholar] [CrossRef]
- Alexakis, C.; Nyongesa, H.O.; Saatchi, R.; Harris, N.D.; Davies, C.; Emery, C.; Ireland, R.H.; Heller, S.R. Feature Extraction and Classification of Electrocardiogram (ECG) signals related to hypoglycaemia. Comput. Cardiol. 2003, 30, 537–540. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar] [CrossRef]
- Yadaw, A.S.; Li, Y.C.; Bose, S.; Iyengar, R.; Bunyavanich, S.; Pandey, G. Clinical features of COVID-19 mortality: Development and validation of a clinical prediction model. Lancet Digit. Health 2020, 2, e516–e525. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Yang, X.; Huang, H.; Peng, C.; Ge, Y.; Wu, H.; Wang, J.; Xiong, G.; Yi, Y. Extreme Gradient Boosting Model Has a Better Performance in Predicting the Risk of 90-Day Readmissions in Patients with Ischaemic Stroke. J. Stroke Cerebrovasc. Dis. 2019, 28, 104441. [Google Scholar] [CrossRef] [PubMed]
- Ogunleye, A.; Wang, Q.G. XGBoost Model for Chronic Kidney Disease Diagnosis. IEEE/ACM Trans. Comput. Biol. Bioinform. 2020, 17, 2131–2140. [Google Scholar] [CrossRef] [PubMed]
- Cichosz, S.L.; Bender, C. Development of Machine Learning Models for the Identification of Elevated Ketone Bodies During Hyperglycemia in Patients with Type 1 Diabetes. Diabetes Technol. Ther. 2024, 26, 403–410. [Google Scholar] [CrossRef]
- Christodoulou, E.; Ma, J.; Collins, G.S.; Steyerberg, E.W.; Verbakel, J.Y.; Van Calster, B. A systematic review shows no performance benefit of machine learning over logistic regression for clinical prediction models. J. Clin. Epidemiol. 2019, 110, 12–22. [Google Scholar] [CrossRef]
- Lerman, P.M. Fitting Segmented Regression Models by Grid Search. J. R. Stat. Soc. Ser. C Appl. Stat. 1980, 29, 77–84. [Google Scholar] [CrossRef]
- Metz, C.E. Basic principles of ROC analysis. Semin. Nucl. Med. 1978, 8, 283–298. [Google Scholar] [CrossRef]
- Lundberg, S.M.; Allen, P.G.; Lee, S.-I. A Unified Approach to Interpreting Model Predictions. arXiv 2017, arXiv:1705.07874. [Google Scholar]
- Ekelund, U.; Brage, S.; Griffin, S.J.; Wareham, N.J. Objectively Measured Moderate- and Vigorous-Intensity Physical Activity but Not Sedentary Time Predicts Insulin Resistance in High-Risk Individuals. Diabetes Care 2009, 32, 1081–1086. [Google Scholar] [CrossRef]
- Kennerly, A.M.; Kirk, A. Physical activity and sedentary behaviour of adults with type 2 diabetes: A systematic review. Pract. Diabetes 2018, 35, 86–89g. [Google Scholar] [CrossRef]
- Lam, B.; Catt, M.; Cassidy, S.; Bacardit, J.; Darke, P.; Butterfield, S.; Alshabrawy, O.; Trenell, M.; Missier, P. Using Wearable Activity Trackers to Predict Type 2 Diabetes: Machine Learning-Based Cross-sectional Study of the UK Biobank Accelerometer Cohort. JMIR Diabetes 2021, 6, e23364. [Google Scholar] [CrossRef]
- Butte, N.F.; Ekelund, U.; Westerterp, K.R. Assessing physical activity using wearable monitors: Measures of physical activity. Med. Sci. Sports Exerc. 2012, 44, S5–S12. [Google Scholar] [CrossRef]




| Feature | Explanation | Definition |
|---|---|---|
| Statistical | ||
| Mean | Mean value of the PAM signal | μ |
| Variance | Spread or dispersion of the signal | σ2 |
| Skewness | Asymmetry of the signal distribution | |
| Kurtosis | Peakedness of the signal distribution | |
| Root Mean Square (RMS) | Square root of the mean of the squared values of the signal | |
| Time Domain | ||
| Zero-Crossing Rate | Number of times the signal crosses a given threshold | |
| Time Above | Fraction of time spent above a given threshold | |
| Signal Energy | Sum of squared values of the signal | |
| Autocorrelation | Similarity between the signal and its delayed versions | |
| Entropy | Signal’s complexity or uncertainty | |
| Waveform Shape | ||
| Peak-to-Peak Amplitude | Range (maximum and minimum values of the signal) | |
| Crest Factor | Ratio of the peak value to the RMS value of the signal | |
| Slope Sign Change | Number of directional changes in the signal | |
| Waveform Length | The total of the absolute differences between each pair of consecutive samples in the signal |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cichosz, S.L.; Bender, C.; Hejlesen, O. Explainable Machine Learning-Based Approach to Identify People at Risk of Diabetes Using Physical Activity Monitoring. BioMedInformatics 2025, 5, 1. https://doi.org/10.3390/biomedinformatics5010001
Cichosz SL, Bender C, Hejlesen O. Explainable Machine Learning-Based Approach to Identify People at Risk of Diabetes Using Physical Activity Monitoring. BioMedInformatics. 2025; 5(1):1. https://doi.org/10.3390/biomedinformatics5010001
Chicago/Turabian StyleCichosz, Simon Lebech, Clara Bender, and Ole Hejlesen. 2025. "Explainable Machine Learning-Based Approach to Identify People at Risk of Diabetes Using Physical Activity Monitoring" BioMedInformatics 5, no. 1: 1. https://doi.org/10.3390/biomedinformatics5010001
APA StyleCichosz, S. L., Bender, C., & Hejlesen, O. (2025). Explainable Machine Learning-Based Approach to Identify People at Risk of Diabetes Using Physical Activity Monitoring. BioMedInformatics, 5(1), 1. https://doi.org/10.3390/biomedinformatics5010001