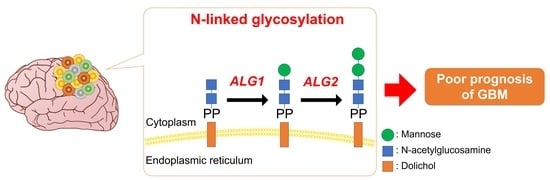
The Bioinformatics Identification of Potential Protein Glycosylation Genes Associated with a Glioma Stem Cell Signature
Abstract
1. Introduction
2. Materials and Methods
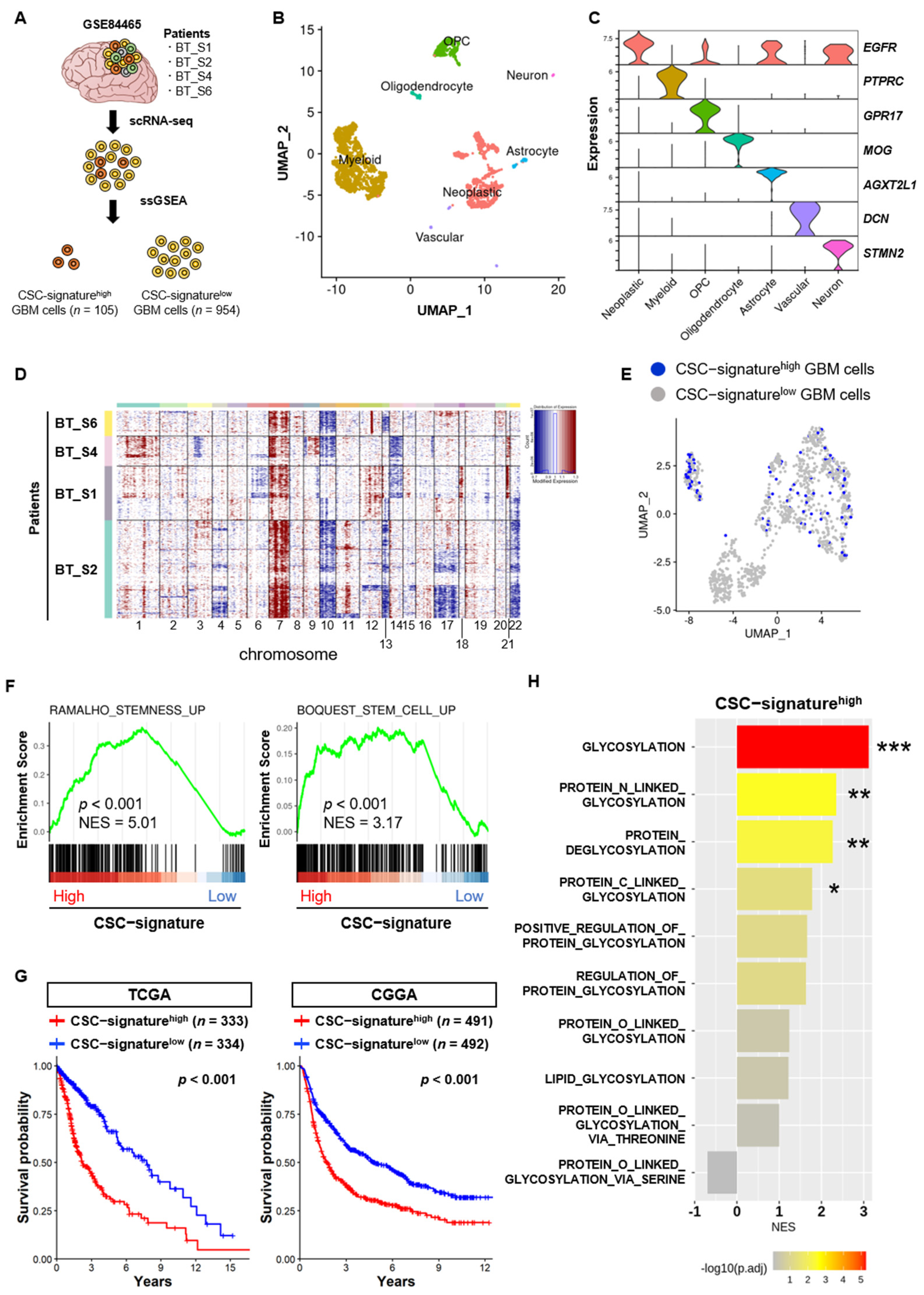
2.1. scRNA-seq Data Analysis
2.2. Bulk RNA-seq Data Analysis
2.3. Survival Analysis
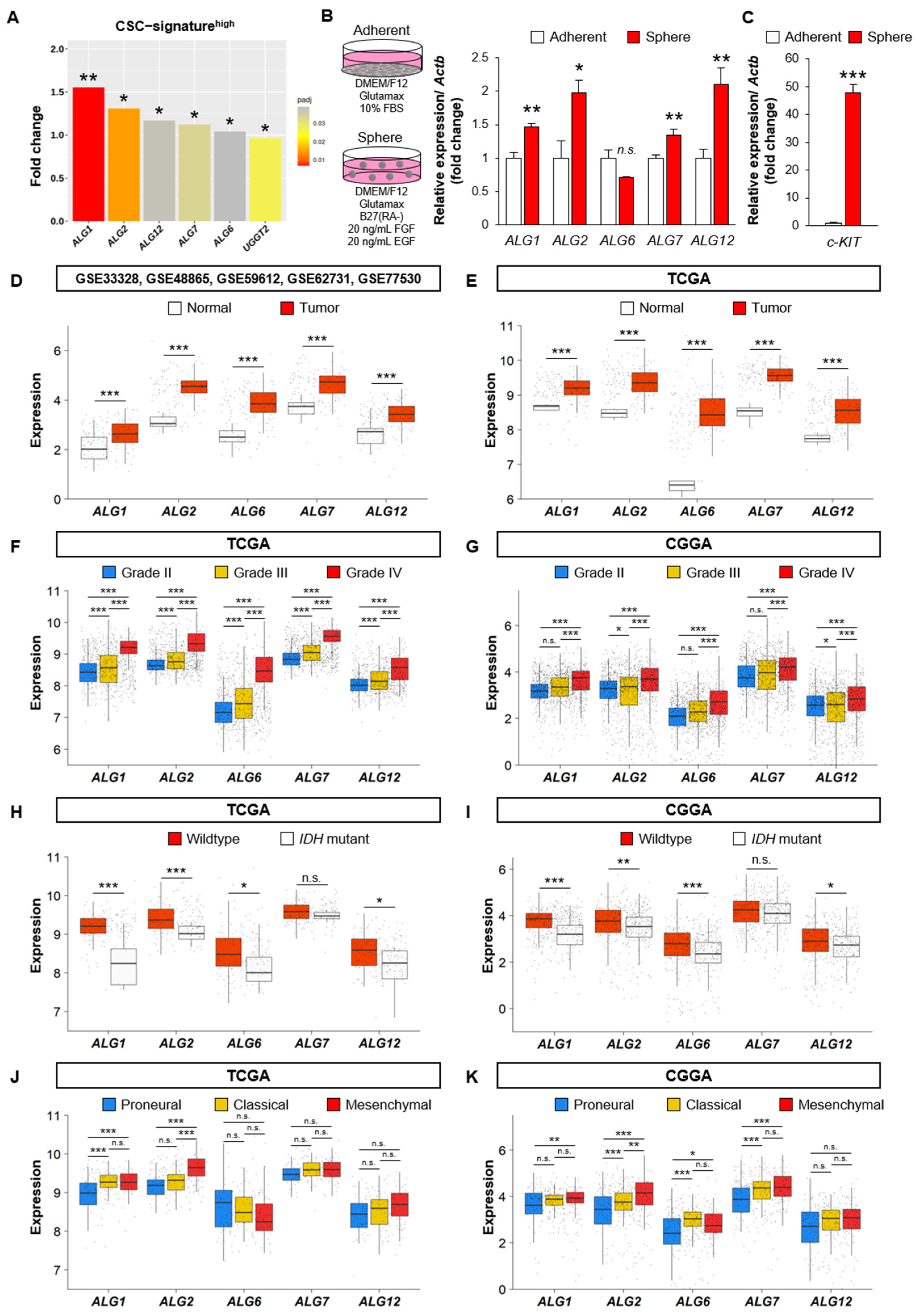
2.4. Cell Culture
2.5. RNA Extraction and Real-Time qPCR
2.6. Statistical Analysis
3. Results
3.1. N- and C-Linked Glycosylation Pathways are Associated with the Stem Cell Properties of GSCs
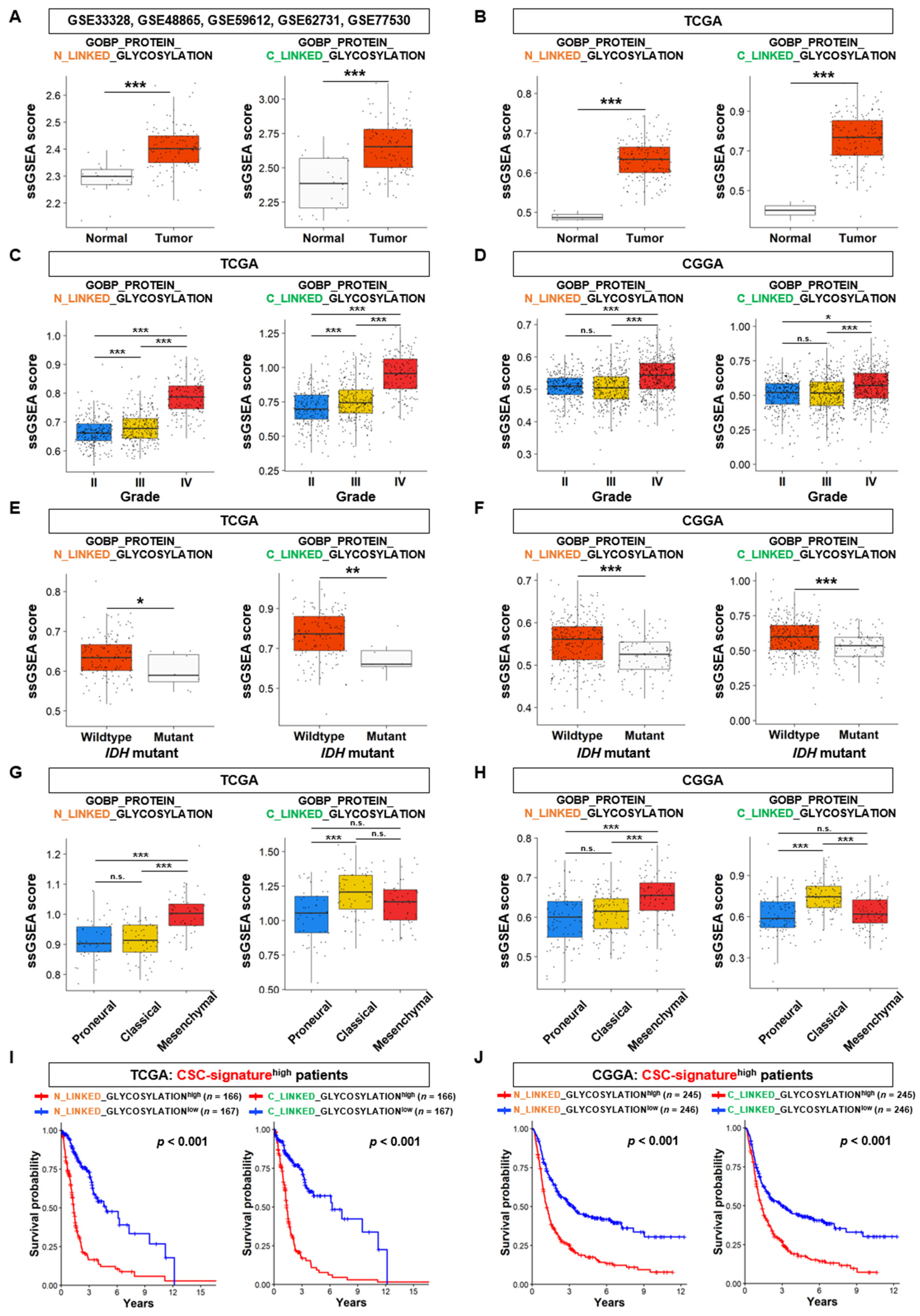
3.2. N-Linked Glycosylation Pathway in GSCs is Linked to the Aggressiveness and Poor Prognosis of GBM
3.3. The Expression Analysis of DEGs Linked to N-Linked Glycosylation Pathway in GSCs
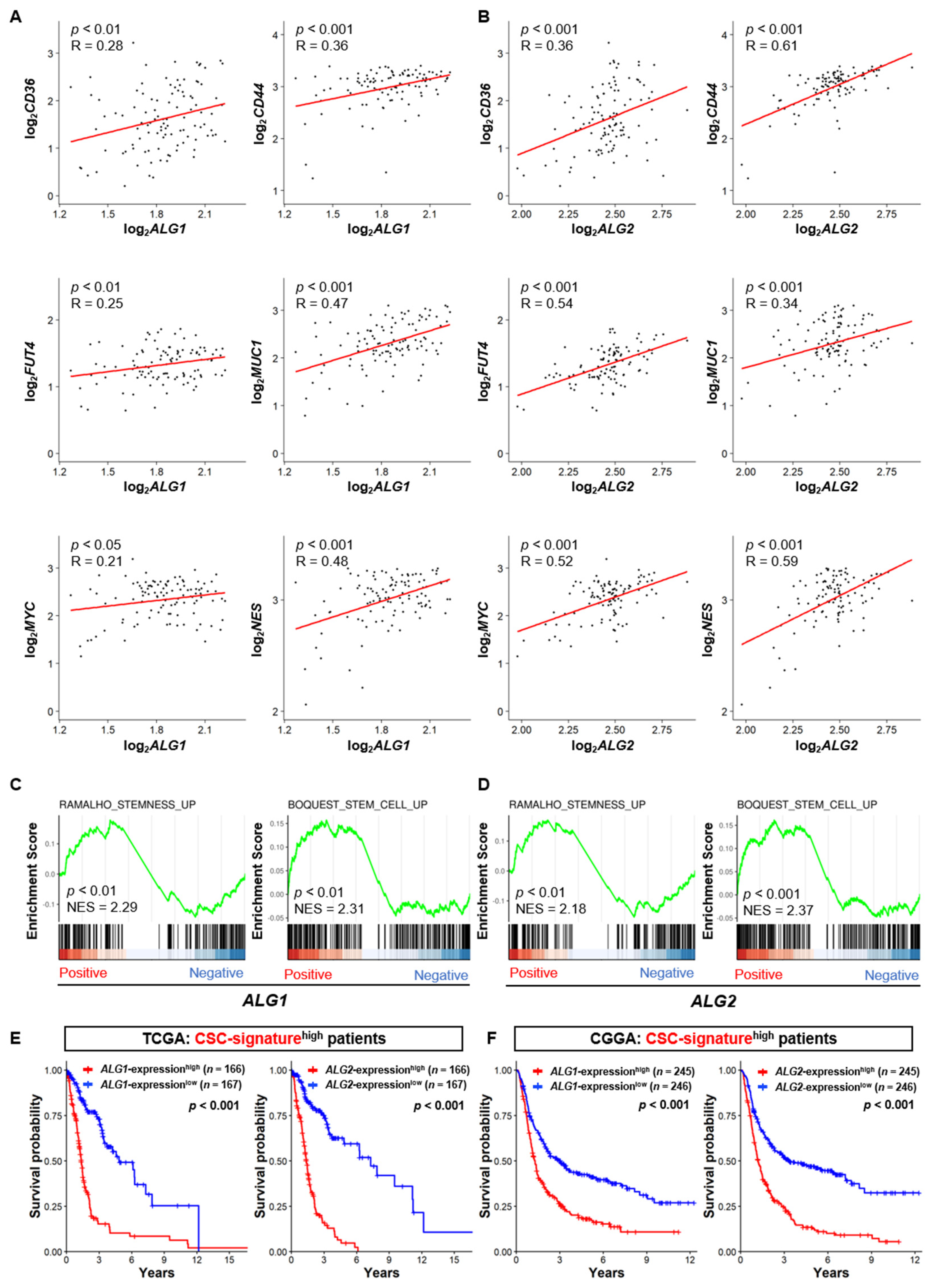
3.4. The Expression of the ALG Enzyme Family is Associated with the Stem Cell Properties of GSCs and Poor Prognosis of GBM
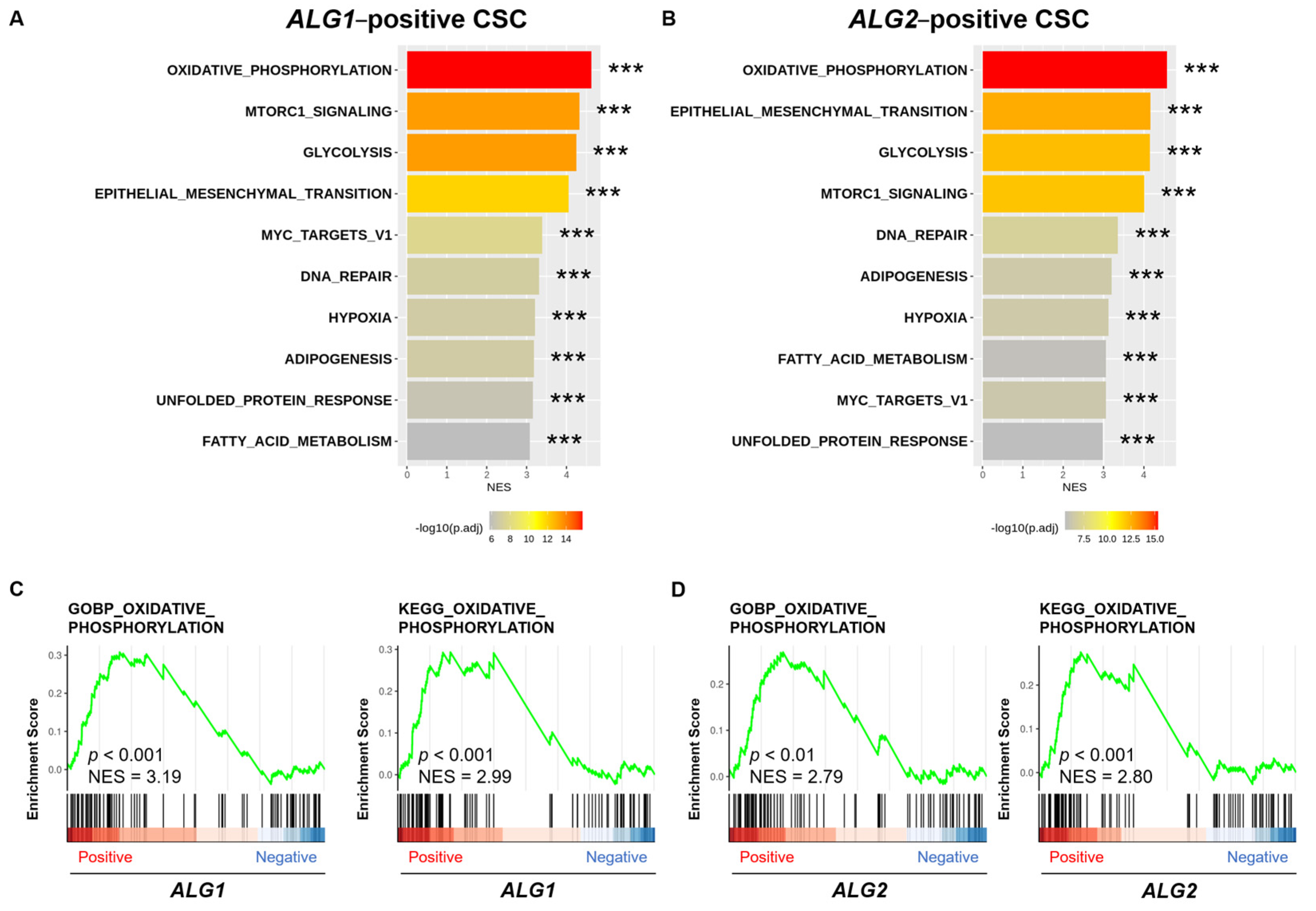
3.5. Oxidative Phosphorylation Pathway Forms a Link Between GSC Properties and the ALG Enzyme Family
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A Summary. Neuro Oncol. 2021, 23, 1231–1251. [Google Scholar] [CrossRef] [PubMed]
- Wen, P.Y.; Packer, R.J. The 2021 WHO Classification of Tumors of the Central Nervous System: Clinical Implications. Neuro Oncol. 2021, 23, 1215–1217. [Google Scholar] [CrossRef] [PubMed]
- Van Solinge, T.S.; Nieland, L.; Chiocca, E.A.; Broekman, M.L.D. Advances in Local Therapy for Glioblastoma—Taking the Fight to the Tumour. Nat. Rev. Neurol. 2022, 18, 221–236. [Google Scholar] [CrossRef] [PubMed]
- Melhem, J.M.; Detsky, J.; Lim-Fat, M.J.; Perry, J.R. Updates in IDH-Wildtype Glioblastoma. Neurotherapeutics 2022, 19, 1705–1723. [Google Scholar] [CrossRef] [PubMed]
- Behnan, J.; Finocchiaro, G.; Hanna, G. The Landscape of the Mesenchymal Signature in Brain Tumours. Brain 2019, 142, 847–866. [Google Scholar] [CrossRef] [PubMed]
- Sidaway, P. CNS Cancer: Glioblastoma Subtypes Revisited. Nat. Rev. Clin. Oncol. 2017, 14, 587. [Google Scholar] [CrossRef] [PubMed]
- Gimple, R.C.; Bhargava, S.; Dixit, D.; Rich, J.N. Glioblastoma Stem Cells: Lessons from the Tumor Hierarchy in a Lethal Cancer. Genes. Dev. 2019, 33, 591–609. [Google Scholar] [CrossRef] [PubMed]
- Prager, B.C.; Bhargava, S.; Mahadev, V.; Hubert, C.G.; Rich, J.N. Glioblastoma Stem Cells: Driving Resilience through Chaos. Trends Cancer 2020, 6, 223–235. [Google Scholar] [CrossRef]
- Schjoldager, K.T.; Narimatsu, Y.; Joshi, H.J.; Clausen, H. Global View of Human Protein Glycosylation Pathways and Functions. Nat. Rev. Mol. Cell Biol. 2020, 21, 729–749. [Google Scholar] [CrossRef]
- Reily, C.; Stewart, T.J.; Renfrow, M.B.; Novak, J. Glycosylation in Health and Disease. Nat. Rev. Nephrol. 2019, 15, 346–366. [Google Scholar] [CrossRef]
- Varki, A. Biological Roles of Glycans. Glycobiology 2017, 27, 3–49. [Google Scholar] [CrossRef] [PubMed]
- Spiro, R.G. Protein Glycosylation: Nature, Distribution, Enzymatic Formation, and Disease Implications of Glycopeptide Bonds. Glycobiology 2002, 12, 43R–56R. [Google Scholar] [CrossRef] [PubMed]
- Breitling, J.; Aebi, M. N-Linked Protein Glycosylation in the Endoplasmic Reticulum. Cold Spring Harb. Perspect. Biol. 2013, 5, a013359. [Google Scholar] [CrossRef] [PubMed]
- Veillon, L.; Fakih, C.; Abou-El-Hassan, H.; Kobeissy, F.; Mechref, Y. Glycosylation Changes in Brain Cancer. ACS Chem. Neurosci. 2018, 9, 51–72. [Google Scholar] [CrossRef] [PubMed]
- Rosa-Fernandes, L.; Oba-Shinjo, S.M.; Macedo-da-Silva, J.; Marie, S.K.N.; Palmisano, G. Aberrant Protein Glycosylation in Brain Cancers, with Emphasis on Glioblastoma. Adv. Exp. Med. Biol. 2022, 1382, 39–70. [Google Scholar] [CrossRef] [PubMed]
- Dusoswa, S.A.; Verhoeff, J.; Abels, E.; Méndez-Huergo, S.P.; Croci, D.O.; Kuijper, L.H.; de Miguel, E.; Wouters, V.M.C.J.; Best, M.G.; Rodriguez, E.; et al. Glioblastomas Exploit Truncated O-Linked Glycans for Local and Distant Immune Modulation via the Macrophage Galactose-Type Lectin. Proc. Natl. Acad. Sci. USA 2020, 117, 3693–3703. [Google Scholar] [CrossRef] [PubMed]
- Ilina, E.I.; Cialini, C.; Gerloff, D.L.; Duarte Garcia-Escudero, M.; Jeanty, C.; Thézénas, M.-L.; Lesur, A.; Puard, V.; Bernardin, F.; Moter, A.; et al. Enzymatic Activity of Glycosyltransferase GLT8D1 Promotes Human Glioblastoma Cell Migration. iScience 2022, 25, 103842. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Jiang, L.; Shi, Y.; Liu, B.; He, Y.; Shen, Q.; Jiang, X.; Nie, Z.; Pu, J.; Yang, C.; et al. Hypoxia-Induced GLT8D1 Promotes Glioma Stem Cell Maintenance by Inhibiting CD133 Degradation through N-Linked Glycosylation. Cell Death Differ. 2022, 29, 1834–1849. [Google Scholar] [CrossRef]
- Marhuenda, E.; Fabre, C.; Zhang, C.; Martin-Fernandez, M.; Iskratsch, T.; Saleh, A.; Bauchet, L.; Cambedouzou, J.; Hugnot, J.-P.; Duffau, H.; et al. Glioma Stem Cells Invasive Phenotype at Optimal Stiffness Is Driven by MGAT5 Dependent Mechanosensing. J. Exp. Clin. Cancer Res. 2021, 40, 139. [Google Scholar] [CrossRef]
- Darmanis, S.; Sloan, S.A.; Croote, D.; Mignardi, M.; Chernikova, S.; Samghababi, P.; Zhang, Y.; Neff, N.; Kowarsky, M.; Caneda, C.; et al. Single-Cell RNA-Seq Analysis of Infiltrating Neoplastic Cells at the Migrating Front of Human Glioblastoma. Cell Rep. 2017, 21, 1399–1410. [Google Scholar] [CrossRef]
- Chen, L.Y.; Wei, K.-C.; Huang, A.C.-Y.; Wang, K.; Huang, C.-Y.; Yi, D.; Tang, C.Y.; Galas, D.J.; Hood, L.E. RNASEQR—A Streamlined and Accurate RNA-Seq Sequence Analysis Program. Nucleic Acids Res. 2012, 40, e42. [Google Scholar] [CrossRef] [PubMed]
- Bao, Z.-S.; Chen, H.-M.; Yang, M.-Y.; Zhang, C.-B.; Yu, K.; Ye, W.-L.; Hu, B.-Q.; Yan, W.; Zhang, W.; Akers, J.; et al. RNA-Seq of 272 Gliomas Revealed a Novel, Recurrent PTPRZ1-MET Fusion Transcript in Secondary Glioblastomas. Genome Res. 2014, 24, 1765–1773. [Google Scholar] [CrossRef] [PubMed]
- Gill, B.J.; Pisapia, D.J.; Malone, H.R.; Goldstein, H.; Lei, L.; Sonabend, A.; Yun, J.; Samanamud, J.; Sims, J.S.; Banu, M.; et al. MRI-Localized Biopsies Reveal Subtype-Specific Differences in Molecular and Cellular Composition at the Margins of Glioblastoma. Proc. Natl. Acad. Sci. USA 2014, 111, 12550–12555. [Google Scholar] [CrossRef] [PubMed]
- Stathias, V.; Pastori, C.; Griffin, T.Z.; Komotar, R.; Clarke, J.; Zhang, M.; Ayad, N.G. Identifying Glioblastoma Gene Networks Based on Hypergeometric Test Analysis. PLoS ONE 2014, 9, e115842. [Google Scholar] [CrossRef] [PubMed]
- Gabrusiewicz, K.; Rodriguez, B.; Wei, J.; Hashimoto, Y.; Healy, L.M.; Maiti, S.N.; Thomas, G.; Zhou, S.; Wang, Q.; Elakkad, A.; et al. Glioblastoma-Infiltrated Innate Immune Cells Resemble M0 Macrophage Phenotype. JCI Insight 2016, 1, e85841. [Google Scholar] [CrossRef] [PubMed]
- Bowman, R.L.; Wang, Q.; Carro, A.; Verhaak, R.G.W.; Squatrito, M. GlioVis Data Portal for Visualization and Analysis of Brain Tumor Expression Datasets. Neuro Oncol. 2017, 19, 139–141. [Google Scholar] [CrossRef] [PubMed]
- Hinoi, E.; Iezaki, T.; Fujita, H.; Watanabe, T.; Odaka, Y.; Ozaki, K.; Yoneda, Y. PI3K/Akt Is Involved in Brown Adipogenesis Mediated by Growth Differentiation Factor-5 in Association with Activation of the Smad Pathway. Biochem. Biophys. Res. Commun. 2014, 450, 255–260. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y.; Hinoi, E.; Iezaki, T.; Takada, S.; Hashizume, S.; Takahata, Y.; Tsuruta, E.; Takahashi, S.; Yoneda, Y. Repression of Adipogenesis through Promotion of Wnt/β-Catenin Signaling by TIS7 up-Regulated in Adipocytes under Hypoxia. Biochim. Biophys. Acta 2013, 1832, 1117–1128. [Google Scholar] [CrossRef]
- Weerapana, E.; Imperiali, B. Asparagine-Linked Protein Glycosylation: From Eukaryotic to Prokaryotic Systems. Glycobiology 2006, 16, 91R–101R. [Google Scholar] [CrossRef]
- Takeda, Y.; Seko, A.; Hachisu, M.; Daikoku, S.; Izumi, M.; Koizumi, A.; Fujikawa, K.; Kajihara, Y.; Ito, Y. Both Isoforms of Human UDP-Glucose:Glycoprotein Glucosyltransferase Are Enzymatically Active. Glycobiology 2014, 24, 344–350. [Google Scholar] [CrossRef]
- Hale, J.S.; Otvos, B.; Sinyuk, M.; Alvarado, A.G.; Hitomi, M.; Stoltz, K.; Wu, Q.; Flavahan, W.; Levison, B.; Johansen, M.L.; et al. Cancer Stem Cell-Specific Scavenger Receptor CD36 Drives Glioblastoma Progression. Stem Cells 2014, 32, 1746–1758. [Google Scholar] [CrossRef] [PubMed]
- Bradshaw, A.; Wickremsekera, A.; Tan, S.T.; Peng, L.; Davis, P.F.; Itinteang, T. Cancer Stem Cell Hierarchy in Glioblastoma Multiforme. Front. Surg. 2016, 3, 21. [Google Scholar] [CrossRef] [PubMed]
- Son, M.J.; Woolard, K.; Nam, D.-H.; Lee, J.; Fine, H.A. SSEA-1 Is an Enrichment Marker for Tumor-Initiating Cells in Human Glioblastoma. Cell Stem Cell 2009, 4, 440–452. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Luo, B.; Pan, M.; Li, M.; Zhao, F.; Dou, J. MUC1 Plays an Essential Role in Tumor Immunity of Colorectal Cancer Stem Cell Vaccine. Int. Immunopharmacol. 2020, 85, 106631. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, H.; Li, Z.; Wu, Q.; Lathia, J.D.; McLendon, R.E.; Hjelmeland, A.B.; Rich, J.N. C-Myc Is Required for Maintenance of Glioma Cancer Stem Cells. PLoS ONE 2008, 3, e3769. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Jin, X.; Jung, J.-E.; Beck, S.; Kim, H. Cell Surface Nestin Is a Biomarker for Glioma Stem Cells. Biochem. Biophys. Res. Commun. 2013, 433, 496–501. [Google Scholar] [CrossRef] [PubMed]
- DeBerardinis, R.J.; Chandel, N.S. Fundamentals of Cancer Metabolism. Sci. Adv. 2016, 2, e1600200. [Google Scholar] [CrossRef] [PubMed]
- Kaur, J.; Bhattacharyya, S. Cancer Stem Cells: Metabolic Characterization for Targeted Cancer Therapy. Front. Oncol. 2021, 11, 756888. [Google Scholar] [CrossRef]
- Sighel, D.; Notarangelo, M.; Aibara, S.; Re, A.; Ricci, G.; Guida, M.; Soldano, A.; Adami, V.; Ambrosini, C.; Broso, F.; et al. Inhibition of Mitochondrial Translation Suppresses Glioblastoma Stem Cell Growth. Cell Rep. 2021, 35, 109024. [Google Scholar] [CrossRef]
- Hay, N. The Akt-mTOR Tango and Its Relevance to Cancer. Cancer Cell 2005, 8, 179–183. [Google Scholar] [CrossRef]
- Francipane, M.G.; Lagasse, E. Therapeutic Potential of mTOR Inhibitors for Targeting Cancer Stem Cells. Br. J. Clin. Pharmacol. 2016, 82, 1180–1188. [Google Scholar] [CrossRef] [PubMed]
- Mecca, C.; Giambanco, I.; Donato, R.; Arcuri, C. Targeting mTOR in Glioblastoma: Rationale and Preclinical/Clinical Evidence. Dis. Markers 2018, 2018, 9230479. [Google Scholar] [CrossRef] [PubMed]
- Sandoval, J.A.; Tomilov, A.; Datta, S.; Allen, S.; O’Donnell, R.; Sears, T.; Woolard, K.; Kovalskyy, D.; Angelastro, J.M.; Cortopassi, G. Novel mTORC1 Inhibitors Kill Glioblastoma Stem Cells. Pharmaceuticals 2020, 13, 419. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.Y.; Sabatini, D.M. mTOR at the Nexus of Nutrition, Growth, Ageing and Disease. Nat. Rev. Mol. Cell Biol. 2020, 21, 183–203. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Ying, H.; Yan, H.; Kimmelman, A.C.; Hiller, D.J.; Chen, A.-J.; Perry, S.R.; Tonon, G.; Chu, G.C.; Ding, Z.; et al. Pten and P53 Converge on C-Myc to Control Differentiation, Self-Renewal, and Transformation of Normal and Neoplastic Stem Cells in Glioblastoma. Cold Spring Harb. Symp. Quant. Biol. 2008, 73, 427–437. [Google Scholar] [CrossRef] [PubMed]
- Lemjabbar-Alaoui, H.; McKinney, A.; Yang, Y.-W.; Tran, V.M.; Phillips, J.J. Glycosylation Alterations in Lung and Brain Cancer. Adv. Cancer Res. 2015, 126, 305–344. [Google Scholar] [CrossRef] [PubMed]
- Fukasawa, K.; Kadota, T.; Horie, T.; Tokumura, K.; Terada, R.; Kitaguchi, Y.; Park, G.; Ochiai, S.; Iwahashi, S.; Okayama, Y.; et al. CDK8 Maintains Stemness and Tumorigenicity of Glioma Stem Cells by Regulating the C-MYC Pathway. Oncogene 2021, 40, 2803–2815. [Google Scholar] [CrossRef]
- Hiraiwa, M.; Fukasawa, K.; Iezaki, T.; Sabit, H.; Horie, T.; Tokumura, K.; Iwahashi, S.; Murata, M.; Kobayashi, M.; Suzuki, A.; et al. SMURF2 Phosphorylation at Thr249 Modifies Glioma Stemness and Tumorigenicity by Regulating TGF-β Receptor Stability. Commun. Biol. 2022, 5, 22. [Google Scholar] [CrossRef]
- Huang, T.; Yang, Y.; Song, X.; Wan, X.; Wu, B.; Sastry, N.; Horbinski, C.M.; Zeng, C.; Tiek, D.; Goenka, A.; et al. PRMT6 Methylation of RCC1 Regulates Mitosis, Tumorigenicity, and Radiation Response of Glioblastoma Stem Cells. Mol. Cell 2021, 81, 1276–1291.e9. [Google Scholar] [CrossRef]
- Zhang, A.; Tao, W.; Zhai, K.; Fang, X.; Huang, Z.; Yu, J.S.; Sloan, A.E.; Rich, J.N.; Zhou, W.; Bao, S. Protein Sumoylation with SUMO1 Promoted by Pin1 in Glioma Stem Cells Augments Glioblastoma Malignancy. Neuro Oncol. 2020, 22, 1809–1821. [Google Scholar] [CrossRef]
- Bieberich, E. Synthesis, Processing, and Function of N-glycans in N-glycoproteins. Adv. Neurobiol. 2014, 9, 47–70. [Google Scholar] [CrossRef]
- Bieberich, E. Synthesis, Processing, and Function of N-Glycans in N-Glycoproteins. Adv. Neurobiol. 2023, 29, 65–93. [Google Scholar] [CrossRef] [PubMed]
- Esmail, S.; Manolson, M.F. Advances in Understanding N-Glycosylation Structure, Function, and Regulation in Health and Disease. Eur. J. Cell Biol. 2021, 100, 151186. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.-X.; Li, S.-T.; Wang, N.; Kitajima, T.; Yoko-O, T.; Fujita, M.; Nakanishi, H.; Gao, X.-D. Structural and Functional Analysis of Alg1 Beta-1,4 Mannosyltransferase Reveals the Physiological Importance of Its Membrane Topology. Glycobiology 2018, 28, 741–753. [Google Scholar] [CrossRef] [PubMed]
- Li, S.-T.; Wang, N.; Xu, X.-X.; Fujita, M.; Nakanishi, H.; Kitajima, T.; Dean, N.; Gao, X.-D. Alternative Routes for Synthesis of N-Linked Glycans by Alg2 Mannosyltransferase. FASEB J. 2018, 32, 2492–2506. [Google Scholar] [CrossRef] [PubMed]
- Ng, B.G.; Shiryaev, S.A.; Rymen, D.; Eklund, E.A.; Raymond, K.; Kircher, M.; Abdenur, J.E.; Alehan, F.; Midro, A.T.; Bamshad, M.J.; et al. ALG1-CDG: Clinical and Molecular Characterization of 39 Unreported Patients. Hum. Mutat. 2016, 37, 653–660. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tokumura, K.; Sadamori, K.; Yoshimoto, M.; Tomizawa, A.; Tanaka, Y.; Fukasawa, K.; Hinoi, E. The Bioinformatics Identification of Potential Protein Glycosylation Genes Associated with a Glioma Stem Cell Signature. BioMedInformatics 2024, 4, 75-88. https://doi.org/10.3390/biomedinformatics4010005
Tokumura K, Sadamori K, Yoshimoto M, Tomizawa A, Tanaka Y, Fukasawa K, Hinoi E. The Bioinformatics Identification of Potential Protein Glycosylation Genes Associated with a Glioma Stem Cell Signature. BioMedInformatics. 2024; 4(1):75-88. https://doi.org/10.3390/biomedinformatics4010005
Chicago/Turabian StyleTokumura, Kazuya, Koki Sadamori, Makoto Yoshimoto, Akane Tomizawa, Yuki Tanaka, Kazuya Fukasawa, and Eiichi Hinoi. 2024. "The Bioinformatics Identification of Potential Protein Glycosylation Genes Associated with a Glioma Stem Cell Signature" BioMedInformatics 4, no. 1: 75-88. https://doi.org/10.3390/biomedinformatics4010005
APA StyleTokumura, K., Sadamori, K., Yoshimoto, M., Tomizawa, A., Tanaka, Y., Fukasawa, K., & Hinoi, E. (2024). The Bioinformatics Identification of Potential Protein Glycosylation Genes Associated with a Glioma Stem Cell Signature. BioMedInformatics, 4(1), 75-88. https://doi.org/10.3390/biomedinformatics4010005