Classification and Explanation of Iron Deficiency Anemia from Complete Blood Count Data Using Machine Learning
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Source
2.2. Variable Selection and Data Cleaning
2.3. Validation Dataset
2.4. Classification of Anemia, Iron Deficiency (ID), and Iron Deficiency Anemia (IDA)
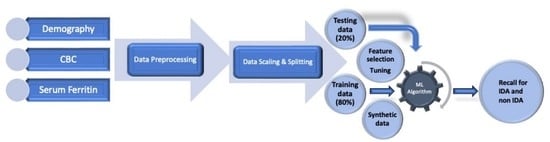
2.5. Data Aggregation and Preprocessing
2.6. Classification of IDA by Machine Learning (ML) Algorithms
2.7. Feature Selection, Cross Validation, Model Explanation
3. Results
3.1. Data Description, Proportion of Anemia, ID, and IDA
3.2. Comparative Analysis of Machine Learning Models
3.3. Optimization of the Gradient Boost Algorithm and Cross Validation with Unseen Data
3.4. Influence of Oversampling on Model Explanations
4. Discussion
5. Limitations and Future Directions
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization (WHO). Haemoglobin Concentrations for the Diagnosis of Anaemia and Assessment of Severity; WHO: Geneva, Switzerland, 2011. [Google Scholar]
- GBD 2021 Anaemia Collaborators. Prevalence, Years Lived with Disability, and Trends in Anaemia Burden by Severity and Cause, 1990–2021: Findings from the Global Burden of Disease Study. Lancet Haematol. 2023, 10, e713–e734. [Google Scholar] [CrossRef] [PubMed]
- Hsia, C.C. Respiratory Function of Hemoglobin. N. Engl. J. Med. 1998, 338, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Sarna, A.; Porwal, A.; Ramesh, S.; Agrawal, P.K.; Acharya, R.; Johnston, R.; Khan, N.; Sachdev, H.P.S.; Nair, K.M.; Ramakrishnan, L.; et al. Characterisation of the Types of Anaemia Prevalent among Children and Adolescents Aged 1-19 Years in India: A Population-Based Study. Lancet Child Adolesc. Health 2020, 4, 515–525. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, M.B.; Hurrell, R.F. Nutritional Iron Deficiency. Lancet 2007, 370, 511–520. [Google Scholar] [CrossRef] [PubMed]
- Uchida, T. Change in Red Blood Cell Distribution Width with Iron Deficiency. Clin. Lab. Haematol. 1989, 11, 117–121. [Google Scholar] [CrossRef] [PubMed]
- van Zeben, D.; Bieger, R.; van Wermeskerken, R.K.A.; Castel, A.; Hermans, J. Evaluation of Microcytosis Using Serum Ferritin and Red Blood Cell Distribution Width. Eur. J. Haematol. 1990, 44, 106–109. [Google Scholar] [CrossRef] [PubMed]
- Burk, M.; Arenz, J.A.; Schneider, W. Erythrocyte Indices as Screening Tests for the Differentiation of Microcytic Anemias. Eur. J. Med. Res. 1995, 1, 33–37. [Google Scholar]
- Cascio, M.J.; DeLoughery, T.G. Anemia: Evaluation and Diagnostic Tests. Med. Clin. 2017, 101, 263–284. [Google Scholar] [CrossRef]
- Kang, M. Machine Learning: Diagnostics and Prognostics. Progn. Health Manag. Electron. 2018, 163–191. [Google Scholar] [CrossRef]
- Al-Zaiti, S.; Martin-Gill, C.; Zègre-Hemsey, J.; Medicine, Z.B. Machine Learning for ECG Diagnosis and Risk Stratification of Occlusion Myocardial Infarction. Nat. Med. 2023, 29, 1804–1813. [Google Scholar] [CrossRef]
- Ayyıldız, H.; Tuncer, S.A. Determination of the Effect of Red Blood Cell Parameters in the Discrimination of Iron Deficiency Anemia and Beta Thalassemia via Neighborhood Component Analysis. Chemom. Intell. Lab. Syst. 2020, 196, 103886. [Google Scholar] [CrossRef]
- Vohra, R.; Hussain, A.; Dudyala, A.K.; Pahareeya, J.; Khan, W. Multi-Class Classification Algorithms for the Diagnosis of Anemia in an Outpatient Clinical Setting. PLoS ONE 2022, 17, e0269685. [Google Scholar] [CrossRef] [PubMed]
- Khan, J.R.; Chowdhury, S.; Islam, H.; Raheem, E. Machine Learning Algorithms to Predict the Childhood Anemia in Bangladesh. J. Data Sci. 2019, 1, 195–218. [Google Scholar] [CrossRef]
- Dejene, B.E.; Abuhay, T.M.; Bogale, D.S. Predicting the Level of Anemia among Ethiopian Pregnant Women Using Homogeneous Ensemble Machine Learning Algorithm. BMC Med. Inform. Decis. Mak. 2022, 22, 247. [Google Scholar] [CrossRef] [PubMed]
- Appiahene, P.; Asare, J.W.; Donkoh, E.T.; Dimauro, G.; Maglietta, R. Detection of Iron Deficiency Anemia by Medical Images: A Comparative Study of Machine Learning Algorithms. BioData Min. 2023, 16, 2. [Google Scholar] [CrossRef]
- Jain, P.; Bauskar, S.; Gyanchandani, M. Neural Network Based Non-Invasive Method to Detect Anemia from Images of Eye Conjunctiva. Int. J. Imaging Syst. Technol. 2020, 30, 112–125. [Google Scholar] [CrossRef]
- Jayakody, J.A.; Edirisinghe, E.A. HemoSmart: A Non-Invasive, Machine Learning Based Device and Mobile App for Anemia Detection. In Proceedings of the 2020 IEEE Region 10 Conference (TENCON), Osaka, Japan, 16–19 November 2020; pp. 1401–1406. [Google Scholar] [CrossRef]
- Asare, J.W.; Appiahene, P.; Donkoh, E.T.; Dimauro, G. Iron Deficiency Anemia Detection Using Machine Learning Models: A Comparative Study of Fingernails, Palm and Conjunctiva of the Eye Images. Eng. Rep. 2023, 5, e12667. [Google Scholar] [CrossRef]
- Sen, B.; Ganesh, A.; Bhan, A.; Dixit, S.; Goyal, A. Machine Learning Based Diagnosis and Classification of Sickle Cell Anemia in Human RBC. In Proceedings of the 2021 Third International Conference on Intelligent Communication Technologies and Virtual Mobile Networks (ICICV), Tirunelveli, India, 4–6 February 2021; pp. 753–758. [Google Scholar] [CrossRef]
- Bellinger, C.; Amid, A.; Japkowicz, N.; Victor, H. Multi-Label Classification of Anemia Patients. In Proceedings of the 2015 IEEE 14th International Conference on Machine Learning and Applications (ICMLA), Miami, FL, USA, 9–11 December 2015; pp. 825–830. [Google Scholar] [CrossRef]
- Saputra, D.C.E.; Sunat, K.; Ratnaningsih, T. A New Artificial Intelligence Approach Using Extreme Learning Machine as the Potentially Effective Model to Predict and Analyze the Diagnosis of Anemia. Healthcare 2023, 11, 697. [Google Scholar] [CrossRef]
- Dogan, S.; Turkoglu, I. Iron-Deficiency Anemia Detection from Hematology Parameters by Using Decision Trees. Int. J. Sci. Technol. 2008, 3, 85–92. [Google Scholar]
- Azarkhish, I.; Raoufy, M.R.; Gharibzadeh, S. Artificial Intelligence Models for Predicting Iron Deficiency Anemia and Iron Serum Level Based on Accessible Laboratory Data. J. Med. Syst. 2012, 36, 2057–2061. [Google Scholar] [CrossRef]
- Yilmaz, A.; Dagli, M.; Allahverdi, N. A Fuzzy Expert System Design for Iron Deficiency Anemia. In Proceedings of the 2013 7th International Conference on Application of Information and Communication Technologies, Baku, Azerbaijan, 23–25 October 2013; pp. 1–4. [Google Scholar] [CrossRef]
- Yıldız, T.K.; Yurtay, N.; Öneç, B. Classifying Anemia Types Using Artificial Learning Methods. Eng. Sci. Technol. Int. J. 2021, 24, 50–70. [Google Scholar] [CrossRef]
- Terzi, E.; Sarıbacak, B.; Sağlam, F.; Cengiz, M.A. A Novel Expert System for Diagnosis of Iron Deficiency Anemia. Comput. Math Methods Med. 2022, 2022, 7352096. [Google Scholar] [CrossRef] [PubMed]
- Kurstjens, S.; De Bel, T.; Van Der Horst, A.; Kusters, R.; Krabbe, J.; Van Balveren, J. Automated Prediction of Low Ferritin Concentrations Using a Machine Learning Algorithm. Clin. Chem. Lab. Med. 2022, 60, 1921–1928. [Google Scholar] [CrossRef] [PubMed]
- Nashwan, A.J.; Alkhawaldeh, I.M.; Shaheen, N.; Albalkhi, I.; Serag, I.; Sarhan, K.; Abujaber, A.A.; Abd-Alrazaq, A.; Yassin, M.A. Using Artificial Intelligence to Improve Body Iron Quantification: A Scoping Review. Blood Rev. 2023, 62, 101133. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.C. Explainable Artificial Intelligence for Predictive Modeling in Healthcare. J. Healthc. Inform Res. 2022, 6, 228–239. [Google Scholar] [CrossRef] [PubMed]
- NHANES—National Health and Nutrition Examination Survey Homepage. Available online: https://www.cdc.gov/nchs/nhanes/index.htm (accessed on 12 February 2024).
- Pandas Documentation—Pandas 2.2.0 Documentation. Available online: https://pandas.pydata.org/docs/ (accessed on 12 February 2024).
- World Health Organization (WHO). WHO Guideline on Use of Ferritin Concentrations to Assess Iron Status in Populations; WHO: Geneva, Switzerland, 2020. [Google Scholar]
- Patel, K. V Epidemiology of Anemia in Older Adults. Semin. Hematol. 2008, 45, 210–217. [Google Scholar] [CrossRef] [PubMed]
- Omuse, G.; Chege, A.; Kawalya, D.E.; Kagotho, E.; Maina, D. Ferritin and Its Association with Anaemia in a Healthy Adult Population in Kenya. PLoS ONE 2022, 17, e0275098. [Google Scholar] [CrossRef] [PubMed]
- Omuse, G.; Maina, D.; Mwangi, J.; Wambua, C.; Radia, K.; Kanyua, A.; Kagotho, E.; Hoffman, M.; Ojwang, P.; Premji, Z.; et al. Complete Blood Count Reference Intervals from a Healthy Adult Urban Population in Kenya. PLoS ONE 2018, 13, e0198444. [Google Scholar] [CrossRef]
- Saito, T.; Rehmsmeier, M. The Precision-Recall Plot Is More Informative than the ROC Plot When Evaluating Binary Classifiers on Imbalanced Datasets. PLoS ONE 2015, 10, e0118432. [Google Scholar] [CrossRef]
- Pasricha, S.R.; Armitage, A.E.; Prentice, A.M.; Drakesmith, H. Reducing Anaemia in Low Income Countries: Control of Infection Is Essential. BMJ 2018, 362. [Google Scholar] [CrossRef]
- Namaste, S.M.; Rohner, F.; Huang, J.; Bhushan, N.L.; Flores-Ayala, R.; Kupka, R.; Mei, Z.; Rawat, R.; Williams, A.M.; Raiten, D.J.; et al. Adjusting Ferritin Concentrations for Inflammation: Biomarkers Reflecting Inflammation and Nutritional Determinants of Anemia (BRINDA) Project. Am. J. Clin. Nutr. 2017, 106 (Suppl. S1), 359S–371S. [Google Scholar] [CrossRef]
- Oda, E.; Kawai, R. Comparison between High-Sensitivity C-Reactive Protein (Hs-CRP) and White Blood Cell Count (WBC) as an Inflammatory Component of Metabolic Syndrome in Japanese. Intern. Med. 2010, 49, 117–124. [Google Scholar] [CrossRef]
- Seo, I.H.; Lee, Y.J. Usefulness of Complete Blood Count (CBC) to Assess Cardiovascular and Metabolic Diseases in Clinical Settings: A Comprehensive Literature Review. Biomedicines 2022, 10, 2697. [Google Scholar] [CrossRef]



| Variable | Description | Units | Range (Min, Max) |
|---|---|---|---|
| RIDAGEYR | Age of the subject at the time of survey | Years | 1 to 60 years |
| RIAGENDR | Gender of the subject | Male 1, Female 2 | None |
| RIDEXPRG | Pregnancy status of the subject | 1 for positive, 0 for negative | None |
| LBXWBCSI | White Blood Cell count | 1000 cells/L | 1.4, 23.4 |
| LBXLYPCT | Lymphocytes percentage | % | 4.2, 84.1 |
| LBXMOPCT | Monocytes percentage | % | 0.7, 40.4 |
| LBXNEPCT | Segmented neutrophils percentage | % | 2.4, 92.3 |
| LBXEOPCT | Eosinophils percentage | % | 5.4 × 10−79, 34.1 |
| LBXBAPCT | Basophils percentage | % | 5.4 × 10−79, 19.7 |
| LBDLYMNO | Lymphocyte count | 1000 cells/µL | 0.2, 12.4 |
| LBDMONO | Monocyte count | 1000 cells/µL | 5.4 × 10−79, 3.8 |
| LBDNENO | Neutrophil count | 1000 cells/µL | 0.2, 16.3 |
| LBDEONO | Eosinophil count | 1000 cells/µL | 5.4 × 10−79, 4.5 |
| LBDBANO | Basophil count | 1000 cells/µL | 5.4 × 10−79, 1.7 |
| LBXRBCSI | Red Blood Cell count | 106 cells/µL | 2.61, 7.33 |
| LBXHGB | Hemoglobin concentration | g/dL | 6.1, 18.1 |
| LBXHCT | Hematocrit | % | 20.5, 54.9 |
| LBXMCVSI | Mean Corpuscular Volume | fL | 35.4, 116.8 |
| LBXMC | Mean Corpuscular Hemoglobin Concentration | g/dL | 25.2, 43.3 |
| LBXMCHSI | Mean Corpuscular Hemoglobin | Pg | 10.2, 56.2 |
| LBXRDW | Red Cell Distribution Width | % | 6.3, 36.5 |
| LBXPLTSI | Platelet count | 1000/µL | 4, 1021 |
| LBXMPSI | Mean Platelet Volume | fL | 5, 13.5 |
| LBXFER | Serum ferritin | µg/L | 1.04, 200 |
| Age | Biological Gender | Hemoglobin Reference (g/100 mL) for Anemia | Serum Ferritin Reference for Iron Deficiency (µg/L) |
|---|---|---|---|
| less than 5 years | Any | 11 | 12 |
| 5–11 years | Any | 11.5 | 15 |
| 12–14 years | Any | 12 | 15 |
| 15 years and above | Male | 13 | 15 |
| 15 years and above | Non pregnant female | 12 | 15 |
| Pregnant female | 11 |
| Group | Anemia | ID | IDA | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Proportion | 5% CI | 95% CI | Proportion | 5% CI | 95% CI | Proportion | 5% CI | 95% CI | |
| All | 8.63 | 8.24 | 9.02 | 13.99 | 13.51 | 14.47 | 4.99 | 4.69 | 5.29 |
| Male | 1.39 | 1.38 | 1.39 | 5.61 | 5.60 | 5.62 | 0.41 | 0.40 | 0.41 |
| Female | 10.86 | 10.85 | 10.86 | 16.57 | 15.56 | 16.57 | 6.40 | 6.40 | 6.41 |
| <5 y | 1.87 | 1.43 | 2.31 | 8.73 | 7.82 | 9.65 | 0.70 | 0.43 | 0.97 |
| 5–9 y | 1.76 | 0.90 | 2.62 | 5.64 | 4.13 | 7.14 | 0.11 | −0.10 | 0.32 |
| 10–19 y | 9.23 | 8.39 | 10.07 | 16.15 | 15.08 | 17.21 | 5.13 | 4.49 | 5.77 |
| >19 y | 11.25 | 10.65 | 11.84 | 15.57 | 14.89 | 16.23 | 6.8 | 6.32 | 7.27 |
| Confusion Matrix | Accuracy | Precision | Recall | ROC AUC | PR AUC | |
|---|---|---|---|---|---|---|
| Logistic regression | [[3769 22] [ 87 121]] | 0.97 | 0.84 | 0.58 | 0.99 | 0.83 |
| Random Forest | [[3761 30] [ 67 141]] | 0.97 | 0.82 | 0.67 | 0.99 | 0.85 |
| K-Nearest Neighbors | [[3759 32] [ 75 133]] | 0.97 | 0.80 | 0.63 | 0.94 | 0.73 |
| Naive Bayes | [[3656 135] [ 33 168]] | 0.95 | 0.56 | 0.84 | 0.97 | 0.72 |
| Gradient Boosting | [[3759 32] [ 61 147]] | 0.97 | 0.82 | 0.70 | 0.99 | 0.87 |
| XGBoost | [[3744 47] [ 55 153]] | 0.97 | 0.76 | 0.73 | 0.99 | 0.85 |
| Confusion Matrix | Accuracy | Precision | Recall | ROC AUC | PR AUC | |
|---|---|---|---|---|---|---|
| With selected features | [[3761 30] [ 59 149]] | 0.97 | 0.83 | 0.71 | 0.99 | 0.87 |
| With selected features and random oversampling | [[3669 122] [ 4 204]] | 0.96 | 0.62 | 0.98 | 0.99 | 0.87 |
| Validation metrics with unseen data | [[479 4] [2 17]] | 0.98 | 0.8 | 0.89 | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pullakhandam, S.; McRoy, S. Classification and Explanation of Iron Deficiency Anemia from Complete Blood Count Data Using Machine Learning. BioMedInformatics 2024, 4, 661-672. https://doi.org/10.3390/biomedinformatics4010036
Pullakhandam S, McRoy S. Classification and Explanation of Iron Deficiency Anemia from Complete Blood Count Data Using Machine Learning. BioMedInformatics. 2024; 4(1):661-672. https://doi.org/10.3390/biomedinformatics4010036
Chicago/Turabian StylePullakhandam, Siddartha, and Susan McRoy. 2024. "Classification and Explanation of Iron Deficiency Anemia from Complete Blood Count Data Using Machine Learning" BioMedInformatics 4, no. 1: 661-672. https://doi.org/10.3390/biomedinformatics4010036
APA StylePullakhandam, S., & McRoy, S. (2024). Classification and Explanation of Iron Deficiency Anemia from Complete Blood Count Data Using Machine Learning. BioMedInformatics, 4(1), 661-672. https://doi.org/10.3390/biomedinformatics4010036