Early Breast Cancer Detection Based on Deep Learning: An Ensemble Approach Applied to Mammograms
Abstract
1. Introduction
2. Related Works
2.1. Transition to Machine Learning and Deep Learning
2.2. Emergence of Ensemble Learning
2.3. Challenges in Dataset Variability and Transfer Learning
2.4. Comparative Analysis of Approaches
3. Research Methodology
3.1. Data Collection and Preprocessing
- INbreast Dataset:
- CBIS-DDSM Dataset:
3.2. INbreast and CBIS-DDSM
3.3. Data Splitting and Justification
3.3.1. INbreast Dataset (80/10/10 Split)
- Maximizing training data:Allocating 80% of the dataset to training ensures the model has sufficient data to learn robust features, reducing the risk of underfitting.
- Validation efficiency:A 10% validation set provides enough samples to tune hyperparameters and monitor model performance without significantly reducing the training data size.
- Testing representativity:The 10% test set ensures the evaluation is conducted on a statistically significant and representative subset, providing reliable insights into the model’s generalization capabilities.
3.3.2. CBIS-DDSM Dataset (70/15/15 Split)
- Robust training:The large size of the training set ensures the model is exposed to a wide variety of cases, enhancing its ability to generalize.
- Comprehensive validation:Allocating 15% of the data for validation reduces variability in performance metrics, providing a more accurate assessment of the model’s hyperparameters and convergence.
- Extensive testing:With 15% reserved for testing, the model’s performance is evaluated on a diverse and substantial subset, simulating real-world conditions.
3.3.3. Stratified Sampling
3.3.4. Comparison with the Literature
3.4. Data Preprocessing
- Step 1: Image Resizing:Mammogram images come in various sizes, which need to be standardized to ensure compatibility with deep learning models. All images were resized to 224 × 224 pixels to match the input size of the pre-trained CNN architectures (VGG16, DenseNet121, InceptionV3). This resizing ensured consistent input dimensions for the network while maintaining the spatial integrity of the mammographic features.
- Step 2: Normalization:Pixel intensities in mammographic images vary widely, which can cause instability during training. To stabilize and accelerate the training process, each pixel’s intensity was normalized to a range between 0 and 1. This scaling ensured that the input values were comparable across all images and helped with faster convergence of the deep learning model.
- Step 3: Data Augmentation:Medical imaging datasets are often limited in size, which can lead to overfitting in deep learning models. To overcome this, we applied data augmentation techniques to artificially increase the diversity of the dataset. Augmentation included random rotations, horizontal and vertical flips, zooming, and shifting, simulating real-world variations in imaging conditions.
- Step 4: Data Splitting:To train and evaluate the deep learning models effectively, the dataset was divided into training, validation, and test subsets. For the INbreast dataset, an 80/10/10 split was used, while a 70/15/15 split was adopted for CBIS-DDSM. Stratified sampling ensured that the class distributions were consistent across the subsets. Algorithm 1 describes the preprocessing pipeline for mammographic datasets.
| Algorithm 1. Preprocessing Pipeline for Mammographic Datasets. |
| Input: Dataset, TargetSize, Augmentations, SplitRatios |
| Output: TrainSet, ValSet, TestSet |
 |
Training/Validation/Test Procedure
- Data Splitting:The INbreast and CBIS-DDSM datasets were split into three subsets: training, validation, and test sets.
- Cross-Validation:To ensure statistical reliability, we implemented k-fold cross-validation with k = 5.
- Training Procedure:Each CNN model (VGG16, DenseNet121, InceptionV3) was pre-trained on the ImageNet dataset and subsequently fine-tuned on the training set of mammographic images.
- Validation Process:The validation set was used to monitor the model’s performance during training. Early stopping was implemented to terminate training if the validation loss ceased improving for a pre-defined number of epochs, thereby preventing overfitting.
- Testing and Evaluation:The test set, containing unseen data, was used exclusively for the final evaluation of each model’s performance.
- Ensemble Model Training:Predictions from the individual CNN models were combined using a soft voting mechanism, where the averaged probabilities determined the final classification.
3.5. Deep Learning Models for Mammogram Analysis
- VGG16: The architecture of VGG16 consists of 16 layers, including 13 convolutional layers and 3 fully connected layers. It uses small 3 × 3 filters in all its convolutional layers, which allows it to capture subtle and fine-grained features from the input images. These filters are critical in mammography as they enable the model to identify microcalcifications and other small, abnormal patterns indicative of early-stage cancer. VGG16 is computationally heavy due to its large number of parameters (approximately 138 million), but it excels in extracting spatial hierarchies of features from the images.
- DenseNet121: The DenseNet121 model is characterized by its dense connectivity, where each layer receives input from all previous layers, allowing for the reuse of features. This dense architecture reduces the number of parameters and mitigates the vanishing gradient problem, which can be particularly helpful in deep networks. DenseNet121’s structure makes it efficient in terms of memory and computational resources, which is essential when dealing with large datasets like CBIS-DDSM.
- InceptionV3: This architecture is known for its Inception modules, which allow it to process features at multiple scales simultaneously. In each Inception module, 1 × 1, 3 × 3, and 5 × 5 convolutions are applied in parallel, and their outputs are concatenated. This design makes InceptionV3 particularly effective in detecting both large and small lesions, which can vary significantly in size. By analyzing features at different scales, InceptionV3 captures the multi-scale nature of abnormalities in mammograms.
3.5.1. Pre-Training of Individual Models
| Algorithm 2. Pre-training of Individual Models |
| Input: Models = {VGG16, DenseNet121, InceptionV3} PretrainedWeights = ImageNet weights |
| Output: PretrainedModels = List of modified models ready for fine-tuning |
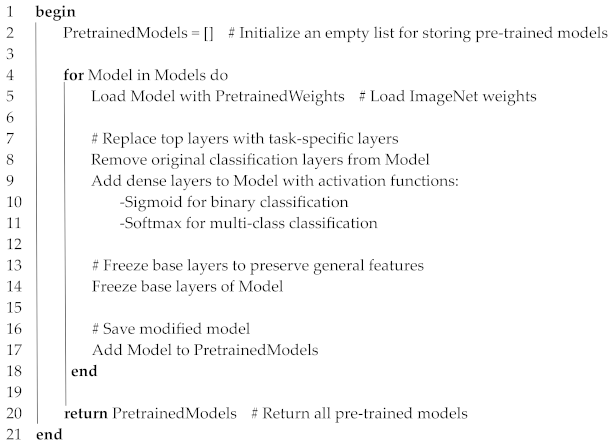 |
3.5.2. Fine-Tuning of Models
| Algorithm 3. Fine-tuning of Models |
| Input: PretrainedModels = List of pre-trained models TrainingData = Training dataset ValidationData = Validation dataset Augmentations = Data augmentation techniques LearningRates = {LowRate, HighRate} |
| Output: FineTunedModels = List of fine-tuned models |
 |
3.5.3. Hyperparameter Optimization
| Algorithm 4. Hyperparameter Optimization |
| Input: Models = {VGG16, DenseNet121, InceptionV3} TrainingData = Training dataset ValidationData = Validation dataset HyperparameterSpace = {LearningRates, BatchSizes, DropoutRates, Epochs} |
| Output: OptimizedModels = List of models with best hyperparameters |
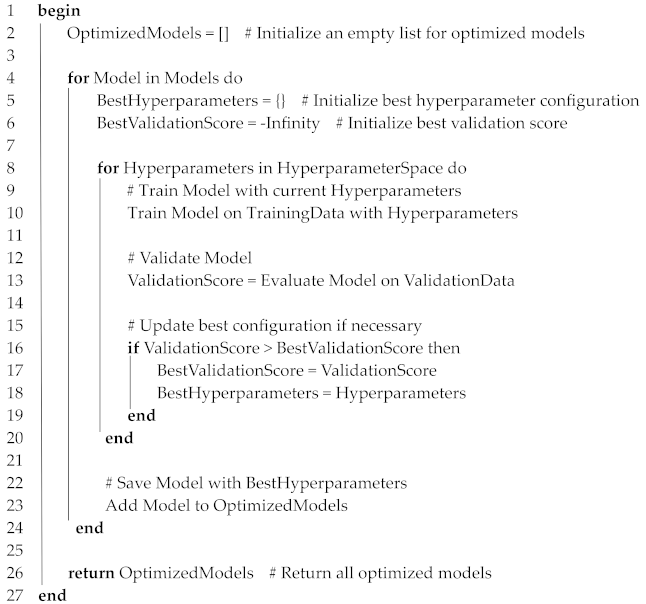 |
3.6. Ensemble CNN Model Design
3.6.1. Ensemble Model Structure
- VGG16 excels at capturing fine details critical for detecting microcalcifications.
- DenseNet121 reuses features efficiently across layers, improving detection in dense tissues.
- InceptionV3 processes features at multiple scales, enabling the detection of lesions with varied sizes.
3.6.2. Voting Mechanism and Optimization
3.6.3. Advanced Ensemble Techniques
- Weighted voting:Assigns higher weights to models with superior validation performance, enhancing their influence on the final decision.
- Stacking:Uses a meta-model (e.g., logistic regression or gradient boosting) to combine predictions from individual CNNs, capturing higher-order dependencies between them.
3.6.4. Ablation Study
- Excluding VGG16 significantly reduced sensitivity, highlighting its role in detecting subtle details.
- Excluding DenseNet121 increased false positives, showing its effectiveness in refining predictions for dense tissues.
- Excluding InceptionV3 lowered specificity, indicating its importance in multi-scale feature extraction.
3.6.5. Evaluation Metrics
- Accuracy:
- 2.
- Sensitivity (recall):
- 3.
- Specificity:
- 4.
- Precision:
- 5.
- F1-Score:
- 6.
- Area Under the Curve (AUC): AUC represents the area under the ROC curve, serving as a comprehensive metric to evaluate a model’s performance. Its value ranges between 0 and 1, where 1 signifies a perfect model, 0.5 indicates no discriminative capability (comparable to random guessing), and 0 reflects a model that consistently makes incorrect predictions.
| Algorithm 5. Pseudocode for the Ensemble Model |
| Input: Models = {VGG16, DenseNet121, InceptionV3} TestData = Mammogram images for testing |
| Output: FinalPredictions = Predicted classes for TestData |
 |
4. Results
4.1. Dataset Distribution
- INbreast dataset (total images: 410)Training: 328 images; validation: 41 images; test: 41 images
- CBIS-DDSM dataset (total images: 10,239)Training: 7167 images; validation: 1536 images; test: 1536 images
4.2. INbreast Dataset
4.2.1. Validation Curves for INbreast
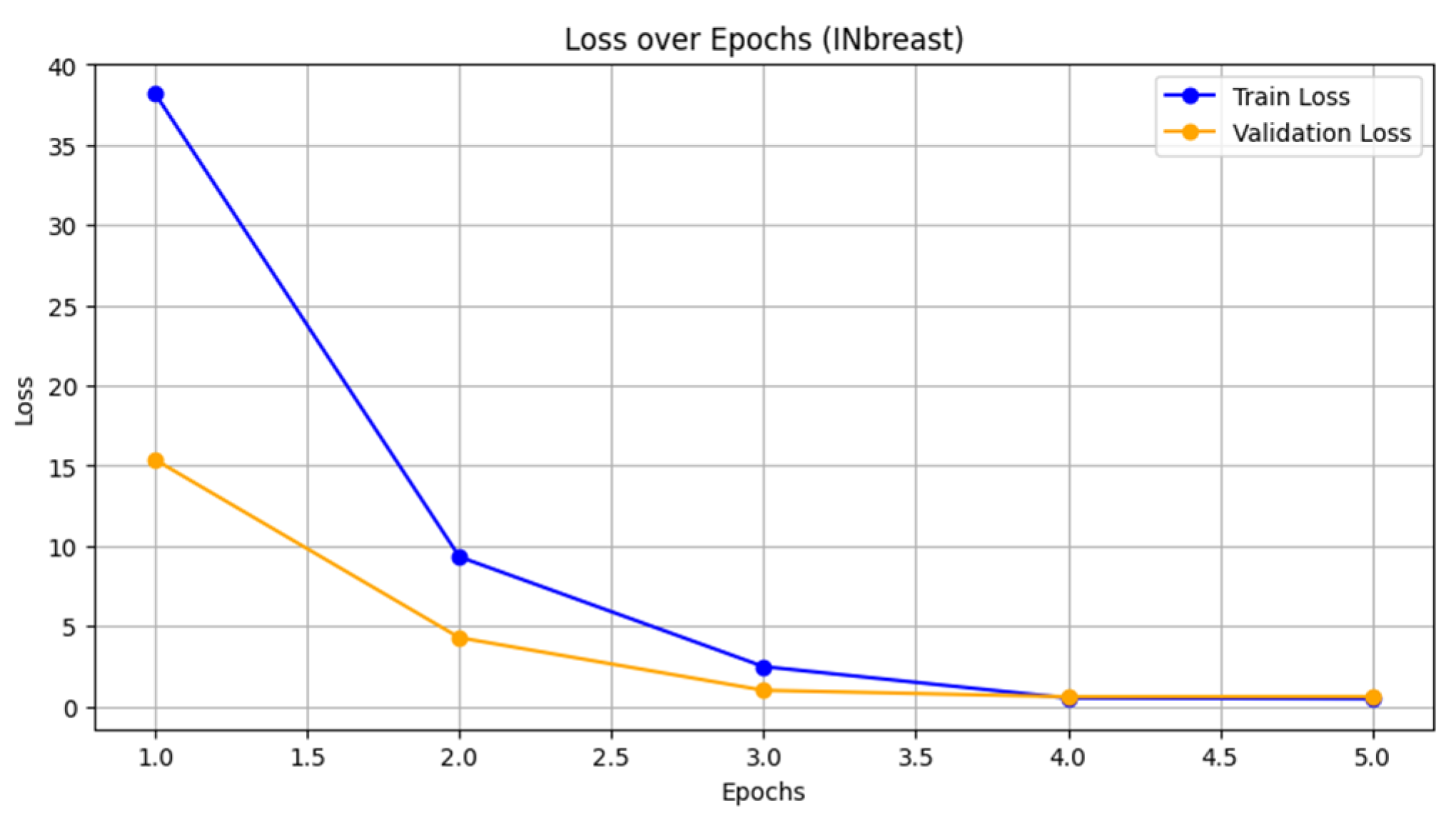
- Loss vs. epoch: Figure 4 shows the loss curves for both training and validation. The training loss decreased rapidly from 38.1931 to 0.4749, indicating effective optimization during the training process. The validation loss also decreased significantly from 15.3724 to approximately 0.6082, demonstrating good convergence. The similarity in trends suggests that the model is not overfitting, even though the validation loss stabilizes in later epochs.
- Accuracy vs. epoch: Figure 5 presents the accuracy curves for both training and validation. The training accuracy improved steadily from 56.66% to 80.39%, while the validation accuracy peaked at 70.73% by the second epoch and remained stable thereafter. This plateau suggests that while the model is learning effectively on the training data, the validation performance may be limited by the dataset’s inherent complexity or noise.
4.2.2. Abnormality Detection Performance
- The individual models VGG16, DenseNet121, and InceptionV3 perform well, with AUCs close to or above 0.90.
- The ensemble model outperforms all individual models, achieving an AUC of 0.96 and an F1-score of 89.1%, which indicates better detection performance, particularly in reducing false positives and false negatives.
- The higher precision and specificity in the ensemble model demonstrate that it is effective in distinguishing between abnormal and normal cases with more reliability.
4.2.3. Malignancy Detection Performance
- The ensemble model again surpasses the individual models, with an AUC of 0.95 and the highest F1-score (89.0%).
- The ensemble model’s superior precision (90.0%) and accuracy (89.3%) reflect its robustness in correctly identifying malignant cases while minimizing false positives.
4.2.4. Detection of Normal, Benign, and Malignant Cases
4.2.5. Accuracy and Precision
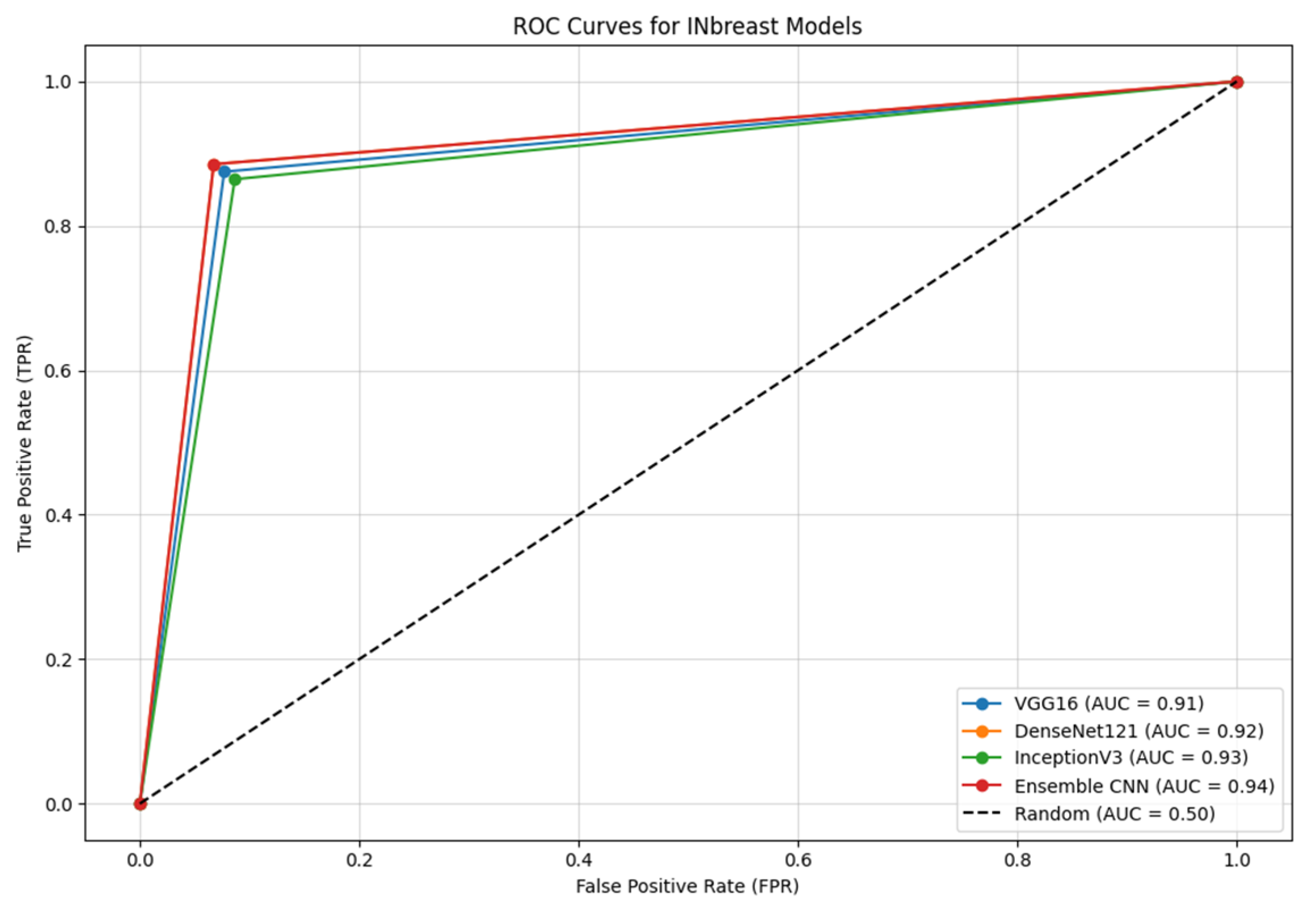
4.2.6. ROC Curves
- Ensemble CNN achieves the highest AUC of 0.94, showcasing its superior ability to balance sensitivity and specificity compared with the individual models.
- DenseNet121 and InceptionV3 follow closely, with AUC values of 0.92 and 0.93, respectively, highlighting their robustness in capturing key features in mammographic images.
- VGG16, while performing slightly below the other models, still demonstrates a strong AUC of 0.91, indicating effective feature extraction and classification capabilities.
4.2.7. Confusion Matrix
4.3. CBIS-DDSM Dataset
4.3.1. Validation Curves for CBIS-DDSM
- Loss vs. epoch: Figure 9 shows the training and validation loss curves for the CBIS-DDSM dataset over 20 epochs. The training loss decreases consistently from 0.60 to 0.29, demonstrating steady optimization during the training process. The validation loss also decreases, starting at 0.62 and stabilizing around 0.35. The convergence between the training and validation loss suggests effective learning with minimal overfitting, despite the dataset’s variability and challenges.
- Accuracy vs. epoch: Figure 10 presents the training and validation accuracy curves. The training accuracy increases steadily, starting at 70% and reaching 92% by epoch 20. The validation accuracy follows a similar trend, starting at 68% and stabilizing at 90%. The alignment between the training and validation accuracy reflects the model’s ability to generalize effectively across diverse imaging conditions in the CBIS-DDSM dataset.
4.3.2. Abnormality Detection Performance
- The performance of the individual CNN models (VGG16, DenseNet121, and InceptionV3) on malignancy detection in the CBIS-DDSM dataset demonstrates moderate results, with AUCs ranging from 0.80 to 0.82. This reflects the complexity of the dataset and the challenge of accurately detecting malignancy in this larger dataset.
- The DenseNet121 model shows the best performance among the individual models, achieving an AUC of 0.82 and an F1-score of 76.2%.
- The ensemble model offers a significant improvement, achieving an AUC of 0.84 and an F1-score of 77.1%. This improvement highlights the advantage of combining the strengths of multiple CNN architectures to reduce false positives and negatives.
- The sensitivity (76.8%) and precision (78.0%) in the ensemble model indicate its ability to detect malignant cases more accurately than the individual models while also reducing the rate of false positives.
4.3.3. Detection of Normal, Benign, and Malignant Cases (CBIS-DDSM Dataset)
| Method | AUC | F1-Score | Sensitivity (%) | Precision (%) | Specificity (%) | Accuracy (%) |
|---|---|---|---|---|---|---|
| VGG16 | 0.82 | 76.5 | 75.9 | 77.1 | 76.2 | 76.0 |
| DenseNet121 | 0.84 | 78.2 | 77.6 | 78.8 | 77.9 | 77.8 |
| InceptionV3 | 0.83 | 77.3 | 76.8 | 77.6 | 77.1 | 77.0 |
| Ensemble | 0.85 | 79.1 | 78.7 | 79.5 | 79.0 | 78.9 |
- The individual models (VGG16, DenseNet121, and InceptionV3) show decent results, with AUCs ranging from 0.82 to 0.84, indicating good performance in detecting normal cases.
- The ensemble model improves these results, achieving an AUC of 0.85 and an F1-score of 79.1%. This shows that the ensemble model is effective in reducing false positives while maintaining high precision and sensitivity.
- The ensemble model’s balanced sensitivity (78.7%) and precision (79.5%) indicate that it can accurately detect normal cases with a low rate of false positives.
- The ensemble model’s specificity (79.0%) and accuracy (78.9%) are higher compared with the individual models, showing greater reliability in distinguishing normal from abnormal cases.
| Method | AUC | F1-Score | Sensitivity (%) | Precision (%) | Specificity (%) | Accuracy (%) |
|---|---|---|---|---|---|---|
| VGG16 | 0.80 | 74.5 | 73.8 | 75.2 | 74.3 | 74.1 |
| DenseNet121 | 0.82 | 76.3 | 75.7 | 76.8 | 76.0 | 75.9 |
| InceptionV3 | 0.81 | 75.5 | 75.0 | 75.9 | 75.4 | 75.3 |
| Ensemble | 0.83 | 77.4 | 77.0 | 78.0 | 77.3 | 77.2 |
- The individual models perform reasonably well for detecting benign cases, with AUCs ranging from 0.80 to 0.82. DenseNet121 outperforms the other models slightly in terms of sensitivity and specificity.
- The ensemble model outperforms the individual models, achieving an AUC of 0.83 and an F1-score of 77.4%, reflecting its improved ability to balance precision and recall when detecting benign cases.
- Sensitivity (77.0%) and precision (78.0%) in the ensemble model are slightly higher than those of the individual models, indicating its capacity to identify benign cases with fewer false positives.
- The ensemble model’s specificity (77.3%) and accuracy (77.2%) are improved compared with the individual models, confirming its ability to distinguish benign cases from malignant or normal ones with greater confidence.
| Method | AUC | F1-Score | Sensitivity (%) | Precision (%) | Specificity (%) | Accuracy (%) |
|---|---|---|---|---|---|---|
| VGG16 | 0.78 | 72.9 | 72.5 | 73.4 | 72.8 | 72.7 |
| DenseNet121 | 0.80 | 74.3 | 73.8 | 74.7 | 74.1 | 74.0 |
| InceptionV3 | 0.79 | 73.6 | 73.1 | 74.0 | 73.5 | 73.4 |
| Ensemble | 0.82 | 75.7 | 75.3 | 76.2 | 75.6 | 75.5 |
- The individual models demonstrate moderate performance for detecting malignant cases, with AUCs ranging from 0.78 to 0.80. DenseNet121 achieves slightly better results in terms of both sensitivity and specificity compared with VGG16 and InceptionV3.
- The ensemble model significantly improves upon the individual models, achieving an AUC of 0.82 and an F1-score of 75.7%. This improvement is particularly important for minimizing false negatives in malignant case detection.
- Sensitivity (75.3%) and precision (76.2%) for the ensemble model are higher than for the individual models, reflecting its capacity to detect more malignant cases while keeping the false negative rate low.
- The ensemble model’s specificity (75.6%) and accuracy (75.5%) further highlight its ability to differentiate between malignant and benign/normal cases more effectively.
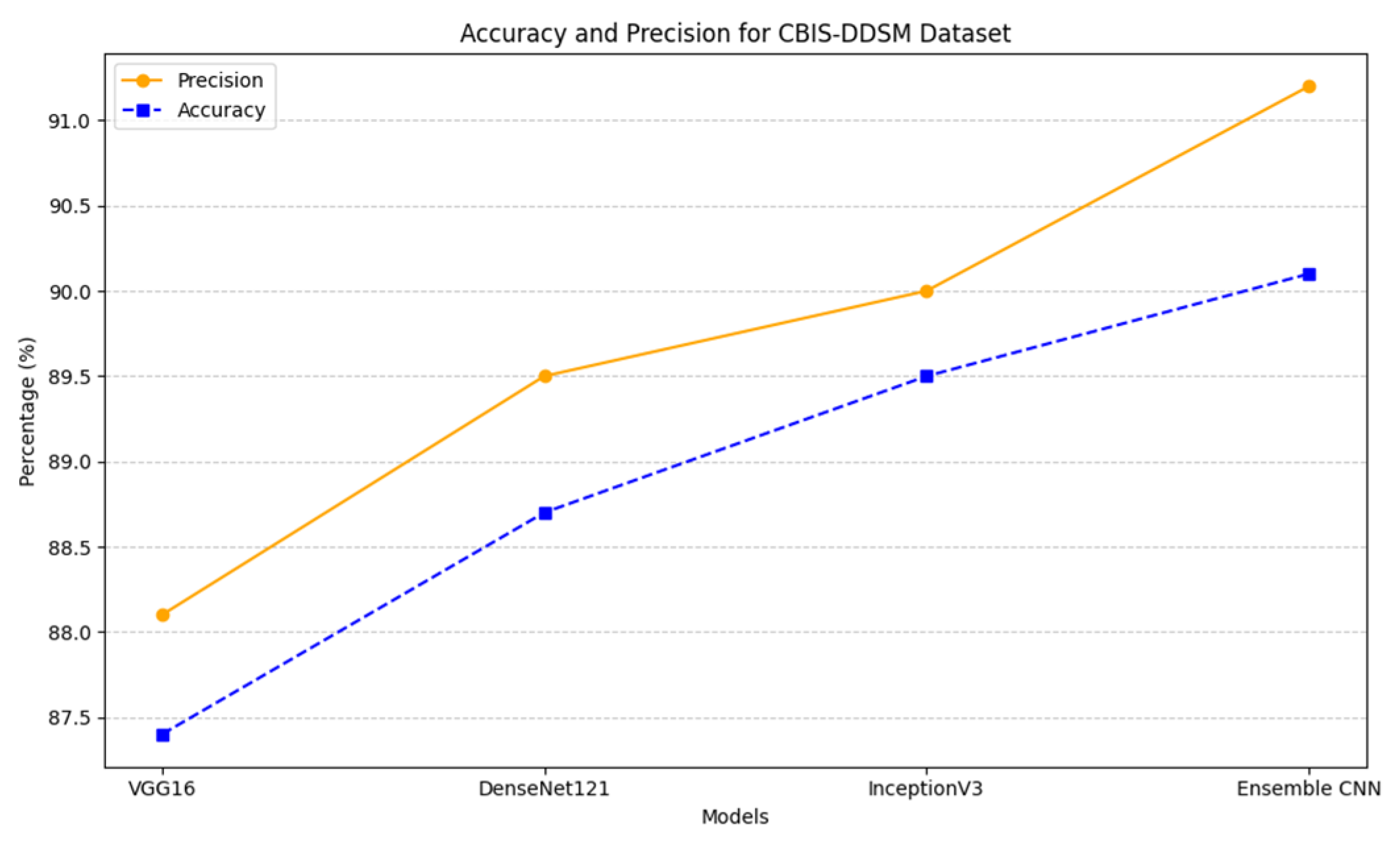
4.3.4. Accuracy and Precision
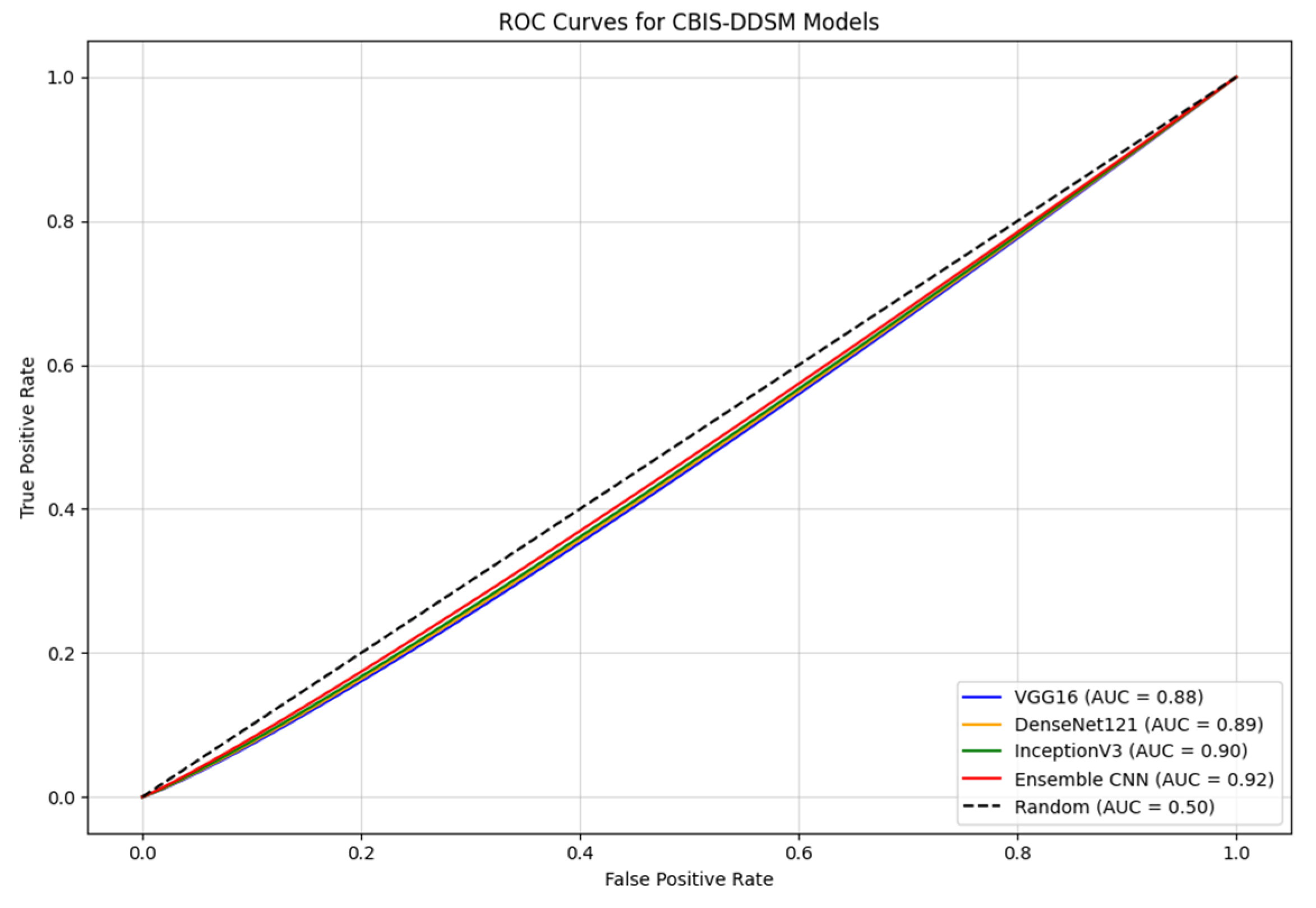
4.3.5. ROC Curves
- The Ensemble CNN achieves the highest AUC of 0.92, showcasing its ability to integrate the strengths of the individual models to achieve a robust and accurate classification.
- InceptionV3 and DenseNet121 demonstrate strong performance, with AUC values of 0.90 and 0.89, respectively. Their advanced architectures contribute to their capacity to effectively extract relevant features from mammographic images.
- VGG16, while slightly trailing with an AUC of 0.88, still shows competitive performance, making it a viable option for feature extraction in this dataset.
4.3.6. Confusion Matrix
4.4. General Observations Across Both Datasets
- Dataset size and diversity: CBIS-DDSM’s larger size and greater variability test the scalability and adaptability of the models, while INbreast’s smaller, homogeneous dataset provides a controlled setting ideal for transfer learning.
- Annotation quality: The precise annotations in INbreast facilitate better feature learning, while the variability in CBIS-DDSM annotations poses additional challenges for training and validation.
- Clinical variability: CBIS-DDSM includes a broader spectrum of cases and imaging conditions, increasing the complexity of classification tasks compared with the more uniform INbreast dataset.
- Model generalization: The ensemble model consistently exhibits the ability to generalize better across both datasets, achieving a balanced trade-off between sensitivity and specificity, particularly crucial in clinical decision making.
4.5. Analysis of Data Augmentation Techniques and Impact on Dataset Balancing
- 1.
- Reduction in Class Imbalance:
- Augmentation significantly increased the availability of underrepresented classes, ensuring a more balanced dataset distribution for training.
- For INbreast, this was particularly effective in compensating for the limited dataset size, amplifying the representation of malignant samples.
- In CBIS-DDSM, augmentation provided robust examples for minority classes across a broader spectrum of imaging conditions.
- 2.
- Improved Generalization:
- By exposing the models to diverse augmented samples, data augmentation reduced overfitting, leading to enhanced validation performance.
- Augmented datasets improved the sensitivity of the models, particularly for detecting malignant cases, as shown in the reduced false negatives in the confusion matrices.
- 3.
- Enhanced Model Performance:
- Both datasets benefited from augmentation, with the models achieving higher F1-scores and accuracy metrics, especially the Ensemble CNN, which capitalized on the enriched data.
5. Discussion
5.1. Experimental Environment
- CPU: Google Compute Engine
- GPU: NVIDIA T4
- RAM: 15 GB
- Storage: 112.6 GB.
5.2. Key Findings
- The ensemble model consistently outperformed individual CNN architectures, achieving higher accuracy, sensitivity, and specificity across both datasets.
- The INbreast dataset provided an ideal testing environment, resulting in higher performance metrics due to its high-resolution images and detailed annotations.
- The CBIS-DDSM dataset posed greater challenges, reflecting the ensemble model’s ability to adapt to more diverse and complex imaging conditions.
- Data augmentation proved essential in balancing the datasets, particularly for underrepresented classes like malignant cases, significantly enhancing model generalization.
- The ensemble approach demonstrated its robustness and scalability, emphasizing its potential for real-world clinical applications.
5.3. Research Limitations
- Dataset Characteristics: The limited size of the INbreast dataset restricted the model’s ability to generalize effectively, despite augmentation techniques.
- Class Imbalance: Both datasets exhibited class imbalances, particularly in the representation of malignant cases. While data augmentation helped mitigate this issue, achieving equitable performance across all classes remains a challenge.
- Ensemble Integration: The soft voting mechanism used for integrating predictions, although effective, may not fully exploit the complementary strengths of the individual CNN architectures. More advanced techniques, such as weighted voting or stacking, could further improve the model’s predictive performance.
- Lack of Domain-Specific Pre-training: The use of ImageNet pre-training provided generalized features for transfer learning. However, pre-training on domain-specific medical datasets could enhance the model’s ability to capture more relevant features for mammographic imaging.
6. Conclusions and Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Breast Cancer. Available online: https://www.who.int/news-room/fact-sheets/detail/breast-cancer (accessed on 29 November 2024).
- Wilkinson, L.; Gathani, T. Understanding breast cancer as a global health concern. Br. J. Radiol. 2022, 95, 20211033. [Google Scholar] [CrossRef] [PubMed]
- Arnold, M.; Morgan, E.; Rumgay, H.; Mafra, A.; Singh, D.; Laversanne, M.; Vignat, J.; Gralow, J.R.; Cardoso, F.; Siesling, S.; et al. Current and future burden of breast cancer: Global statistics for 2020 and 2040. Breast 2022, 66, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Barclay, J.; Ernster, V.; Kerlikowske, K.; Grady, D.; Sickles, E.A. Comparison of risk factors for ductal carcinoma in situ and invasive breast cancer. J. Natl. Cancer Inst. 1997, 89, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Kerlikowske, K. Epidemiology of ductal carcinoma in situ. J. Natl. Cancer Inst. Monogr. 2010, 2010, 139–141. [Google Scholar] [CrossRef]
- Goh, C.W.; Wu, J.; Ding, S.; Lin, C.; Chen, X.; Huang, O.; Chen, W.; Li, Y.; Shen, K.; Zhu, L. Invasive ductal carcinoma with coexisting ductal carcinoma in situ (IDC/DCIS) versus pure invasive ductal carcinoma (IDC): A comparison of clinicopathological characteristics, molecular subtypes, and clinical outcomes. J. Cancer Res. Clin. Oncol. 2019, 145, 1877–1886. [Google Scholar] [CrossRef]
- Biglia, N.; Maggiorotto, F.; Liberale, V.; Bounous, V.; Sgro, L.; Pecchio, S.; D’Alonzo, M.; Ponzone, R. Clinical-pathologic features, long term-outcome and surgical treatment in a large series of patients with invasive lobular carcinoma (ILC) and invasive ductal carcinoma (IDC). Eur. J. Surg. Oncol. 2013, 39, 455–460. [Google Scholar] [CrossRef]
- Barroso-Sousa, R.; Metzger-Filho, O. Differences between invasive lobular and invasive ductal carcinoma of the breast: Results and therapeutic implications. Ther. Adv. Med. Oncol. 2016, 8, 261–266. [Google Scholar] [CrossRef]
- Wang, P.; Chen, J.; Zhao, W. Overview of Early Detection for Breast Cancer: Current Status and Future Perspectives. J. Mod. Med Oncol. 2023, 3, 10. [Google Scholar] [CrossRef]
- Ekpo, E.U.; Alakhras, M.; Brennan, P. Errors in mammography cannot be solved through technology alone. Asian Pac. J. Cancer Prev. APJCP 2018, 19, 291–301. [Google Scholar]
- Das, D.K. Artificial intelligence technologies for breast cancer screening. Oncol. Times 2021, 43, 20–21. [Google Scholar] [CrossRef]
- Drukteinis, J.S.; Mooney, B.P.; Flowers, C.I.; Gatenby, R.A. Beyond mammography: New frontiers in breast cancer screening. Am. J. Med. 2013, 126, 472–479. [Google Scholar] [CrossRef] [PubMed]
- Pesapane, F.; Rotili, A.; Raimondi, S.; Aurilio, G.; Lazzeroni, M.; Nicosia, L.; Latronico, A.; Pizzamiglio, M.; Cassano, E.; Gandini, S. Evolving paradigms in breast cancer screening: Balancing efficacy, personalization, and equity. Eur. J. Radiol. 2024, 172, 111321. [Google Scholar] [CrossRef] [PubMed]
- Abdelhafiz, D.; Yang, C.; Ammar, R.; Nabavi, S. Deep convolutional neural networks for mammography: Advances, challenges and applications. BMC Bioinform. 2019, 20, 281. [Google Scholar] [CrossRef] [PubMed]
- Bhowmik, A.; Eskreis-Winkler, S. Deep learning in breast imaging. BJR|Open 2022, 4, 20210060. [Google Scholar] [CrossRef]
- Yan, K.; Wang, X.; Lu, L.; Summers, R.M. DeepLesion: Automated mining of large-scale lesion annotations and universal lesion detection with deep learning. J. Med. Imaging 2018, 5, 036501. [Google Scholar] [CrossRef]
- Garrucho, L.; Kushibar, K.; Jouide, S.; Diaz, O.; Igual, L.; Lekadir, K. Domain generalization in deep learning based mass detection in mammography: A large-scale multi-center study. Artif. Intell. Med. 2022, 132, 102386. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, S.; Liu, L.; Hickman, S.; Gilbert, F.J.; Schönlieb, C.B.; Aviles-Rivero, A.I. MammoDG: Generalisable Deep Learning Breaks the Limits of Cross-Domain Multi-Center Breast Cancer Screening. arXiv 2023, arXiv:2308.01057. [Google Scholar]
- Nguyen, H.T.; Lam, T.B.; Tran, Q.T.D.; Nguyen, M.T.; Chung, D.T.; Dinh, V.Q. In-context cross-density adaptation on noisy mammogram abnormalities detection. In Proceedings of the 2023 IEEE Statistical Signal Processing Workshop (SSP), Hanoi, Vietnam, 2–5 July 2023; pp. 383–387. [Google Scholar]
- Liu, F.; Panagiotakos, D. Real-world data: A brief review of the methods, applications, challenges and opportunities. BMC Med. Res. Methodol. 2022, 22, 287. [Google Scholar] [CrossRef]
- Murphy, G.; Singh, R. Comparative Analysis and Ensemble Enhancement of Leading CNN Architectures for Breast Cancer Classification. arXiv 2024, arXiv:2410.03333. [Google Scholar]
- Nie, K.; Chen, J.-H.; Hon, J.Y.; Chu, Y.; Nalcioglu, O.; Su, M.-Y. Quantitative analysis of lesion morphology and texture features for diagnostic prediction in breast MRI. Acad. Radiol. 2008, 15, 1513–1525. [Google Scholar] [CrossRef]
- Albashish, D.; Al-Sayyed, R.; Abdullah, A.; Ryalat, M.H.; Almansour, N.A. Deep CNN model based on VGG16 for breast cancer classification. In Proceedings of the 2021 International Conference on Information Technology (ICIT), Amman, Jordan, 14–15 July 2021; pp. 805–810. [Google Scholar]
- Bello, A.; Ng, S.-C.; Leung, M.-F. Skin Cancer Classification Using Fine-Tuned Transfer Learning of DENSENET-121. Appl. Sci. 2024, 14, 7707. [Google Scholar] [CrossRef]
- Pattanaik, R.K.; Mishra, S.; Siddique, M.; Gopikrishna, T.; Satapathy, S. Breast Cancer Classification from Mammogram Images Using Extreme Learning Machine-Based DenseNet121 Model. J. Sens. 2022, 2022, 2731364. [Google Scholar] [CrossRef]
- Al Husaini, M.A.S.; Habaebi, M.H.; Gunawan, T.S.; Islam, M.R.; Elsheikh, E.A.A.; Suliman, F.M. Thermal-based early breast cancer detection using inception V3, inception V4 and modified inception MV4. Neural Comput. Appl. 2022, 34, 333–348. [Google Scholar] [CrossRef] [PubMed]
- Al Husaini, M.A.S.; Habaebi, M.H.; Gunawan, T.S.; Islam, M.R.; Hameed, S.A. Automatic breast cancer detection using inception V3 in thermography. In Proceedings of the 2021 8th International Conference on Computer and Communication Engineering (ICCCE), Kuala Lumpur, Malaysia, 22–23 June 2021; pp. 255–258. [Google Scholar]
- Petrini, D.G.P.; Shimizu, C.; Roela, R.A.; Valente, G.V.; Folgueira, M.A.A.K.; Kim, H.Y. Breast cancer diagnosis in two-view mammography using end-to-end trained efficientnet-based convolutional network. IEEE Access 2022, 10, 77723–77731. [Google Scholar] [CrossRef]
- Leighton, T.G. What is ultrasound? Prog. Biophys. Mol. Biol. 2007, 93, 3–83. [Google Scholar] [CrossRef]
- Dalecki, D. Mechanical bioeffects of ultrasound. Annu. Rev. Biomed. Eng. 2004, 6, 229–248. [Google Scholar] [CrossRef]
- Whitney, H.M.; Li, H.; Ji, Y.; Liu, P.; Giger, M.L. Comparison of breast MRI tumor classification using human-engineered radiomics, transfer learning from deep convolutional neural networks, and fusion methods. Proc. IEEE 2019, 108, 163–177. [Google Scholar] [CrossRef]
- Mann, R.M.; Kuhl, C.K.; Kinkel, K.; Boetes, C. Breast MRI: Guidelines from the European society of breast imaging. Eur. Radiol. 2008, 18, 1307–1318. [Google Scholar] [CrossRef]
- Khourdifi, Y.; Bahaj, M. Applying Best Machine Learning Algorithms for Breast Cancer Prediction and Classification. In Proceedings of the 2018 International Conference on Electronics, Control, Optimization and Computer Science, Kenitra, Morocco, 5–6 December 2018; pp. 1–5. [Google Scholar] [CrossRef]
- Khourdifi, Y.; Bahaj, M. Selecting Best Machine Learning Techniques for Breast Cancer Prediction and Diagnosis. In Information Systems and Technologies to Support Learning: Proceedings of EMENA-ISTL 2018, Fez, Morocco, 25–27 October 2018; Springer: Cham, Switzerland, 2019; pp. 565–571. [Google Scholar]
- Debelee, T.G.; Schwenker, F.; Ibenthal, A.; Yohannes, D. Survey of deep learning in breast cancer image analysis. Evol. Syst. 2020, 11, 143–163. [Google Scholar] [CrossRef]
- Rao, C.N.; Chatrapathy, K.; Fathima, A.J.; Sathish, G.; Mukherjee, S.; Reddy, P.C.S. Intelligent Deep Learning Framework for Breast Cancer Prediction using Feature Ensemble Learning. In Proceedings of the 2023 4th IEEE Global Conference for Advancement in Technology (GCAT), Bangalore, India, 6–8 October 2023; pp. 1–5. [Google Scholar]
- Munshi, R.M.; Cascone, L.; Alturki, N.; Saidani, O.; Alshardan, A.; Umer, M. A novel approach for breast cancer detection using optimized ensemble learning framework and XAI. Image Vis. Comput. 2024, 142, 104910. [Google Scholar] [CrossRef]
- Eldin, S.N.; Hamdy, J.K.; Adnan, G.T.; Hossam, M.; Elmasry, N.; Mohammed, A. Deep learning approach for breast cancer diagnosis from microscopy biopsy images. In Proceedings of the 2021 International Mobile, Intelligent, and Ubiquitous Computing Conference (MIUCC), Cairo, Egypt, 26–27 May 2021; pp. 216–222. [Google Scholar]
- Amgad, N.; Ahmed, M.; Haitham, H.; Zaher, M.; Mohammed, A. A robust ensemble deep learning approach for breast cancer diagnosis. In Proceedings of the 2023 Intelligent Methods, Systems, and Applications (IMSA), Giza, Egypt, 15–16 July 2023; pp. 452–457. [Google Scholar]
- Kumar, D.; Batra, U. Breast cancer histopathology image classification using soft voting classifier. In Proceedings of the 3rd International Conference on Computing Informatics and Networks: ICCIN 2020, Delhi, India, 29–30 July 2020; Springer: Singapore, 2021; pp. 619–631. [Google Scholar]
- Montaha, S.; Azam, S.; Rafid, A.K.M.R.H.; Ghosh, P.; Hasan, Z.; Jonkman, M.; De Boer, F. BreastNet18: A high accuracy fine-tuned VGG16 model evaluated using ablation study for diagnosing breast cancer from enhanced mammography images. Biology 2021, 10, 1347. [Google Scholar] [CrossRef] [PubMed]
- Al-Haija, Q.A.; Manasra, G.F. Development of breast cancer detection model using transfer learning of residual neural network (resnet-50). Am. J. Sci. Eng. 2020, 1, 30–39. [Google Scholar] [CrossRef]
- Ali, S.; Akhlaq, F.; Imran, A.S.; Kastrati, Z.; Daudpota, S.M.; Moosa, M. The enlightening role of explainable artificial intelligence in medical & healthcare domains: A systematic literature review. Comput. Biol. Med. 2023, 166, 107555. [Google Scholar]
- Rane, N.; Choudhary, S.; Rane, J. Explainable Artificial Intelligence (XAI) in healthcare: Interpretable Models for Clinical Decision Support. SSRN 4637897. 2023. Available online: https://ssrn.com/abstract=4637897 (accessed on 15 November 2023).
- González-Alday, R.; García-Cuesta, E.; Kulikowski, C.A.; Maojo, V. A scoping review on the progress, applicability, and future of explainable artificial intelligence in medicine. Appl. Sci. 2023, 13, 10778. [Google Scholar] [CrossRef]
- van der Velden, B.H.; Kuijf, H.J.; Gilhuijs, K.G.; Viergever, M.A. Explainable artificial intelligence (XAI) in deep learning-based medical image analysis. Med. Image Anal. 2022, 79, 102470. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, Y.; Han, E.Y.; Jacobs, N.; Han, Q.; Wang, X.; Liu, J. Classification of whole mammogram and tomosynthesis images using deep convolutional neural networks. IEEE Trans. NanoBiosci. 2018, 17, 237–242. [Google Scholar] [CrossRef]
- Prasad, C.R.; Arun, B.; Amulya, S.; Abboju, P.; Kollem, S.; Yalabaka, S. Breast cancer classification using CNN with transfer learning models. In Proceedings of the 2023 International Conference for Advancement in Technology (ICONAT), Goa, India, 24–26 January 2023; pp. 1–5. [Google Scholar]
- Mohapatra, S.; Abhishek, N.V.S.; Bardhan, D.; Ghosh, A.A.; Mohanty, S. Comparison of MobileNet and ResNet CNN Architectures in the CNN-Based Skin Cancer Classifier Model. In Machine Learning for Healthcare Applications; John Wiley & Sons: Hoboken, NJ, USA, 2021; pp. 169–186. [Google Scholar]
- INbreast Database. Available online: https://www.kaggle.com/datasets/tommyngx/inbreast2012 (accessed on 5 September 2024).
- Moreira, I.C.; Amaral, I.; Domingues, I.; Cardoso, A.; Cardoso, M.J.; Cardoso, J.S. Inbreast: Toward a full-field digital mammographic database. Acad. Radiol. 2012, 19, 236–248. [Google Scholar] [CrossRef]
- Huang, M.-L.; Lin, T.-Y. Dataset of breast mammography images with masses. Data Brief 2020, 31, 105928. [Google Scholar] [CrossRef]
- CBIS-DDSM Dataset. Available online: https://www.kaggle.com/datasets/awsaf49/cbis-ddsm-breast-cancer-image-dataset (accessed on 5 September 2024).
- Lee, R.S.; Gimenez, F.; Hoogi, A.; Miyake, K.K.; Gorovoy, M.; Rubin, D.L. A curated mammography data set for use in computer-aided detection and diagnosis research. Sci. Data 2017, 4, 170177. [Google Scholar] [CrossRef]

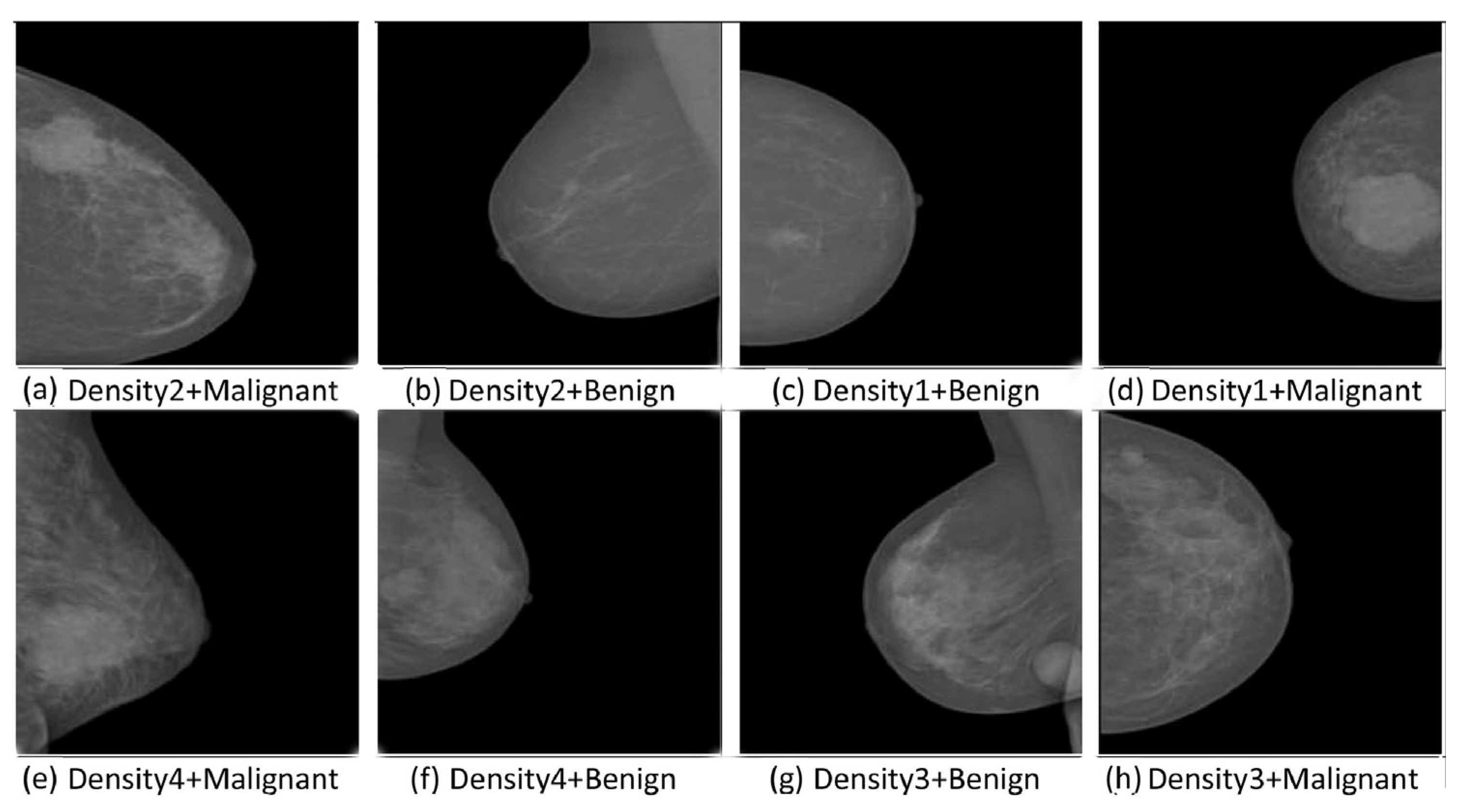







| Authors | Publisher/Year | Algorithms Used | Dataset(s) | Advantages | Limitations |
|---|---|---|---|---|---|
| Chintala et al. [36] | IEEE Access, 2023 | Optimized DRNN | Multiple datasets | High accuracy (99.16%); robust feature selection and optimization | Limited scalability to larger datasets; computational complexity |
| Munshi et al. [37] | ScienceDirect, 2023 | Custom CNN + RF + SVM (ensemble) | Wisconsin Breast Cancer Data | Achieved 99.99% accuracy; integration of XAI improves interpretability | Dataset-specific approach; generalizability untested |
| Noor Eldin et al. [38] | Springer, 2022 | DenseNet169, ResNet50, ResNet101 (ensemble) | Biopsy microscopy images | Improved accuracy to 92.5% with ensemble and augmentation | Limited dataset diversity; moderate baseline performance |
| Amgad et al. [39] | Elsevier, 2023 | Multi-stage ensemble (voting, weighted, meta-learning) | IHC images | Boosted F1-score to 89.2%; surpasses baseline CNN models by 22.2% | High computational cost; requires balanced datasets |
| Kumar and Batra [40] | Springer, 2023 | Soft voting (7 CNNs) | H&E histopathology images | Achieved 96.91% accuracy; effective combination of multiple CNNs | Focused on histopathology; lacks generalizability |
| Montaha et al. [41] | Springer, 2023 | BreastNet18 (fine-tuned VGG16) | Augmented mammography dataset | High test accuracy (98.02%); ablation studies improve robustness | Specific to mammography; overfitting risks without augmentation |
| Al-Haija et al. [42] | IEEE Access, 2023 | Transfer learning (ResNet50) | BreakHis | Exceptional accuracy (99%); simple yet effective architecture | Limited model diversity; lacks ensemble integration |
| Our Model | - | VGG16, DenseNet121, InceptionV3 (ensemble) | INbreast, CBIS-DDSM | Combines multi-scale, fine-grained, and reused features; achieved 90.1% accuracy on INbreast and 89.5% on CBIS-DDSM | Slightly higher computational cost due to ensemble complexity |
| Method | AUC | F1-Score | Sensitivity (%) | Precision (%) | Specificity (%) | Accuracy (%) |
|---|---|---|---|---|---|---|
| VGG16 | 0.93 | 87.2 | 86.7 | 88.1 | 87.8 | 87.4 |
| DenseNet121 | 0.95 | 88.5 | 89.0 | 89.5 | 88.5 | 88.7 |
| InceptionV3 | 0.94 | 87.7 | 88.0 | 88.7 | 88.3 | 88.0 |
| Ensemble | 0.96 | 89.1 | 88.3 | 90.1 | 90.2 | 89.5 |
| Method | AUC | F1-Score | Sensitivity (%) | Precision (%) | Specificity (%) | Accuracy (%) |
|---|---|---|---|---|---|---|
| VGG16 | 0.92 | 86.5 | 86.0 | 87.5 | 86.9 | 86.2 |
| DenseNet121 | 0.94 | 88.2 | 88.7 | 89.0 | 88.2 | 88.4 |
| InceptionV3 | 0.93 | 87.4 | 87.6 | 88.5 | 87.7 | 87.3 |
| Ensemble | 0.95 | 89.0 | 88.5 | 90.0 | 89.5 | 89.3 |
| Method | AUC | F1-Score | Sensitivity (%) | Precision (%) | Specificity (%) | Accuracy (%) |
|---|---|---|---|---|---|---|
| VGG16 | 0.94 | 88.2 | 87.8 | 88.5 | 88.0 | 88.3 |
| DenseNet121 | 0.96 | 89.7 | 89.0 | 90.2 | 89.5 | 89.6 |
| InceptionV3 | 0.95 | 88.5 | 88.1 | 89.0 | 88.3 | 88.4 |
| Ensemble | 0.97 | 90.5 | 90.1 | 90.8 | 90.3 | 90.4 |
| Method | AUC | F1-Score | Sensitivity (%) | Precision (%) | Specificity (%) | Accuracy (%) |
|---|---|---|---|---|---|---|
| VGG16 | 0.90 | 86.4 | 85.9 | 86.8 | 86.2 | 86.0 |
| DenseNet121 | 0.92 | 87.8 | 87.3 | 88.2 | 87.5 | 87.4 |
| InceptionV3 | 0.91 | 87.1 | 86.7 | 87.5 | 86.9 | 86.8 |
| Ensemble | 0.93 | 88.5 | 88.1 | 88.9 | 88.3 | 88.2 |
| Method | AUC | F1-Score | Sensitivity (%) | Precision (%) | Specificity (%) | Accuracy (%) |
|---|---|---|---|---|---|---|
| VGG16 | 0.92 | 85.8 | 85.4 | 86.2 | 85.6 | 85.5 |
| DenseNet121 | 0.94 | 87.4 | 87.0 | 88.0 | 87.2 | 87.1 |
| InceptionV3 | 0.93 | 86.5 | 86.2 | 87.0 | 86.4 | 86.3 |
| Ensemble | 0.95 | 88.8 | 88.4 | 89.2 | 88.6 | 88.5 |
| Method | AUC | F1-Score | Sensitivity (%) | Precision (%) | Specificity (%) | Accuracy (%) |
|---|---|---|---|---|---|---|
| VGG16 | 0.82 | 75.5 | 74.8 | 76.2 | 75.1 | 74.7 |
| DenseNet121 | 0.83 | 77.0 | 76.5 | 77.8 | 76.3 | 76.1 |
| InceptionV3 | 0.81 | 74.5 | 74.0 | 75.0 | 74.2 | 74.1 |
| Ensemble | 0.85 | 78.2 | 77.5 | 79.0 | 78.0 | 77.8 |
| Method | AUC | F1-Score | Sensitivity (%) | Precision (%) | Specificity (%) | Accuracy (%) |
|---|---|---|---|---|---|---|
| VGG16 | 0.81 | 74.0 | 73.5 | 75.0 | 74.1 | 73.8 |
| DenseNet121 | 0.82 | 76.2 | 75.5 | 77.0 | 75.7 | 75.4 |
| InceptionV3 | 0.80 | 73.7 | 73.0 | 74.5 | 73.5 | 73.2 |
| Ensemble | 0.84 | 77.1 | 76.8 | 78.0 | 77.3 | 76.9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khourdifi, Y.; El Alami, A.; Zaydi, M.; Maleh, Y.; Er-Remyly, O. Early Breast Cancer Detection Based on Deep Learning: An Ensemble Approach Applied to Mammograms. BioMedInformatics 2024, 4, 2338-2373. https://doi.org/10.3390/biomedinformatics4040127
Khourdifi Y, El Alami A, Zaydi M, Maleh Y, Er-Remyly O. Early Breast Cancer Detection Based on Deep Learning: An Ensemble Approach Applied to Mammograms. BioMedInformatics. 2024; 4(4):2338-2373. https://doi.org/10.3390/biomedinformatics4040127
Chicago/Turabian StyleKhourdifi, Youness, Alae El Alami, Mounia Zaydi, Yassine Maleh, and Omar Er-Remyly. 2024. "Early Breast Cancer Detection Based on Deep Learning: An Ensemble Approach Applied to Mammograms" BioMedInformatics 4, no. 4: 2338-2373. https://doi.org/10.3390/biomedinformatics4040127
APA StyleKhourdifi, Y., El Alami, A., Zaydi, M., Maleh, Y., & Er-Remyly, O. (2024). Early Breast Cancer Detection Based on Deep Learning: An Ensemble Approach Applied to Mammograms. BioMedInformatics, 4(4), 2338-2373. https://doi.org/10.3390/biomedinformatics4040127










