Simple Summary
Protein structure prediction using computer algorithms has long been a challenge; however, the recent introduction of algorithms like AlphaFold2 and ESMFold to predict protein structure has raised the hope for in silico drug discovery, a long sought-after breakthrough. Since the release of these algorithms, it has not been realized whether these algorithms apply if the structure is already reported to be available to the algorithms. Still, the confidence in the predicted structure varies from very low to very high, which is an observation that is unrelated to any physicochemical or biological property of the protein. Any amino acid chain sequence change fails to predict the structure, limiting the utility of these algorithms to an academic exercise. Still, researchers continue to search for the utility of the confidence scores and, despite failing, continue to suggest possible applications, resulting from the logical belief that if the confidence scores are different and reproducible, this must relate to the protein structure. To end this misconception, we predicted the structures of 204 FDA-approved therapeutic proteins, with a wishful thought that the confidence scores, if correlated on this large database, can assist in rank-ordering these proteins for their possible batch-to-batch variability, which could help to reduce testing when these molecules are developed as biosimilars. We also studied modified structures that were not predicted since no reference structure was available for the algorithms to function. This conclusion applies to the two tested algorithms, which showed comparable and proportional confidence intervals. This conclusion is controversial but deserves the attention of researchers who continue to hope to find any drug discovery utility for these algorithms.
Abstract
The three-dimensional protein structure is pivotal in comprehending biological phenomena. It directly governs protein function and hence aids in drug discovery. The development of protein prediction algorithms, such as AlphaFold2, ESMFold, and trRosetta, has given much hope in expediting protein-based therapeutic discovery. Though no study has reported a conclusive application of these algorithms, the efforts continue with much optimism. We intended to test the application of these algorithms in rank-ordering therapeutic proteins for their instability during the pre-translational modification stages, as may be predicted according to the confidence of the structure predicted by these algorithms. The selected molecules were based on a harmonized category of licensed therapeutic proteins; out of the 204 licensed products, 188 that were not conjugated were chosen for analysis, resulting in a lack of correlation between the confidence scores and structural or protein properties. It is crucial to note here that the predictive accuracy of these algorithms is contingent upon the presence of the known structure of the protein in the accessible database. Consequently, our conclusion emphasizes that these algorithms primarily replicate information derived from existing structures. While our findings caution against relying on these algorithms for drug discovery purposes, we acknowledge the need for a nuanced interpretation. Considering their limitations and recognizing that their utility may be constrained to scenarios where known structures are available is important. Hence, caution is advised when applying these algorithms to characterize various attributes of therapeutic proteins without the support of adequate structural information. It is worth noting that the two main algorithms, AlfphaFold2 and ESMFold, also showed a 72% correlation in their scores, pointing to similar limitations. While much progress has been made in computational sciences, the Levinthal paradox remains unsolved.
1. Introduction
The first protein structure prediction algorithm was reported in the late 1960s, yet protein structure prediction has remained a paradox for a long time. Levinthal’s dilemma was put forth in a seminal study by Cyrus Levinthal in 1969, titled “How to Fold Graciously”, in Science [1]. The paradox demonstrated the enormous range of potential conformations a protein might adopt, indicating that it would be unrealistic to sample all potential configurations and determine the native structure of the protein through a random positioning of the amino acids. Even though Levinthal did not suggest using a precise algorithm to predict protein structures, his study generated considerable interest and served as a springboard for further investigation.
A few years later, in 1973, Christian B. Anfinsen postulated that proteins do not follow a random configuration process and that the amino acid sequence is theoretically sufficient to determine the three-dimensional structure of a protein within certain limits [2]. This postulate posed a significant challenge to the structural bioinformaticians of predicting, with high accuracy, the structure of proteins based only on amino acid sequence data [3]. Until recently, such ab initio models were lagging behind the accuracy of template-based approaches, where an experimentally determined, homologous protein structure drives the modeling of the protein of interest [4].
The creation of the first successful protein folding algorithm, named ‘DREIDING’, developed by Richard Corey and Irving Kuntz in 1974, represents a significant turning point in the history of protein structure prediction algorithms. DREIDING predicted the folding of tiny proteins using a distance geometry technique [5].
In 2020, two parallel breakthroughs were achieved in ab initio modeling, relying on applying artificial intelligence techniques. AlphaFold demonstrated that predicting protein structures with high accuracy and at an unprecedented scale is possible and has already been achieved [6]. Shortly after the publication of the AlphaFold methodology, DeepMind and EMBL-EBI developed and launched a data resource, the AlphaFold Protein Structure Database (AlphaFold DB) [7]. Other AI-based structure prediction tools include ESMFold [8], trRosetta, Robetta, RoseTTA Fold [9], RaptorX [10], and OmegaFold [11]; the field of protein structure prediction witnessed a groundbreaking advancement.
Specific novel proteins and therapeutics have unique structures that can be predicted using computational methods like I-TASSER [12], SWISS-MODEL [13], MODELLER [14], Rosetta [15], Phyre2 [16], etc., which are template-based homology modeling, protein threading, and ab initio approaches. While prediction methods for protein structure exhibit substantial variations in their specific procedures, there are fundamental steps that remain consistent across different approaches. These steps typically involve selecting templates, reconstructing the structure, refining the predictions, and conducting a subsequent analysis.
Since AF2 depends upon Multiple Sequence Alignment (MSA), it is limited by the availability of sequential and spatial data and experimentally derived structures present in the databases, i.e., PDB. By training the network to predict the distances between pairs of residues in a protein sequence, AF2 infers the 3D spatial arrangement of the protein by incorporating a combination of convolutional layers, residual connections, and attention mechanisms within the Evoformer architecture. Further optimization is guided by a scoring function that considers the various physical and geometric properties of proteins. This refinement stage helps to improve the accuracy of the final predicted structures. However, AF2 is trained on PDB, which may not necessarily have the structures of proteins in their natural fold states (i.e., some of the PDB structures are documented in the presence of other proteins or conjugates during the solvation process). This limitation is most clearly observable for proteins with multiple native structures. Regardless, by analyzing the vast amount of known protein structures from various databases, AF2 has shown the ability to generate highly reliable structural predictions for proteins, even without close homologous structures [17,18,19].
AlphaFold can predict the structures of proteins even when there are no known structures for a particular amino acid sequence in the databases. It does not rely solely on having an exact match in the database. Still, it leverages the information from similar or related protein sequences and structures to predict the structure of the new protein. The level of accuracy in such predictions can vary depending on the uniqueness of the sequence and the availability of related protein information. AlphaFold begins by creating an MSA, aligning the target sequence with thousands of similar sequences found in large sequence databases like UniProt. This process helps even when there is no exact match or known structure for the target sequence. The algorithm identifies patterns of amino acid co-evolution in the MSA. These patterns reveal which amino acid residues will likely be near the 3D structure. AlphaFold uses these co-evolution patterns to predict contacts between amino acid residues, which are critical constraints for predicting the 3D structure.
Like AF2, ESMFold employs transformer models to encode protein sequences 60 times faster than AF2, eliminating the MSA while maintaining high-quality predictions and using as many as 15 billion parameters. Its most significant advantage is its ability to predict structures many times faster than any other tools that are available, making it excellent for identifying remote homology and conservation in an extensive collection of novel sequences. ESMFold generates structure predictions using only one sequence as an input by leveraging the internal representations of the language model. By examining the co-evolutionary patterns among the amino acid residues in a protein family to capture valuable information about residue interactions and structural constraints, ESMF makes accurate predictions. The strength of ESMF lies in its ability to integrate diverse sources of information and incorporate predicted secondary structure information and contact maps. These additional inputs provide valuable insights into local structural elements and the spatial proximity of amino acids, further refining the accuracy of the predicted protein structures. ESMFold produces a more accurate atomic-level prediction than AlphaFold2 or RoseTTAFold. However, similar to AlphaFold2, it is limited by the training data that require significant computational resources to run, which can limit its accessibility.
AF2 and ESMF, along with 3D structures, also generate model confidence prediction scores as predicted local distance difference test (pLDDT) scores and predicted template modeling (pTM) scores. The predicted local distance difference test (pLDDT) measures confidence or reliability assigned to each residue in the predicted protein structure. It represents the predicted accuracy of the local distance difference, the difference between the predicted and actual distances in the experimentally determined protein structure. The pLDDT score ranges from 0 to 100, with higher scores indicating higher confidence in the predicted local structure. Regions with high pLDDT scores (e.g., >80) are considered to have accurate predictions, while lower scores (e.g., <50) indicate regions where the predictions may be less reliable. Lower pLDDT scores may also indicate that the fold is in intrinsically disordered protein regions (IDPRs). On the other hand, the predicted template modeling, or TM scores (pTM), is the global metric of structure assessment and evaluates the overall quality of the predicted protein structure by comparing it to experimentally determined structures of similar proteins available in the Protein Data Bank (PDB). The pTM score assesses how well the predicted structure aligns with the known structure of a related protein template. It ranges from 0 to 1, and a higher pTM score signifies a better alignment and a higher likelihood of the predicted structure being accurate [20,21,22].
2. Finding Applications
A quest among researchers continued with the hope of finding applications of these tools on multiple fronts, such as in predicting the structural context of mutations associated with a disease or an escape from an immune response. The PubMed database reported over 825 peer-reviewed articles published on AlphaFold [23] since the 2021 publication detailing AlphaFold, and only 10 mention ESMFold [24]. Multiple studies have shown conclusive analyses, with some questioning the utility of these algorithms. The most robust and reliable protein structures are based on experimentally derived data [25]. These structures are archived and made publicly available through the Protein Data Bank (PDB), the global resource of experimentally determined protein and nucleic acid models [26]. However, due to the difficulties in solving the structures of proteins using experimental techniques, the gap between known protein sequences and experimentally determined structures continues to grow [27].
With the development of AlphaFold, various applications could be suggested and are currently underway. AlphaFold has been applied to decipher complex structures and the mechanisms of binding of multiple proteins in vast domains. Recently, it aided in unveiling the structure of the human Nuclear Pore Complex (NPC) [28] model using nucleoporin structure models. It was also employed to solve complicated structures of the ATP-dsDNA-SMC5/6 protein based on the combination of cryo-EM density maps and AlphaFold-generated models [29]. Furthermore, it has been used to identify a new distinct fold in rotavirus group B, revealing its functionality and the predicted stress-inducible phosphoprotein 1 (STIP1) structure. This study further revealed its role as a neuroprotective factor against Parkinson’s disease [30,31,32]. It has also helped predict the structure of a whole host adhesion device from the Lactobacillus casei bacteriophage J-1. As the human gut phagosome exemplifies, these AF2-based structure predictions can be further used to revisit phage genome annotations and efficiently characterize newly discovered phages [33]. Besides this, AF2 has been used to identify the descriptors of variant pathogenicity, aids in discriminating disorder regions, helps in characterizing local dynamics, differentiates strong and weak binders, and aids in inferring the binding transitions of apo-holo pairs of proteins and ligands [34,35,36,37,38]. In contrast, the AF2 model remains relatively stable to point mutations, and the scores do not vary. Since the potential of AF2 predictions in the designability of new therapeutics and structural stability testing through mutagenesis analysis failed, a direct way to use AF2 for predicting ΔΔG upon mutation in the sequence has not yet been identified [39,40,41].
The plethora of these studies presents the need to determine the interpretability of the prediction scores and their applicability to real-world problems. Even though the creators of these algorithms have said that no importance should be given to the predictability scores, various studies, as discussed previously, remain proof of the scores’ appropriation and use as the descriptors of variant pathogenicity and the discriminants of intrinsically disordered regions, for the characterization of the local dynamics of proteins, to identify strong vs. weak binders, to infer ligand binding transitions in apo-holo pairs, to analyze the effect of specific mutations on proteins’ structures, and much more. A vast space requires searching to determine the further applications of these predicted structures and scores.
3. Testing a New Application
Given the consistent reproducibility of prediction scores (pLDDT and pTM) in AlphaFold 2 (AF2) and ESMF, an inference could be made that lower confidence scores may suggest a higher degree of structural variability. It is important to note that this assumption is made within the context of the inherent limitations of the algorithms. While the correlation between a lower confidence score and increased structural variability is a logical observation based on our analysis, it should be considered that this inference is contingent upon the specific constraints and capabilities of the algorithms under consideration. One application of this argument can be made in rank-ordering proteins for their structural instability, which might show up as structural variability when a recombinant protein is expressed in a culture medium. It can be suggested that a protein with a low pLDDT score (e.g., pLDDT < 50) is more likely to have structural variability. Thus, when developing biosimilars, this rank order of susceptibility to variation can be utilized to create testing protocols for biosimilars.
It is also interesting to further explore the factors determining confidence scores. Still, the AF2 developers have suggested that such correlations are not possible despite the difference in the physicochemical properties of proteins. These two considerations prompted the study of the confidence scores of the FDA-approved therapeutic proteins to explore if any conclusions can be drawn from these scores that might help compare a biosimilar product with its reference product.
4. Materials and Methods
4.1. Data Collection
The investigation into the hypothesis involved retrieving data on FDA-approved therapeutic proteins with established safety and efficacy. After the removal of redundancies, a total of 223 molecules were initially gathered from the THPdb database [42] along with the FDA’s Purple and Orange Books [43,44]. At the outset, any molecule categorized as a ‘Therapeutic Protein’ was documented. Subsequently, 11 molecules characterized by conjugation, modification, pegylation, or combination protein features were omitted from the analysis, resulting in a final set of 204 molecules for hypothesis testing. Given the focus on predicting protein structures using an AI-based tool, excluding post-translationally or artificially modified molecules was deemed logical to facilitate a more accurate comparison.
The therapeutic protein’s amino acid sequences were obtained from the FDA’s Purple Book, Orange Book, patents, and regulatory filings [45] and the Inxight Drug [46], Kegg Pathway [47], and DrugBank [48] databases. The sequences in the UniProt database were found to have residual differences compared to the amino acid sequences in patents; hence, the resources mentioned above were used and cross-checked through these references for similarity. A total of 8 molecules falling outside the cut-off range of 5 to 1000 amino acids were removed, leaving 204 molecules behind. This list of 204 products included 188 protein molecules and 16 molecules with amino acid sequence lengths less than 40, classified as polypeptides and not treated as biological drugs by the FDA [49]. The final dataset contained two classes: ‘peptides’ for 16 polypeptides and ‘proteins’ for 188 products (Supplementary File).
Each category’s file contains information on the therapeutics name (both generic and brand name), accession number from Inxight, KeggDrug, or DrugBank databases, and Biologics License Application (BLA) number or New Drug Application (NDA) number acquired from FDA approval documentation. Furthermore, amino acid sequence, sequence length along with molecular weight computed by the Cusabio tool [50], and the type of therapeutic molecule (i.e., enzyme, monoclonal antibody, blood factor, cytokine, growth factor, hormone, inhibitors, fusion protein, recombinant human protein, etc.) are part of the data.
4.2. Structure Prediction and Scores
The amino acid sequences were subjected to 3D structure prediction using two different tools: ColabFold using the UCSF ChimeraX (version 1.5) [51] software for AF2 and an independent Google Colaboratory notebook for ESMF [52,53]. The confidence scores obtained from the predictions are referred to as ‘AlphaFold pLDDT Score’, ‘AlphaFold pTM Score’, ‘ESMFold pLDDT Score’, and ‘ESMFold pTM Score’ for all of the molecules analyzed. These scores provided insights into the predicted accuracy and reliability of the 3D structures generated by each respective prediction tool and were observed to be highly correlating.
4.3. Physiochemical Properties
The physicochemical parameters, including hydrophobicity, isoelectric point, extinction coefficients, and instability index, for all 204 molecules were computed through a Python script employing the Expasy ProParam package (Supplementary File) [54]. The hydrophobicity was calculated using the GRAVY (grand average of hydropathy) index to measure the aggregation of hydropathy of amino acid residues. The isoelectric point (pI) was used to account for the pH of the protein at a net neutral charge. In addition, the theoretical molecular extinction coefficients for both reduced and non-reduced cysteine residue structures were calculated to determine the protein concentration by measuring its absorbance at ~280 nm wavelength [55]. Finally, the proteins’ instability index values were calculated based on the compositions of their amino acids, with higher values indicating greater instability and more propensity for protein degradation. These attributes were analogized with the pLDDT/pTM scores to deduce dependence on amino acid sequence, if any.
4.4. Protein Interactions
LZerD [56], a web server for multiple protein–protein docking, was used to acquire the interactions of cytokines, hormones, and fusion proteins with their targets, identified through the DrugBank database. The chosen complexes included one therapeutic protein with a high pLDDT score and one with a low pLDDT score, as ranked by AF2 and ESMF, respectively. All target molecules were retrieved from the PDB database. PDB structures often have non-standard chain names and residue numbering that can cause compatibility issues with the docking tools. For standardization, chains were renamed, and residues were renumbered using UCSF Chimera (version 1.17) [57] software. Docked complexes with the highest rank-sum from GOAP [58], DFIRE [59], and ITScore scores [60] from the LZerD server were given to the PRODIGY [61] server, and their Gibbs free energy/binding affinity (ΔG), dissociation constant (Kd), Interfacial Contacts (ICs) and Non-Interacting Surfaces (NIS) values were computed. Combining these methods provided a comprehensive assessment of protein–protein interactions and improved the accuracy of the docking predictions acquired from the LZerD server. As discussed later, these physiological, chemical, and functional parameters were employed to analyze their relationships with prediction scores.
5. Results
5.1. Orthogonal Comparison—AF2 vs. ESMF
Evidence from the literature shows the Pearson correlation of the pLDDT scores between the AF2 and ESMF on a random subset of around 4000 metagenomic sequences to be ~0.79 [62]. The Pearson correlation for the pLDDT scores from our data of 204 molecules was found to be ~0.72, whereas for the pTM scores, it was ~0.88.
For a comparison among the peptide and protein classes, we used two cut-offs: AA < 40 (amino acid count less than 40) and 40 < AA < 1000 (amino acid number between 40 and 1000), respectively. The first was used to understand the predictability of polypeptides below 40 amino acids from AF2 and ESMF, which resulted in a correlation of ~0.83 using the pLDDT score and ~0.95 using the pTM score. The second cut-off was of all of the proteins above 40 amino acids and below 1000, which resulted in a correlation of ~0.69 using the pLDDT score and ~0.84 using the pTM score. The Pearson correlation (corr.) and correlation coefficient (R2) for the pTM scores from both algorithms agreed better than the pLDDT scores. Recently, an AF2-based mode, AFDistill, estimated the structural consistency measured by the pTM or pLDDT scores for a given protein sequence. Interestingly, the experimental results demonstrated that the pTM-based structural consistency scores positively impacted the model’s performance more than the pLDDT-based scores. This indicates that pTM scores might be more reliable than pLDDT scores for evaluating protein structure [63].
5.2. Complexity of Structures and Prediction Scores
It was anticipated that the pLDDT and pTM scores would decrease with an increase in the complexity of a structure. An increase in sequence length can increase the complexity of a structure; keeping this in view, no significant correlation (significance threshold: R2 > 0.5) between the complexity of the protein structure and the pLDDT or pTM scores was observed (Figure 1 and Figure 2). The peptides showed a weak positive correlation (R2~0.40) with pLDDT from AF2 only, whereas a significant correlation from the AF2 and ESMF was seen for the pTM scores (R2~0.61 and R2~0.63). In conclusion, the pTM scores were shown to better agree with the pLDDT from AF2 and ESMF.
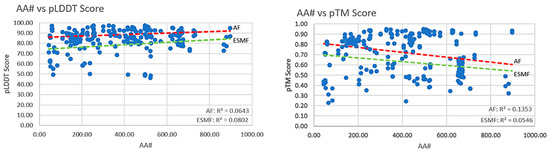
Figure 1.
Comparison of the increasing complexity of therapeutic protein with the pLDDT and pTM scores from AF2 and ESMF.
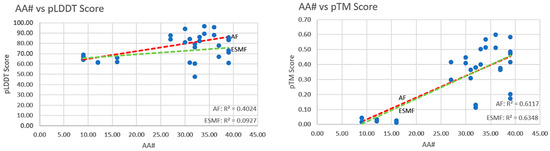
Figure 2.
Comparison of the complexity of therapeutic peptides with the pLDDT and pTM scores from AF2 and ESMF.
5.3. Physiochemical Attributes and 3D Structure
The properties of the constituent amino acids determine the physicochemical properties of proteins [64]. Since the relationship between the protein sequence and structure arises entirely from the amino acids’ physical properties, their activities and properties result from interactions among their constitutive amino acids [65]. Understanding the relationship between the position-specific properties of amino acid sequences and how these physiochemical properties influence the structure formation is vital. The amino acid sequence forms the secondary fold of a protein that plays a critical role in determining its 3D structure, which, in turn, governs its therapeutic potential, especially in the case of therapeutic proteins and biosimilars [66,67]. Henceforth, there is a strong interdependence of the physicochemical properties of proteins on amino acid sequences, secondary structures, and 3D structures. Understanding this relationship is essential for predicting protein stability and its implicit dependence on the amino acid sequence.
The prediction scores and physiochemical properties were compared to gain insights into therapeutic proteins’ physiological and functional properties to enable rank-ordering proteins for the risk of structural variability that might be used to establish biosimilarities [68]. There was a weak correlation between the proteins and the peptide, but no correlation was found in the hydrophobicity, isoelectric point, EC, and I-index for the proteins and peptides.
5.4. Protein Interactions—Effects of Structural Folds
The atomic pLDDT by AF2 and ESMF measures the atomic-level prediction accuracy based on the degree of agreement between the predicted model and the experimental structure. In principle, certain portions of a protein hold therapeutic potential with residues responsible for binding.
Parathyroid (PTH) structures predicted from AF2 and ESMF, when docked to the PTHR1 receptor (PDB: Q03431) through the LZerD server and evaluated through the PRODIGY server, produced ΔG (binding energy) values of −11.1 and −10.3, respectively. Despite the ability of AF2 to misfold structures, as observed in other studies [69,70], the predicted structure has a higher prediction score (pLDDT: ~71.00, pTM: ~0.37) as well as binding energy (ΔG: −11.1). Compared to the PTH predicted from AF2, the ESMF-predicted structure has lower values (pLDDT: ~58.50, pTM: ~0.25, ΔG: −10.3). The evidence from the literature [71] and residue–residue pair file (.ic) produced by the PRODIGY server indicated that residues 1 to 37 of PTH contributed to the binding with PTHR1. Few of the residues involved in the binding—Ser1, Ser3, Glu4, Ile5, Leu7, Met8, Leu11, His14, Leu15, Ser17, Met18, Glu19, Arg20, and Phe34—of the PTH structure predicted from ESMF had low pLDDT values, but when predicted from AF2, they had high pLDDT values. It can be inferred that the lower confidence residues lead to lost interactions, lowering the binding affinity for ESMF-PTH (Table 1).

Table 1.
Interacting PTH-PTHR1 residues of pLDDT from AF2 and ESMF; although they have different pLDDT scores, they produced similar binding interactions and scores.
In some cases, proteins, by nature, can retain their functional properties regardless of the conformational variation, given that the domains were predicted confidently and the remaining structure does not hinder binding. To explore any correlation among cytokines, hormones, and fusion proteins, the binding affinity values were calculated from the PRODIGY server. No statistically significant difference was recorded across the class protein structure comparisons.
The structural differences in proteins can influence the binding affinity. The ICs, NIS of proteins, and residue pairs with charged and aromatic side chains are essential for binding. These residues influence the formation of cationic, electrostatic, and aromatic interactions between the protein and target molecule, helping to explain the drastic variance in the binding affinity [72,73]. The ESMF-predicted structures had more robust interactions and higher binding affinity with their targets regardless of a lower pLDDT (68.90) value.
These contrasting results led us to conclude that the prediction scores do not relate to the binding value. Thus, these algorithms cannot be correlated with proteins’ chemical and functional attributes.
6. Discussion
The bioavailability, pharmacokinetics, and pharmacodynamics of a therapeutic drug are greatly influenced by its structural elements, as well as the concentration and dosage of the drug. The extinction coefficient is often used in protein purification, quantification, and structural studies where accurate protein concentration determination is required for therapeutics [74,75]. Proteins must be folded into their native stable states to perform their function, which typically involves binding to their respective targets. They have the inherent ability of stable fold formation and strong binding interactions, acquired through adaptation and conservation, even when these changes do not directly increase the organism’s fitness [76]. The distribution of polar and apolar residues on the surface mediates protein–target interactions, influencing their specificity and affinity.
Multiple studies have concluded that the charged residues interact with targets through the exposed surfaces rather than the interface to affect the binding ability of the interacting proteins. Enhanced intra-molecular electrostatic interactions lower the desolvation penalty. In contrast, the inter-molecular interactions with charged residues on the target molecule enable better complementarity and electrostatic steering, resulting in increased solubility and bioavailability of these proteins in living systems [77]. A study evaluated the effects of five processing strategies of coordinates generated by AF2 (i.e., spatial filtering, the singular value decomposition of a distance map, secondary structure feature, and relatively accessible surface area (rASA)) on the proteins. This study concluded that all of the strategies predicted novel features that could aid in some deep learning-based prediction of the binding sites of proteins through primary sequences only [78].
Physicochemical parameters like hydrophobicity and the isoelectric point also play crucial roles in these interactions, contributing to the stability of the formation of 3D folds. The computed list of physicochemical parameters was analyzed to gain insights into therapeutic proteins’ physiological and functional properties; however, no significant correlation was found, which could be used as a metric. Proteins with a higher abundance of residues with a lower half-life tend to have a relatively higher instability index. Therefore, they may have a shorter lifespan in vivo, and may be more prone to degradation. However, even when the pLDDT scores are high, few proteins have higher instability index values (more susceptible to degradation), i.e., choriogonadotropin alfa has an AF2 pLDDT score of ~83.40, and the chances of its degradation in vivo are high (instability index 67.46).
Similarly, Sargramostim has a confidence-predicted structure from AF2 with a pLDDT score of 90.10. Still, the instability index is 63.87, indicating that a reliable structure prediction cannot vouch for the structure’s stability in vivo. This eliminates the possibility of correlating pLDDT scores with instability indexes; hence, a predicted structure cannot vouch for the stability of a protein in in vivo systems.
AF2 has been used for protein–protein docking in various studies as a structural template generator alongside physics-based docking algorithms to predict protein–protein interactions [79,80]. It has also been used to improve peptide–protein docking by predicting which peptides and proteins interact and by modeling the resulting interaction in combination with Rosetta. However, studies have also concluded that the accuracy of AF2 in reproducing protein topology and binding site anatomy is insufficient to ensure that its models can be reliably used for molecular docking purposes. Consequently, post-modeling refinement techniques have been suggested to be necessary to improve the accuracy of AF2 models for docking [81,82].
Our study aimed to find interlinking attributes among the AF2 predictions and binding of proteins through the scores generated for cytokines, hormones, and fusion proteins to their respective targets. Supposedly, it can be said that if the domains are strongly predicted, and the rest of the structure does not produce hindrance, strong binding can be obtained. Therefore, it is, in fact, possible for a protein residue to have low atomic pLDDT scores and contribute towards a strong binding affinity with its target, and vice versa. However, the lack of correlation of prediction scores with the binding affinity value leads to inconclusive results.
It was also observed that nearly all of the multidomain molecules (mAbs, fusion proteins, etc.) had higher prediction scores, directing that both AF2 and ESMF perform well on multi-domain proteins, making single- and multiple-domain molecules equally likely to have lower scores. Multi-domain molecules with longer sequence lengths tend to have larger radii of gyration, resulting in increased complexity [83,84]. Generally, a larger radius of gyration indicates a more extended or less compact structure; therefore, it might add up to the challenge of structure prediction for prediction tools to model a structure accurately, resulting in lower scores. However, our results negated this hypothesis.
Since it was demonstrated through previous studies and a comparative analysis in this study that the pTM-based structural scores were better metrics of comparison than the pLDDT-based scores, 13 proteins with lower pTM scores were selected (Table 2). Among these, the proteins with larger radii of gyration and multiple domains resulted in relatively better prediction scores (i.e., Aflibercept and Tositumomab), while the proteins with smaller radii of gyration and single domains had relatively lower pTM scores (i.e., Lepirudin, Parathyroid, and Lixisenatide). The larger molecules might have features that make them easier to model accurately, such as distinctive folds, recognizable structural motifs, or simply better MSA, resulting in relatively better scores.

Table 2.
AF2 and ESMF prediction score comparison for mutated single and multiple domains.
Furthermore, the prediction power of these AI-based tools also plays a vital role in determining the quality of the predicted structure. Extending the analysis, monomer proteins with lower pTM scores were predicted through Yang Servers trRosetta [85,86], and momentous improvements in the pTM scores were seen, hence backing up the inference that the accuracy of predicted protein structures increases with the prediction power of an AI-based tool and the algorithm and data used during its training. The drastic increase in pTM scores with prediction power and better training data indicates that it might be possible to predict complex protein structures with accuracy closer to the experimentally driven structures.
With the observed improvements in the pTM scores from trRosetta, the dependence of AI tools on data availability was significantly evident. Furthermore, to test the prediction models’ extent of dependence on the training data, the domains identified from the NCBI-CDD database of a few sequences were ‘shuffled’ using Molbiotool’s Random Sequence Generator [87] to ensure the highest mutation rate. First, a single domain was randomized through shuffling, followed by shuffling/randomizing all of the domains of Trastuzumab, Etanercept, Coagulation Factor VIIa, and Darbepoetin alfa to produce novel molecules. These mutated sequences, which resulted in novel molecules, were run through BLAST PDB, and their query coverage and percentage identity scores were retrieved. These data were generated and collected to identify similarities between the randomized sequence combinations and folds in the UniProt and PDB databases. If the data were available, the AF2 and ESMF models must have learned these folds and sequence patterns during the training phase. However, if the coverage and identity scores were extremely low or zero, the AF2 and ESMF models would rely solely on their trained models to predict the structure. Since ESMF does not use MSA, it can be anticipated that this model would perform better than the MSA-dependent model, AF2.
7. Conclusions
The newer structure prediction methods include improving pairwise and higher-order residue distance constraints from multiple sequence alignments and understanding how this information is eventually encoded into a predicted 3D structure. These developments have been reviewed recently, showing how the increasing use of neural network models forms the backbone of predicting protein structures from their primary sequence. This is supported by the rise of protein sequence and structure databases, critical resources for input, and training sophisticated prediction methods [88,89].
The structural complexity of proteins depends on the number of amino acids, resulting in less confidence in the structure prediction, as predicted by Levinthal. This work shows that for the category of therapeutic proteins above 40 amino acids, there is a weak or no correlation between the number of amino acids and their pLDDT or pTM scores. In the case of polypeptides, a reverse observation showed that a smaller number provides more complexity in prediction and less confidence in structure predictability, as applied to polypeptides [90,91]. Both tools correlate significantly positively, representing their orthogonality. The finding of this paper suggests that the predictions based on sequence alone cannot be used to describe the folding of a structure and its accuracy. The pLDDT and pTM scores do not correlate with any structural or functional parameters, and hence, they cannot be used to determine protein stability in in vivo systems or propose concentration and dosing for better efficacy, nor can they be correlated with the binding properties of proteins even though they are all dependent upon the amino acid sequence. It can be concluded that the surface elements responsible for the physicochemical properties and binding are not necessarily involved in the folding process to a degree that correlates with structure prediction; therefore, they do not affect the pharmacology or toxicology of the protein [92,93,94].
Even though AF2 slightly tends to misfold structures, it performs reliable predictions on multidomain molecules. However, these predictions can be significantly improved with better prediction power tools, concluding that it might soon be possible to predict complex protein structures with an accuracy closer to the experimental structures with more robust prediction tools. Few FDA- and EMA-approved biosimilars demonstrated pLDDT scores greater than 80 using the AF2 predictions; thus, it can be concluded that there is less variability in the 3D structure, and these molecules may not require extensive testing to establish molecular biosimilarity. Extending this argument, 188 proteins (excluding peptides) were rank-ordered in the context of structural variability using the AF2 pLDDT and pTM scores. Biosimilars with high pLDDT and pTM scores were concluded to have the highest stability and are less prone to variations in the 3D structure. This assumption was proven wrong.
In conclusion, while the current study primarily focused on prediction scores, recognizing the multifaceted nature of protein structure prediction, we acknowledge the importance of incorporating additional defining factors for a more thorough evaluation of prediction algorithms. Beyond prediction scores, factors such as accuracy in secondary structure prediction, robustness to sequence variability, speed and efficiency, consistency across protein families, handling large protein complexes, capability in predicting binding sites, and sensitivity to input data quality are essential dimensions that could further enrich our understanding. It is evident that the structural complexity of proteins, as highlighted by Levinthal’s paradox, is not solely dictated by the number of amino acids. Our findings underscore that the pLDDT and pTM scores, while essential metrics, do not directly correlate with structural or functional parameters. Notably, this study highlights the orthogonality of the two tools and suggests that predictions based on sequence alone may not fully describe the folding process. Although AlphaFold 2 exhibits reliability in predicting multidomain molecules, improvements with more robust prediction tools are anticipated for accurate predictions closer to experimental structures. The evaluation of biosimilars further emphasizes the need for a nuanced understanding of structural variability.
Additionally, the dependency of the AlphaFold 2 and ESMF models on data from databases like PDB and UniProt underscores the crucial role of training data availability. Future investigations should identify influential amino acids and address challenges in loop modeling, ensuring a comprehensive and informed approach to advancing protein structure prediction methodologies. While ongoing advancements may enhance predictability, maintaining high confidence in structure predictability is essential for establishing biosimilarity and guiding drug development decisions.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biomedinformatics4010007/s1.
Author Contributions
S.K.N.: conceptualization, study design, analysis; Z.M.: data analysis, writing and supplementary data collection; R.Z.P.: supervision, coordination, review. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not Applicable.
Informed Consent Statement
Not Applicable.
Data Availability Statement
All analytical data are available on GitHub: https://github.com/zamaram12/Molecules_data/blob/1df3e12e955c49c3531312ecc79a084cfe0ba208/proteins_data.csv, (accessed on 30 November 2023).
Acknowledgments
The authors are thankful for the comments provided by John Jumper, the developer of the AF2 algorithm.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Cyrus, L. How to Fold Graciously. In Mossbauer Spectroscopy in Biological Systems: Proceedings of a Meeting Held at Allerton House; University of Illinois Bulletin: Monticello, IL, USA, 1969; pp. 22–24. [Google Scholar]
- Hirata, F.; Sugita, M.; Yoshida, M.; Akasaka, K. Perspective: Structural fluctuation of protein and Anfinsen’s thermodynamic hypothesis. J. Chem. Phys. 2018, 148, 020901. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Pearce, R.; Li, Y.; Omenn, G.S.; Zhang, Y. Fast and accurate Ab Initio Protein structure prediction using deep learning potentials. PLoS Comput. Biol. 2022, 18, e1010539. [Google Scholar] [CrossRef]
- Corey, R.B.; Kuntz, I.D. ENCEPP: A program for predicting the conformational geometry of organic molecules. J. Comput. Chem. 1974, 2, 287–303. [Google Scholar]
- Pereira, J.; Simpkin, A.J.; Hartmann, M.D.; Rigden, D.J.; Keegan, R.M.; Lupas, A.N. High-accuracy protein structure prediction in CASP14. Proteins 2021, 89, 1687–1699. [Google Scholar] [CrossRef] [PubMed]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef] [PubMed]
- Rives, A.; Meier, J.; Sercu, T.; Goyal, S.; Lin, Z.; Liu, J.; Zitnick, C.L. Meta’s Genomics AI ESMFold Predicts Protein Structure 6x Faster Than AlphaFold2. InfoQ. 2022. Available online: https://www.infoq.com/news/2022/08/meta-genomic-ai-esmfold/ (accessed on 11 May 2023).
- Baek, M.; DiMaio, F.; Anishchenko, I.; Dauparas, J.; Ovchinnikov, S.G.; Lee, G.R.; Wang, J.; Cong, Q.; Kinch, L.N.; Schaeffer, R.D.; et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science 2021, 373, 871–876. [Google Scholar] [CrossRef]
- Peng, J.; Xu, J. Raptorx: Exploiting structure information for protein alignment by statistical inference. Proteins Struct. Funct. Bioinform. 2011, 79 (Suppl. 10), 161–171. [Google Scholar] [CrossRef]
- Wu, R.; Ding, F.; Wang, R.; Shen, R.; Zhang, X.; Luo, S.; Su, C.; Wu, Z.; Xie, Q.; Berger, B.; et al. High-resolution de novos tructure prediction from primary sequence. bioRxiv, 2022; preprint. [Google Scholar] [CrossRef]
- Zhang, Y. I-TASSER: Fully automated protein structure prediction in CASP8. Proteins Struct. Funct. Bioinform. 2008, 77 (Suppl. 9), 100–113. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Schwede, T. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Webb, B.; Sali, A. Comparative Protein Structure Modeling Using MODELLER. Curr. Protoc. Bioinform. 2016, 54, 5.6.1–5.6.37. [Google Scholar] [CrossRef] [PubMed]
- Leaver-Fay, A.; Tyka, M.; Lewis, S.M.; Lange, O.F.; Thompson, J.; Jacak RBradley, P. ROSETTA3: An object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 2011, 487, 545–574. [Google Scholar] [PubMed]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [PubMed]
- Montanucci, L.; Capriotti, E.; Frank, Y.; Ben-Tal, N.; Fariselli, P. DDGun: An untrained method for the prediction of protein stability changes upon single and multiple point variations. BMC Bioinform. 2019, 20, S14. [Google Scholar] [CrossRef] [PubMed]
- Savojardo, C.; Fariselli, P.; Martelli, P.L.; Casadio, R. INPS-MD: A web server to predict stability of protein variants from sequence and structure. Bioinformatics 2016, 32, 2542–2544. [Google Scholar] [CrossRef]
- Lv, X.; Chen, J.; Lu, Y.; Chen, Z.; Xiao, N.; Yang, Y. Accurately Predicting Mutation-Caused Stability Changes from Protein Sequences Using Extreme Gradient Boosting. J. Chem. Inf. Model. 2020, 60, 2388–2395. [Google Scholar] [CrossRef]
- Yin, J.; Lei, J.; Yu, J.; Cui, W.; Satz, A.L.; Zhou, Y.; Feng, H.; Deng, J.; Su, W.; Kuai, L. Assessment of AI-Based Protein Structure Prediction for the NLRP3 Target. Molecules 2022, 27, 5797. [Google Scholar] [CrossRef]
- Gao, M.; An, D.N.; Parks, J.M.; Skolnick, J. AF2Complex predicts direct physical interactions in multimeric proteins with deep learning. Nat. Commun. 2022, 13, 1744. [Google Scholar] [CrossRef]
- Yin, R.; Feng, B.Y.; Varshney, A.; Pierce, B.G. Benchmarking AlphaFold for Protein Complex Modeling Reveals Accuracy Determinants. Protein Sci. 2022, 31, e4379. [Google Scholar] [CrossRef]
- Available online: https://pubmed.ncbi.nlm.nih.gov/?term=alphafold (accessed on 11 May 2023).
- Available online: https://pubmed.ncbi.nlm.nih.gov/?term=ESMFold (accessed on 11 May 2023).
- Velankar, S.; Burley, S.K.; Kurisu, G.; Hoch, J.C.; Markley, J.L. The protein data bank archive. Methods Mol. Biol. 2021, 2305, 3–21. [Google Scholar]
- Burley, S.K.; Berman, H.M.; Kleywegt, G.J.; Markley, J.L.; Nakamura, H.; Velankar, S. Protein Data Bank (PDB): The single global macromolecular structure archive. Methods Mol. Biol. 2017, 1607, 627–641. [Google Scholar] [PubMed]
- Dana, J.M.; Gutmanas, A.; Tyagi, N.; Qi, G.; O’donovan, C.; Martin, M.; Velankar, S. SIFTS: Updated structure integration with function, taxonomy and sequences resource allows 40-fold increase in coverage of structure-based annotations for proteins. Nucleic Acids Res. 2019, 47, D482–D489. [Google Scholar] [CrossRef] [PubMed]
- Mosalaganti, S.; Obarska-Kosinska, A.; Siggel, M.; Taniguchi, R.; Turoňová, B.; Zimmerli, C.E.; Buczak, K.; Schmidt, F.H.; Margiotta, E.; Mackmull, M.-T.; et al. AI-based structure prediction empowers integrative structural analysis of human nuclear pores. Science 2022, 376, eabm9506. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Li, S.; Ser, Z.; Kuang, H.; Than, T.; Guan, D.; Zhao, X.; Patel, D.J. Cryo-EM structure of DNA-bound Smc5/6 reveals DNA clamping enabled by multi-subunit conformational changes. Proc. Natl. Acad. Sci. USA 2022, 119, e2202799119. [Google Scholar] [CrossRef]
- Hötzel, I. Deep-Time Structural Evolution of Retroviral and Filoviral Surface Envelope Proteins. J. Virol. 2022, 96, e0006322. [Google Scholar] [CrossRef]
- Huang, P.S.; Boyken, S.E.; Baker, D. The coming of age of de novo protein design. Nature 2016, 537, 320–327. [Google Scholar] [CrossRef]
- Caldararu, O.; Blundell, T.L.; Kepp, K.P. A base measure of precision for protein stability predictors: Structural sensitivity. BMC Bioinform. 2021, 22, 88. [Google Scholar] [CrossRef]
- Goulet, A.; Cambillau, C. Present Impact of AlphaFold2 Revolution on Structural Biology, and an Illustration with the Structure Prediction of the Bacteriophage J-1 Host Adhesion Device. Front. Mol. Biosci. 2022, 9, 907452. [Google Scholar] [CrossRef]
- Anbo, H.; Sakuma, K.; Fukuchi, S.; Ota, M. How AlphaFold2 Predicts Conditionally Folding Regions Annotated in an Intrinsically Disordered Protein Database, IDEAL. Biology 2023, 12, 182. [Google Scholar] [CrossRef]
- Saldaño, T.; Escobedo, N.; Marchetti, J.; Zea, D.J.; Mac Donagh, J.; Velez Rueda, A.J.; Gonik, E.; García Melani, A.; Novomisky Nechcoff, J.; Salas, M.N.; et al. Impact of protein conformational diversity on AlphaFold predictions. Bioinformatics 2022, 38, 2742–2748. [Google Scholar] [CrossRef]
- Roney, J.P.; Ovchinnikov, S. State-of-the-Art Estimation of Protein Model Accuracy Using AlphaFold. Phys. Rev. Lett. 2022, 129, 238101. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Perez, A. AlphaFold encodes the principles to identify high affinity peptide binders. bioRxiv, 2022; preprint. [Google Scholar] [CrossRef]
- Chakravarty, D.; Porter, L.L. AlphaFold2 fails to predict protein fold switching. Protein Sci. 2022, 31, e4353. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://alphafold.ebi.ac.uk/faq (accessed on 11 May 2023).
- Pak, M.A.; Markhieva, K.A.; Novikova, M.S.; Petrov, D.S.; Vorobyev, I.S.; Maksimova, E.S.; Kondrashov, F.A.; Ivankov, D.N. Using AlphaFold to predict the impact of single mutations on protein stability and function. PLoS ONE 2023, 18, e0282689. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://torchmetrics.readthedocs.io/en/stable/classification/auroc.html (accessed on 11 May 2023).
- Usmani, S.S.; Bedi, G.; Samuel, J.S.; Singh, S.; Kalra, S.; Kumar, P.; Ahuja, A.A.; Sharma, M.; Gautam, A.; SRaghava, G.P. THPdb: Database of FDA-approved peptide and protein therapeutics. PLoS ONE 2017, 12, e0181748. [Google Scholar] [CrossRef] [PubMed]
- FDA Purplebook. (n.d.-b). Available online: https://purplebooksearch.fda.gov/ (accessed on 28 January 2023).
- Orange Book: Approved Drug Products with Therapeutic Equivalence Evaluations. (n.d.); FDA: Rockville, MD, USA, 2023. Available online: https://www.accessdata.fda.gov/scripts/cder/ob/index.cfm (accessed on 28 January 2023).
- Available online: https://webs.iiitd.edu.in/raghava/thpdb/length.php (accessed on 11 May 2023).
- NCATS Inxight Drugs. (n.d.). Available online: https://drugs.ncats.io/ (accessed on 11 May 2023).
- KEGG Pathways Database. Available online: https://www.genome.jp/kegg/pathway.html (accessed on 11 May 2023).
- DrugBank Online|Database for Drug and Drug Target Info. (n.d.). DrugBank. Available online: https://go.drugbank.com/ (accessed on 11 May 2023).
- FDA. ANDAs for Certain Highly Purified Synthetic Peptide Drug Products That Refer to Listed Drugs of rDNA Origin. Available online: https://www.fda.gov/media/107622/download (accessed on 10 July 2023).
- Available online: https://www.cusabio.com/m-299.html#a03 (accessed on 11 May 2023).
- Goddard, T.D.; Huang, C.C.; Meng, E.C.; Pettersen, E.F.; Couch, G.S.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 2018, 27, 14–25. [Google Scholar] [CrossRef] [PubMed]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making protein folding accessible to all. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef] [PubMed]
- Google Colaboratory. (n.d.). Available online: https://colab.research.google.com/github/sokrypton/ColabFold/blob/main/ESMFold.ipynb#scrollTo=CcyNpAvhTX6q (accessed on 11 May 2023).
- Expasy-ProtParam tool. (n.d.). Available online: https://web.expasy.org/protparam/ (accessed on 15 April 2023).
- Structural Characterization Methods for Biosimilars: Fit-for-Purpose, Qualified or Validated-GaBI Journal. (n.d.). Available online: http://gabi-journal.net/structural-characterization-methods-for-biosimilars-fit-for-purpose-qualified-or-validated.html (accessed on 11 May 2023).
- LZerD Web Server. (n.d.). Available online: https://lzerd.kiharalab.org/ (accessed on 11 May 2023).
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Skolnick, J. GOAP: A generalized orientation-dependent, all-atom statistical potential for protein structure prediction. Biophys. J. 2011, 101, 2043–2052. [Google Scholar] [CrossRef]
- Zhou, H.; Zhou, Y. Distance-scaled, finite ideal-gas reference state improves structure-derived potentials of mean force for structure selection and stability prediction. Protein Sci. 2002, 11, 2714–2726. [Google Scholar]
- Huang, S.Y.; Zou, X. ITScorePro: An efficient scoring program for evaluating the energy scores of protein structures for structure prediction. Protein Struct. Predict. 2014, 71–81. [Google Scholar]
- Prodigy Webserver. (n.d.). Available online: https://wenmr.science.uu.nl/prodigy/ (accessed on 11 May 2023).
- Lin, Z.; Akin, H.; Rao, R.; Hie, B.; Zhu, Z.; Lu, W.; Smetanin, N.; Verkuil, R.; Kabeli, O.; Shmueli, Y.; et al. Evolutionary-scale prediction of atomic-level protein structure with a language model. Science 2023, 379, 1123–1130. [Google Scholar] [CrossRef] [PubMed]
- Melnyk, I. AlphaFold Distillation for Improved Inverse Protein Folding. OpenReview. Available online: https://openreview.net/forum?id=brk7Ct4Tb1M (accessed on 29 September 2022).
- Campo, D.S.; Dimitrova, Z.; Khudyakov, Y. Physicochemical Correlation between Amino Acid Sites in Short Sequences under Selective Pressure. In Proceedings of the Bioinformatics Research and Applications: Fourth International Symposium, ISBRA 2008, Atlanta, GA, USA, 6–9 May 2008; pp. 146–158. [Google Scholar]
- He, Y.; Rackovsky, S.; Yin, Y.; Scheraga, H.A. Alternative approach to protein structure prediction based on sequential similarity of physical properties. Proc. Natl. Acad. Sci. USA 2015, 112, 5029–5032. [Google Scholar] [CrossRef] [PubMed]
- Pok, G.; Jin, C.; Ryu, K.H. Correlation of Amino Acid Physicochemical Properties with Protein Secondary Structure Conformation. In Proceedings of the 2008 International Conference on BioMedical Engineering and Informatics, Sanya, China, 27–30 May 2008. [Google Scholar]
- Saghapour, E.; Sehhati, M. Physicochemical Position-Dependent Properties in the Protein Secondary Structures. Iran. Biomed. J. 2019, 23, 253–261. [Google Scholar] [CrossRef] [PubMed]
- Nupur, N.; Joshi, S.; Gulliarme, D.; Rathore, A.S. Analytical Similarity Assessment of Biosimilars: Global Regulatory Landscape, Recent Studies and Major Advancements in Orthogonal Platforms. Front. Bioeng. Biotechnol. 2022, 10, 832059. [Google Scholar]
- Rigi, G.; Kardar, G.; Hajizade, A.; Zamani, J.; Ahmadian, G. The effects of a truncated form of Staphylococcus aureus protein A (SpA) on the expression of cytokines of autoimmune patients and healthy individuals. Res. Sq. 2022. [Google Scholar]
- Stevens, A.O.; He, Y. Benchmarking the Accuracy of AlphaFold 2 in Loop Structure Prediction. Biomolecules 2022, 12, 985. [Google Scholar] [CrossRef]
- Cheloha, R.W.; Gellman, S.H.; Vilardaga, J.-P.; Gardella, T.J. PTH receptor-1 signalling—mechanistic insights and therapeutic prospects. Nat. Rev. Endocrinol. 2015, 11, 712–724. [Google Scholar]
- Kastritis, P.L.; Rodrigues, J.D.; Folkers, G.E.; Boelens, R.; Bonvin AM, J.J. Proteins Feel More Than They See: Fine-Tuning of Binding Affinity by Properties of the Non-Interacting Surface. J. Mol. Biol. 2014, 426, 2632–2652. [Google Scholar] [CrossRef]
- Gromiha, M.M.; Yokota, K.; Fukui, K. Energy based approach for understanding the recognition mechanism in protein–protein complexes. Mol. Biosyst. 2009, 5, 1779–1786. [Google Scholar] [CrossRef]
- Hilario, E.C.; Stern, A.; Wang, C.H.; Vargas, Y.W.; Morgan, C.J.; Swartz, T.E.; Patapoff, T.W. An Improved Method of Predicting Extinction Coefficients for the Determination of Protein Concentration. PDA J. Pharm. Sci. Technol. 2017, 71, 127–135. [Google Scholar] [CrossRef]
- Yuan, H.; Li, Z.; Wang, X.; Qi, R. Photodynamic Antimicrobial Therapy Based on Conjugated Polymers. Polymers 2022, 14, 3657. [Google Scholar] [CrossRef] [PubMed]
- Manhart, M.; Morozov, A.V. Protein folding and binding can emerge as evolutionary spandrels through structural coupling. Proc. Natl. Acad. Sci. USA 2015, 112, 1797–1802. [Google Scholar] [CrossRef] [PubMed]
- Patil, A.; Nakamura, H. The role of charged surface residues in the binding ability of small hubs in protein-protein interaction networks. Biophysics 2007, 3, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Pan, W.; Li, W.; Zhen, X.; Liang, J.; Cai, W.; Xu, F.; Yuan, K.; Lin, G.N. Evaluation of the Effectiveness of Derived Features of AlphaFold2 on Single-Sequence Protein Binding Site Prediction. Biology 2022, 11, 1454. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Chen, Z.; Zhang, C.; Xie, Y.; Ovchinnikov, S.G.; Gao, Y.Q.; Liu, S. ColabDock: Inverting AlphaFold structure prediction model for protein-protein docking with experimental restraints. bioRxiv 2023. [Google Scholar] [CrossRef]
- Bryant, P.; Pozzati, G.; Elofsson, A. Improved prediction of protein-protein interactions using AlphaFold2. Nat. Commun. 2022, 13, 1265. [Google Scholar] [CrossRef]
- Scardino, V.; Di Filippo, J.I.; Cavasotto, C.N. How good are AlphaFold models for docking-based virtual screening? iScience 2022, 26, 105920. [Google Scholar] [CrossRef]
- Johansson-Åkhe, I.; Wallner, B. Improving peptide-protein docking with AlphaFold-Multimer using forced sampling. Front. Bioinform. 2022, 2, 85. [Google Scholar] [CrossRef]
- Tang, Q.; Ren, W.; Wang, J.; Kaneko, K. The Statistical Trends of Protein Evolution: A Lesson from AlphaFold Database. Mol. Biol. Evol. 2022, 39, msac197. [Google Scholar] [CrossRef]
- Lobanov, M.Y.; Bogatyreva, N.S.; Galzitskaya, O.V. Radius of gyration as an indicator of protein structure compactness. Mol. Biol. 2008, 42, 623–625. [Google Scholar] [CrossRef]
- Available online: https://yanglab.nankai.edu.cn/trRosetta/ (accessed on 12 August 2023).
- Available online: https://predictioncenter.org/casp15/zscores_final.cgi (accessed on 12 August 2023).
- Random Sequence Generator-Random DNA, RNA or Protein Sequences. (n.d.). Available online: https://molbiotools.com/randomsequencegenerator.php (accessed on 29 April 2023).
- Thomas, J.; Ramakrishnan, N.; Bailey-Kellogg, C. Graphical models of residue coupling in protein families. In Proceedings of the 5th International Workshop on Bioinformatics, Chicago, IL, USA, 7 May 2008; pp. 12–20. [Google Scholar]
- Senior, A.W.; Evans, R.; Jumper, J.; Kirkpatrick, J.; Sifre, L.; Green THassabis, D. Improved protein structure prediction using potentials from deep learning. Nature 2020, 577, 706–710. [Google Scholar] [CrossRef] [PubMed]
- AlQuraishi, M. End-to-End Differentiable Learning of Protein Structure. Cell Syst. 2019, 8, 292–301.e3. [Google Scholar] [CrossRef] [PubMed]
- Ismi, D.P.; Pulungan, R. Deep learning for protein secondary structure prediction: Pre and post-AlphaFold. Comput. Struct. Biotechnol. J. 2022, 20, 6271–6286. [Google Scholar] [CrossRef] [PubMed]
- Godzik, A. Metagenomics and the protein universe. Curr. Opin. Struct. Biol. 2011, 21, 398–403. [Google Scholar] [CrossRef]
- Protein Data Bank: The single global archive for 3D macromolecular structure data. Nucleic Acids Res. 2019, 47, D520–D528. [CrossRef]
- Laurents, D.V. AlphaFold 2 and NMR Spectroscopy: Partners to understand protein structure, dynamics and function. Front. Mol. Biosci. 2022, 9, 906437. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).