1. Introduction
Skin cancer, a widespread and potentially life-threatening disease, impacts millions globally. Its harmful effects can range from disfigurement to significant medical expenses, and even mortality if not diagnosed and treated early. Approximately one in five Americans are projected to develop skin cancer in their lifetime, with around 9500 daily diagnoses in the U.S. [
1]. Beyond physical consequences, skin cancer can induce emotional distress due to invasive treatments and visible scars.
Skin cancer is a prevalent malignancy linked to prolonged exposure to ultraviolet (UV) radiation, either from the sun or artificial sources [
2]. UV radiation causes DNA damage, leading to genetic mutations and abnormal cell growth. Fair-skinned individuals with a history of sunburns, especially in childhood, are more susceptible. Genetic factors, including familial cases and specific conditions like xeroderma pigmentosum, elevate risk. Aging, immune system suppression (in transplant recipients or HIV/AIDS patients), and certain chemical exposures also contribute. Individuals with prior skin cancer require vigilant follow-up and skin checks due to an increased risk of recurrence.
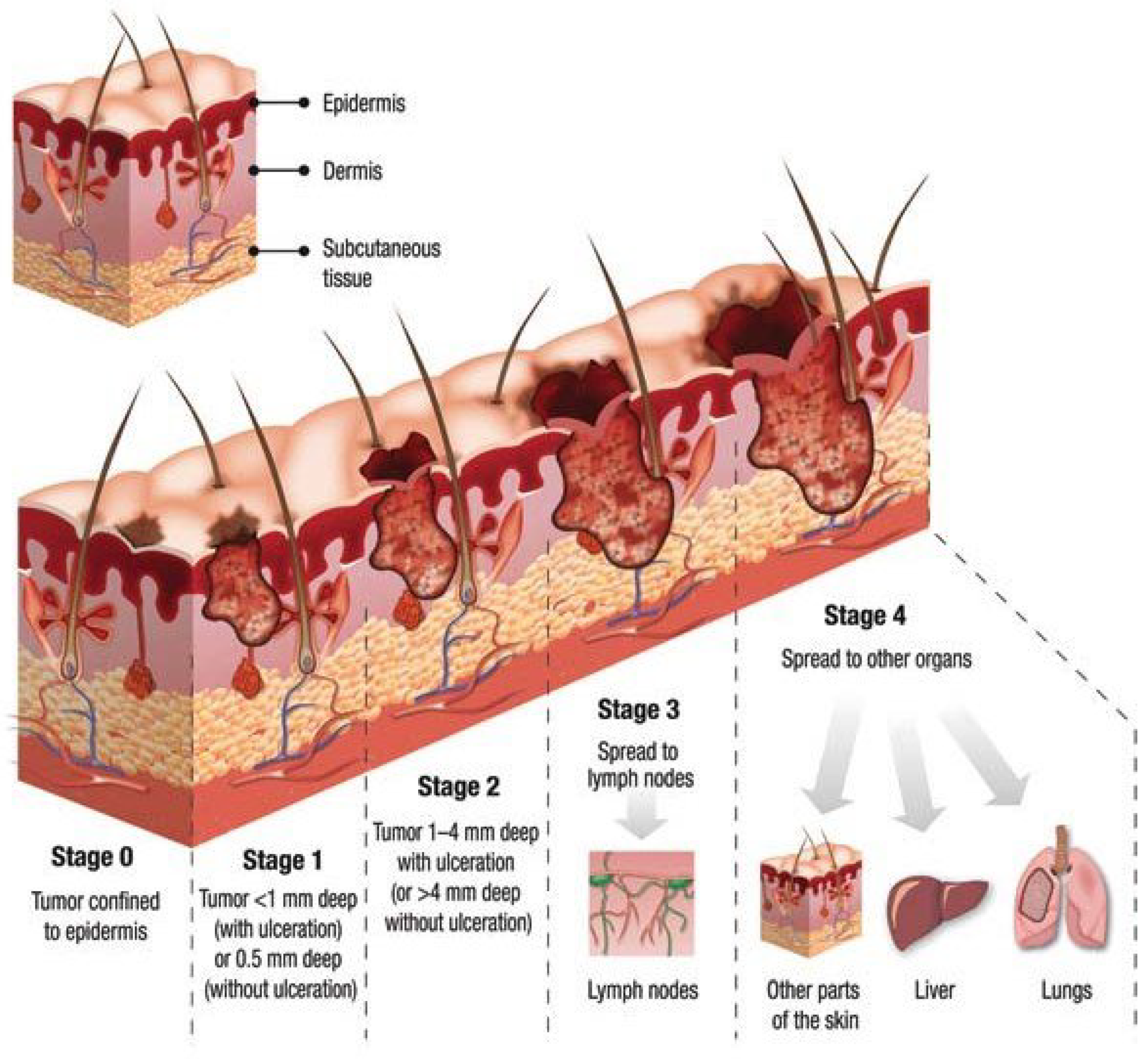
Figure 1 [
3] shows different skin cancer stages from stage 0 to stage 4 and its corresponding severity.
The ISIC 2019 dataset [
4] is a significant compilation within the International Skin Imaging Collaboration (ISIC) series, specifically curated for advancing research in dermatology, particularly in the field of computer-aided diagnosis (CAD) for skin cancer detection and classification. This dataset [
4], released in 2019, is a continuation of the effort to provide a comprehensive collection of high-quality dermoscopic images accompanied by annotations and metadata. It consists of thousands of images showcasing various skin lesions, including melanomas, nevi, and other types of benign and malignant skin conditions.
One of the primary objectives of the ISIC 2019 dataset is to facilitate the development and evaluation of machine learning algorithms, computer vision models, and Artificial Intelligence systems geared towards accurate and early detection of skin cancers. Researchers, data scientists, and developers leverage this dataset to train, validate, and test their algorithms for automated skin lesion analysis, classification, and diagnosis. The availability of annotated images within the ISIC 2019 dataset [
4] allows for supervised learning approaches, enabling algorithms to learn patterns and features associated with different types of skin lesions. By utilizing this dataset, researchers aim to improve the accuracy and efficiency of diagnostic tools, potentially aiding dermatologists and healthcare professionals in making more precise and timely diagnoses.
In recent years, deep learning [
5] has brought about a transformative revolution in the field of machine learning. It stands out as the most advanced subfield, centering on artificial neural network algorithms inspired by the structure and function of the human brain. Deep learning techniques find extensive application in diverse domains, including but not limited to speech recognition, pattern recognition, and bioinformatics. Notably, in comparison to traditional machine learning methods, deep learning systems have demonstrated remarkable achievements in these domains. Recent years have witnessed the adoption of various deep learning strategies for computer-based medical applications [
6], such as skin cancer detection. This paper delves comprehensively into the examination and evaluation of deep learning-based skin cancer classification techniques.
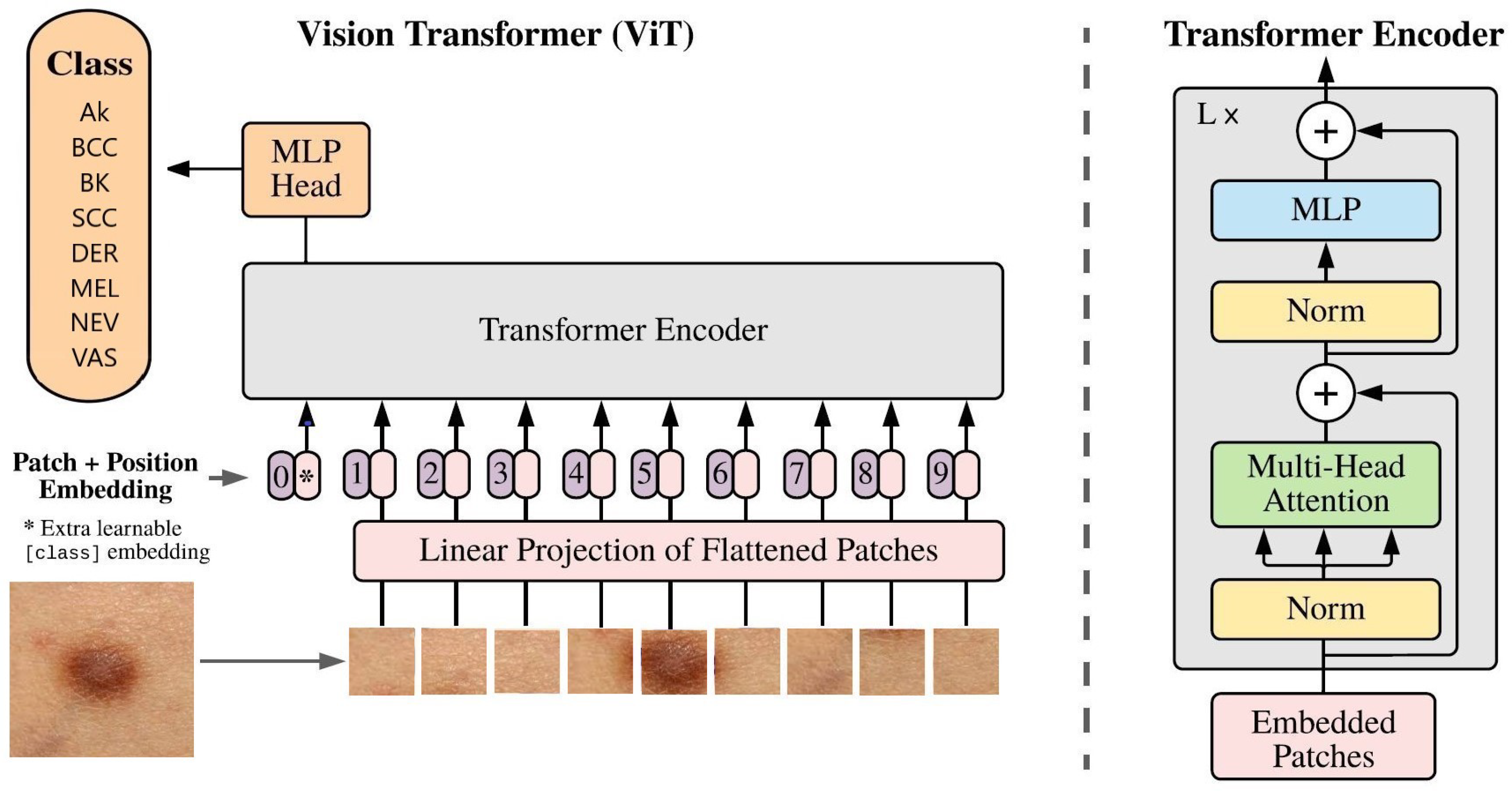
Our approach incorporates state-of-the-art deep learning models, including ImageNet ConvNets [
7] and Vision Transformer (ViT) [
8], through techniques like transfer learning, and fine-tuning. Evaluation encompasses quantitative assessments using confusion matrices, classification reports, and visual evaluations using tools like Score-CAM [
9].
The integration of “Naturalize” techniques, as referenced in [
10], alongside these strides represents significant headway in automating the analysis of skin cancer classification.
A consequence of employing the Naturalize technique is the establishment of two well-balanced datasets, namely Naturalized 2.4K and 7.2K datasets, encompassing 2400 and 7200 images, respectively, for each of the eight types of skin cancer. This paper extensively explores the methodologies and outcomes derived from these state-of-the-art approaches, shedding light on their transformative capacity within the realm of skin cancer.
After this introduction, the rest of the paper will continue as follows:
Section 2 highlights the relevant literature related to the detection and classification of skin cancer using pre-trained CNNs, and
Section 3 describes the methodology used in this study. In addition,
Section 4 presents the experimental results obtained using pre-trained models and Google ViT for the skin cancer classification; an in-depth analysis of the results is performed. Finally, the paper is concluded in
Section 5.
2. Related Works
Recent advancements in deep learning models for skin lesion classification have showcased significant progress. This review consolidates findings from notable studies employing diverse convolutional neural network (CNN) architectures for this purpose. These studies explore methodologies and performances using the ISIC2019 dataset.
Kassem et al. [
11] utilized a GoogleNet (Inception V1) model with transfer learning on the ISIC2019 dataset, achieving 94.92% accuracy. They demonstrated commendable performance in recall (79.80%), precision (80.36%), and F1-score (80.07%).
Sun et al. [
12] employed an Ensemble CNN-EfficientNet model on the ISIC2019 dataset, achieving an accuracy of 89.50%. Additionally, the authors investigated the integration of extra patient information to improve the precision of skin lesion classification. They presented performance metrics with recall (89.50%), precision (89.50%), and F1-score (89.50%).
Singh et al. [
13] utilized the Ensemble Inception-ResNet model on the ISIC2019 dataset, achieving an accuracy of 96.72%. Their results showcased notable performance in recall (95.47%), precision (84.70%), and F1-score (89.76%).
In 2022, Li et al. [
14] introduced the Quantum Inception-ResNet-V1, achieving 98.76% accuracy on the same ISIC2019 dataset. Their model exhibited substantial improvements in recall (98.26%), precision (98.40%), and F1-score (98.33%), signifying a significant leap in accuracy.
Mane et al. [
15] leveraged MobileNet with transfer learning, achieving an accuracy of 83% on the ISIC2019 dataset. Despite relatively lower results compared to other models, their consistent performance across recall, precision, and F1-score at 83% highlighted robust classification.
Hoang et al. [
16] introduced the Wide-ShuffleNet combined with segmentation techniques, achieving an accuracy of 84.80%. However, their model showed comparatively lower metrics for recall (70.71%), precision (75.15%), and F1-score (72.61%) than prior studies.
In 2023, Fofanah et al. [
17] introduced a four-layer DCNN model, achieving an accuracy of 84.80% on a modified dataset split. Their model showcased well-rounded performance with a recall of 83.80%, precision of 80.50%, and an F1-score of 81.60%.
Similarly, Alsahaf et al. [
18] proposed a Residual Deep CNN model in the same year, attaining an impressive accuracy of 94.65% on a different dataset split. They maintained equilibrium across metrics, with a recall of 70.78%, precision of 72.56%, and an F1-score of 71.33%.
Venugopal et al. [
19] presented a modified version of the EfficientNetV2 model in 2023, achieving a high accuracy of 95.49% on a different dataset split. They demonstrated balance in key metrics, including recall (95%), precision (96%), and an F1-score of 95%.
Tahir et al. [
20] proposed a DSCC-Net model with SMOTE Tomek in 2023, achieving an accuracy of 94.17% on a different dataset split. Their model exhibited well-balanced metrics, with a recall of 94.28%, precision of 93.76%, and an F1-score of 93.93%.
Radhika et al. [
21] introduced an MSCDNet Model in 2023, achieving an outstanding accuracy of 98.77% on a different dataset split. Their model maintained a harmonious blend of metrics, with a recall of 98.42%, precision of 98.56%, and an F1-score of 98.76%.
These studies collectively showcase the evolution of skin lesion classification models, indicating significant progress in accuracy and performance metrics. Comparative analysis highlights the strengths and weaknesses of each model, laying the groundwork for further advancements in dermatological image classification.
The literature review focuses on a series of studies (
Table 1), concentrating on automating skin cancer classification using the ISIC2019 dataset, offering a summarized view of these endeavors.
Our groundbreaking research presents the novel augmentation technique “Naturalize”, specifically designed to tackle the challenges posed by data scarcity and class imbalance within deep learning. Through the implementation of “Naturalize”, we have successfully overcome these hurdles, achieving an unprecedented 100% average testing accuracy, precision, recall, and F1-score in our skin cancer classification model. This groundbreaking technique revolutionizes the landscape of deep learning, offering a solution that not only elevates classification performance but also redefines the potential for accurate and reliable diagnosis across various imbalanced skin cancer classes.
3. Materials and Methods
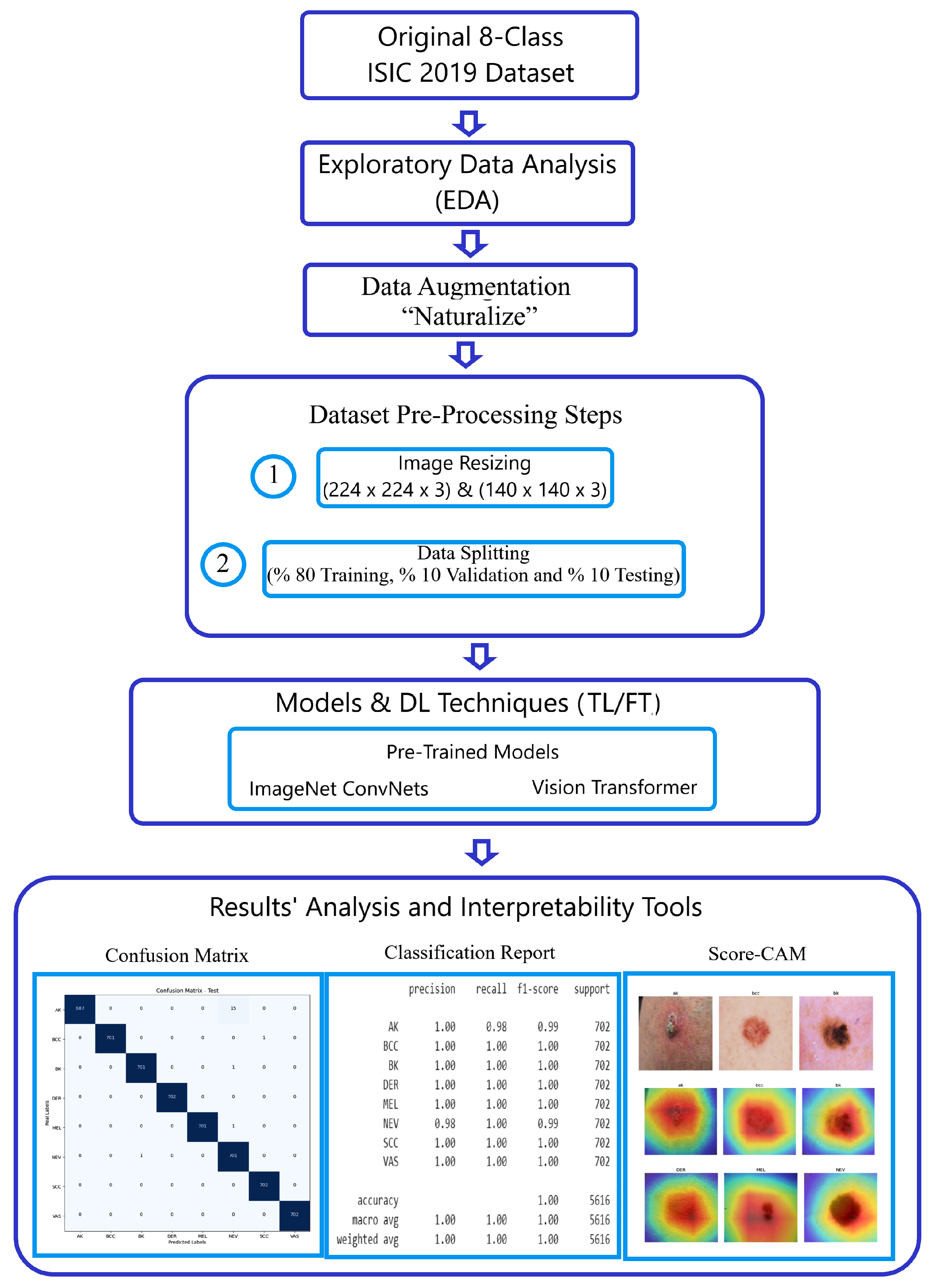
In this section, we offer an in-depth explanation of our methodology for classifying skin cancer images using the challenging ISIC2019 dataset. The steps of our approach are visually depicted in
Figure 2.
Author Contributions
Conceptualization, M.A.A. and F.D.; methodology, M.A.A., F.D. and H.A.; software, M.A.A.; validation, M.A.A.; formal analysis, M.A.A., F.D. and I.A.-C.; investigation, M.A.A.; resources, M.A.A. and F.D.; data curation, M.A.A.; writing—original draft preparation, M.A.A., F.D., H.A. and M.K.; writing—review and editing, M.A.A., F.D. and I.A.-C.; supervision, F.D. and I.A.-C.; project administration, F.D. and I.A.-C.; funding acquisition, F.D. and I.A.-C. All authors have read and agreed to the published version of the manuscript.
Funding
This work is partially supported by grant GIU23/022 unded by the University of the Basque Country (UPV/EHU), and grant PID2021-126701OB-I00, funded by the Ministerio de Ciencia, Innovación y Universidades, AEI, MCIN/AEI/10.13039/501100011033, and by “ERDF A way of making Europe” (to I.A-C.)
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data usedin this paper are publicly available.
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Garrubba, C.; Donkers, K. Skin cancer. JAAPA J. Am. Acad. Physician Assist. 2020, 33, 49–50. [Google Scholar] [CrossRef] [PubMed]
- Moqadam, S.M.; Grewal, P.K.; Haeri, Z.; Ingledew, P.A.; Kohli, K.; Golnaraghi, F. Cancer detection based on electrical impedance spectroscopy: A clinical study. J. Electr. Bioimpedance 2018, 9, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Codella, N.; Rotemberg, V.; Tschandl, P.; Celebi, M.E.; Dusza, S.; Gutman, D.; Helba, B.; Kalloo, A.; Liopyris, K.; Marchetti, M.; et al. Skin lesion analysis toward melanoma detection 2018: A challenge hosted by the International Skin Imaging Collaboration (ISIC). arXiv 2019, arXiv:1902.03368. [Google Scholar]
- Taye, M.M. Understanding of machine learning with deep learning: Architectures, workflow, applications and future directions. Computers 2023, 12, 91. [Google Scholar] [CrossRef]
- Kufel, J.; Bargieł-Łączek, K.; Kocot, S.; Koźlik, M.; Bartnikowska, W.; Janik, M.; Czogalik, Ł.; Dudek, P.; Magiera, M.; Lis, A.; et al. What Is Machine Learning, Artificial Neural Networks and Deep Learning?—Examples of Practical Applications in Medicine. Diagnostics 2023, 13, 2582. [Google Scholar] [CrossRef]
- Stock, P.; Cisse, M. ConvNets and ImageNet beyond accuracy: Understanding mistakes and uncovering biases. In Computer Vision–ECCV 2018; Springer International Publishing: Cham, Switzerland, 2018; pp. 504–519. [Google Scholar] [CrossRef]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An image is worth 16 × 16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Wang, H.; Wang, Z.; Du, M.; Yang, F.; Zhang, Z.; Ding, S.; Mardziel, P.; Hu, X. Score-CAM: Score-weighted visual explanations for convolutional neural networks. arXiv 2019. [Google Scholar] [CrossRef]
- Ali, M.A.; Dornaika, F.; Arganda-Carreras, I. Blood Cell Revolution: Unveiling 11 Distinct Types with ‘Naturalize’ Augmentation. Algorithms 2023, 16, 562. [Google Scholar] [CrossRef]
- Kassem, M.A.; Hosny, K.M.; Fouad, M.M. Skin Lesions Classification Into Eight Classes for ISIC 2019 Using Deep Convolutional Neural Network and Transfer Learning. IEEE Access 2020, 8, 114822–114832. [Google Scholar] [CrossRef]
- Sun, Q.; Huang, C.; Chen, M.; Xu, H.; Yang, Y. Skin Lesion Classification Using Additional Patient Information. BioMed Res. Int. 2021, 2021, 6673852. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.K.; Abolghasemi, V.; Anisi, M.H. Skin cancer diagnosis based on neutrosophic features with a deep neural network. Sensors 2022, 22, 6261. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Chen, Z.; Che, X.; Wu, Y.; Huang, D.; Ma, H.; Dong, Y. A classification method for multi-class skin damage images combining quantum computing and Inception-ResNet-V1. Front. Phys. 2022, 10, 1–11. [Google Scholar] [CrossRef]
- Mane, D.; Ashtagi, R.; Kumbharkar, P.; Kadam, S.; Salunkhe, D.; Upadhye, G. An Improved Transfer Learning Approach for Classification of Types of Cancer. Trait. Signal 2022, 39, 2095–2101. [Google Scholar] [CrossRef]
- Hoang, L.; Lee, S.-H.; Lee, E.-J.; Kwon, K.-R. Multiclass Skin Lesion Classification Using a Novel Lightweight Deep Learning Framework for Smart Healthcare. Appl. Sci. 2022, 12, 2677. [Google Scholar] [CrossRef]
- Fofanah, A.B.; Özbilge, E.; Kirsal, Y. Skin cancer recognition using compact deep convolutional neural network. Cukurova Univ. J. Fac. Eng. 2023, 38, 787–797. [Google Scholar] [CrossRef]
- Alsahafi, Y.S.; Kassem, M.A.; Hosny, K.M. Skin-Net: A novel deep residual network for skin lesions classification using multilevel feature extraction and cross-channel correlation with detection of outlier. J. Big Data 2023, 10, 1–23. [Google Scholar] [CrossRef]
- Venugopal, V.; Raj, N.I.; Nath, M.K.; Stephen, N. A deep neural network using modified EfficientNet for skin cancer detection in dermoscopic images. Decis. Anal. J. 2023, 8, 100278. [Google Scholar] [CrossRef]
- Tahir, M.; Naeem, A.; Malik, H.; Tanveer, J.; Naqvi, R.A.; Lee, S.W. DSCCNet: Multiclassification deep learning models for diagnosing of skin cancer using dermoscopic images. Cancers 2023, 15, 2179. [Google Scholar] [CrossRef]
- Radhika, V.; Chandana, B.S. MSCDNet-based multi-class classification of skin cancer using dermoscopy images. PeerJ Comput. Sci. 2023, 9, e1520. [Google Scholar] [CrossRef]
- Kirillov, A.; Mintun, E.; Ravi, N.; Mao, H.; Rolland, C.; Gustafson, L.; Xiao, T.; Whitehead, S.; Berg, A.C.; Lo, W.-Y.; et al. Segment Anything. arXiv 2023, arXiv:2304.02643. [Google Scholar]
- Shorten, C.; Khoshgoftaar, T.M. A survey on image data augmentation for deep learning. J. Big Data 2019, 6, 60. [Google Scholar] [CrossRef]
- Khalifa, N.E.; Loey, M.; Mirjalili, S. A comprehensive survey of recent trends in deep learning for digital images augmentation. Artif. Intell. Rev. 2021, 55, 2351–2377. [Google Scholar] [CrossRef]
- Zhang, A.; Lipton, Z.C.; Li, M.; Smola, A.J. Dive into deep learning. arXiv 2021, arXiv:2106.11342. [Google Scholar]
- Liu, Z.; Mao, H.; Wu, C.-Y.; Feichtenhofer, C.; Darrell, T.; Xie, S. A ConvNet for the 2020s. arXiv 2022, arXiv:2201.03545. [Google Scholar]
- Huang, G.; Liu, Z.; Maaten, L.V.; Weinberger, K.Q. Densely connected convolutional networks. arXiv 2016, arXiv:1608.06993. [Google Scholar]
- Tan, M.; Le, Q.V. EfficientNetV2: Smaller models and faster training. arXiv 2021, arXiv:2104.00298. [Google Scholar]
- Szegedy, C.; Ioffe, S.; Vanhoucke, V.; Alemi, A. Inception-v4, Inception-ResNet and the impact of residual connections on learning. arXiv 2016, arXiv:1602.07261. [Google Scholar] [CrossRef]
- Chollet, F. Xception: Deep learning with depthwise separable convolutions. arXiv 2016, arXiv:1610.02357. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Kim, H.E.; Cosa-Linan, A.; Santhanam, N.; Jannesari, M.; Maros, M.E.; Ganslandt, T. Transfer learning for medical image classification: A literature review. BMC Med. Imaging 2022, 22, 69. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Chen, W.; Wu, X.; Yue, H. Fine-tuning and visualization of convolutional neural networks. In Proceedings of the 2017 12th IEEE Conference on Industrial Electronics and Applications (ICIEA), Siem Reap, Cambodia, 18–20 June 2017; pp. 1310–1315. [Google Scholar] [CrossRef]
- Dalianis, H. Evaluation Metrics and Evaluation. In Clinical Text Mining; Springer International Publishing: Cham, Switzerland, 2018; pp. 45–53. [Google Scholar] [CrossRef]
Figure 1.
Skin cancer stages and severity.
Figure 2.
Methodology workflow using the ISIC 2019 dataset.
Figure 3.
The 8 types of skin cancer [
14].
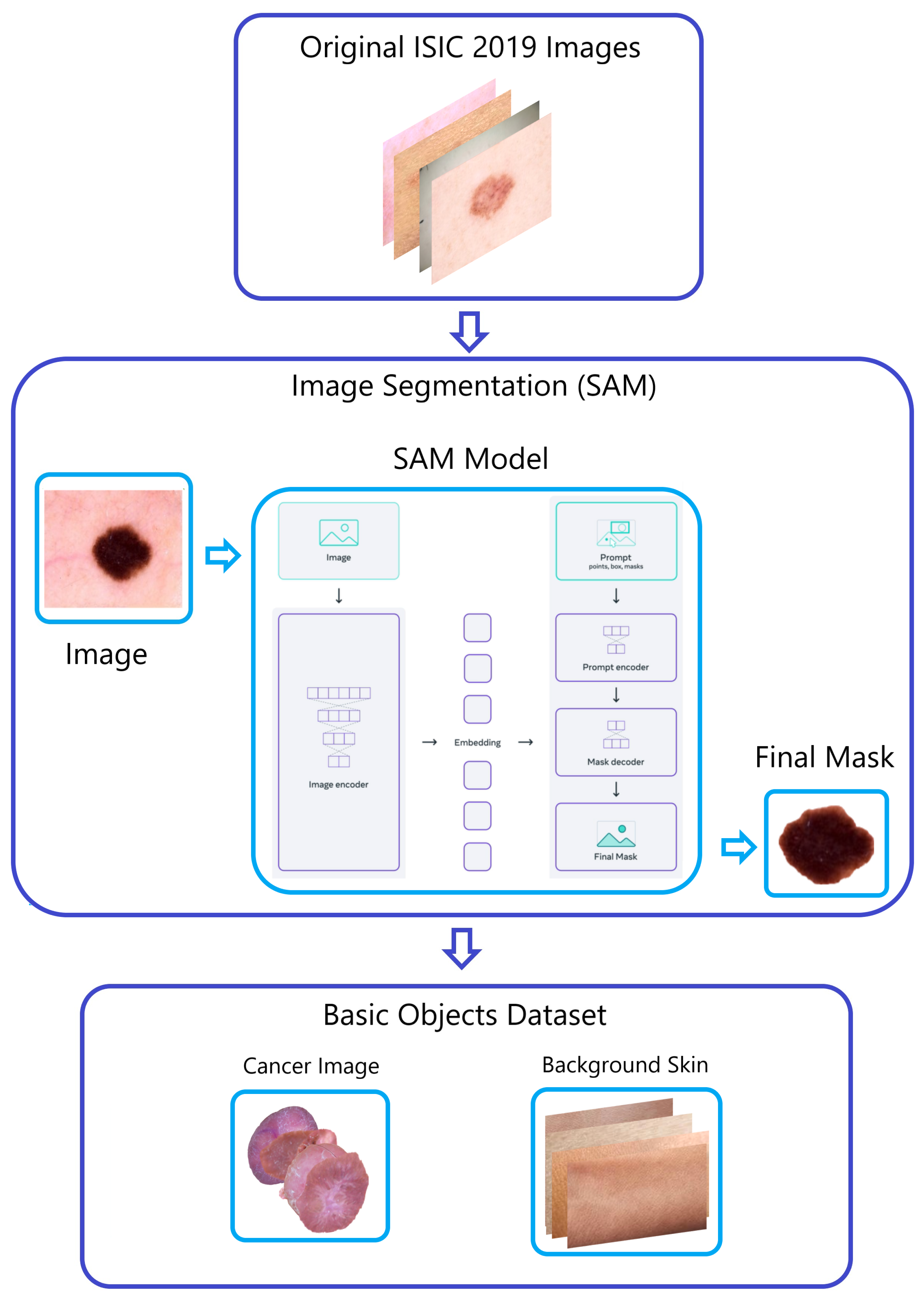
Figure 4.
The “Naturalize” first step—segmentation.
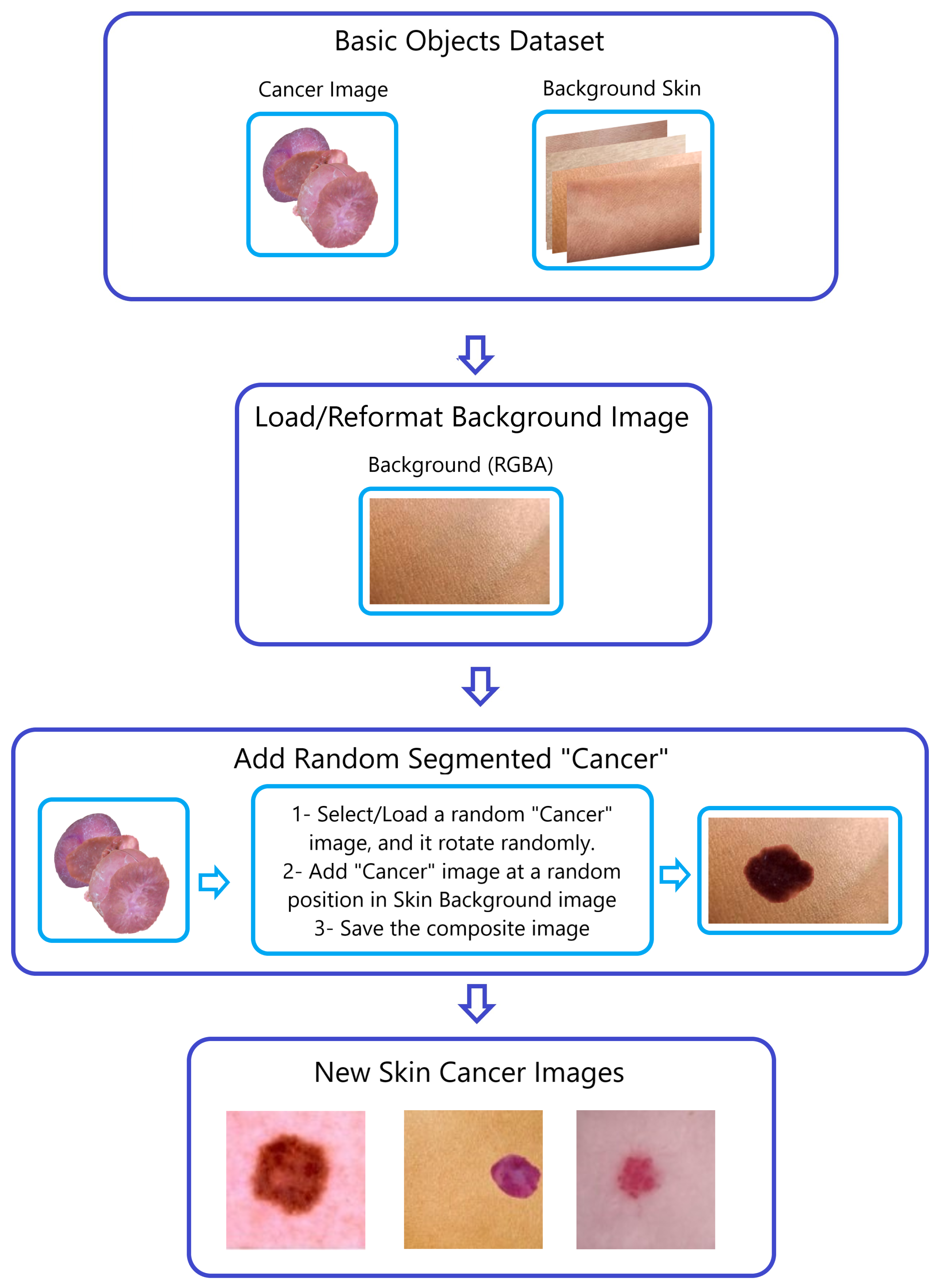
Figure 5.
The “Naturalize” second step—composite image generation.
Figure 6.
Architecture of VGG-19 model classifying a skin cancer image.
Figure 7.
Architecture of ViT classifying a skin cancer image.
Figure 8.
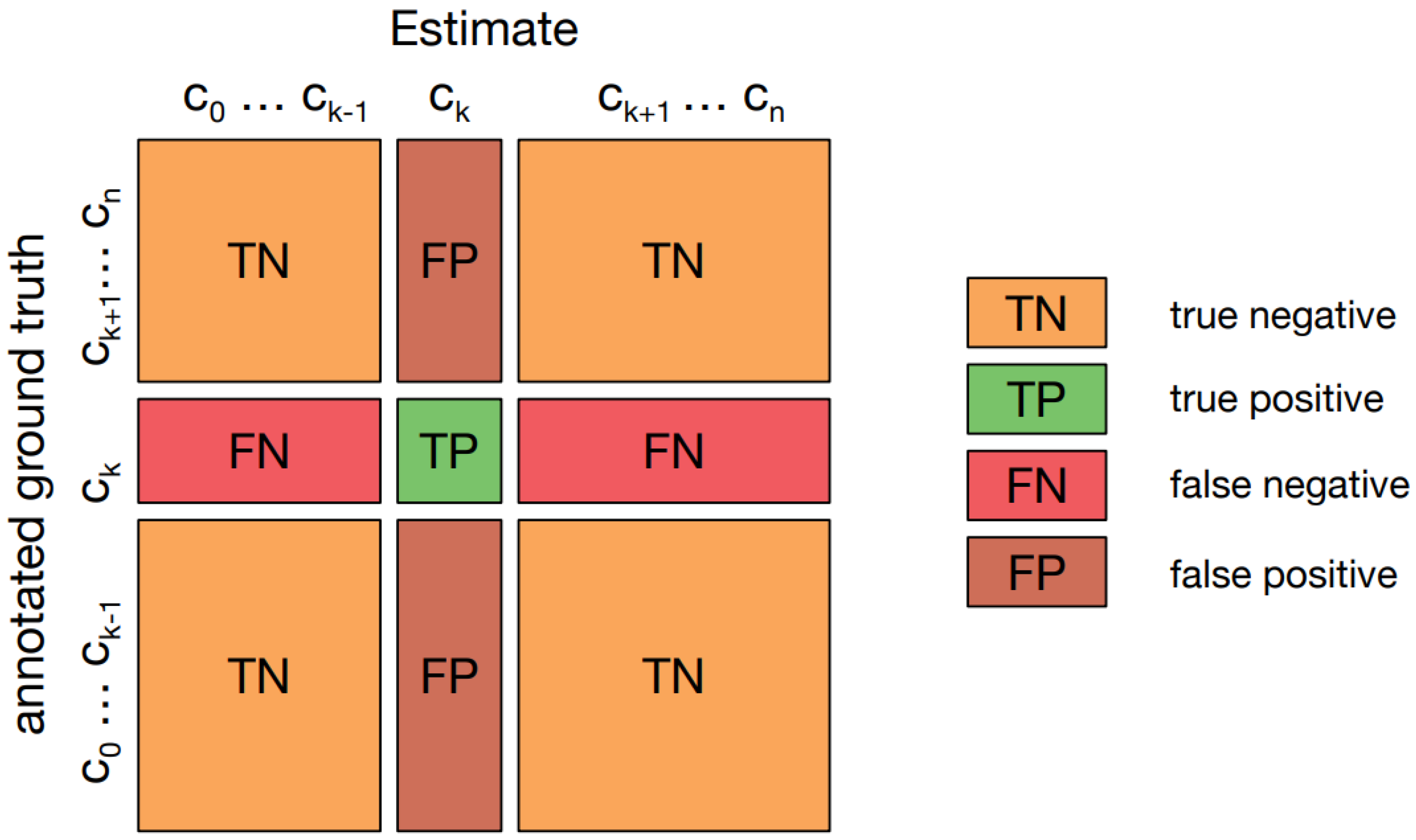
Confusion matrix for multiclass classification.
Figure 9.
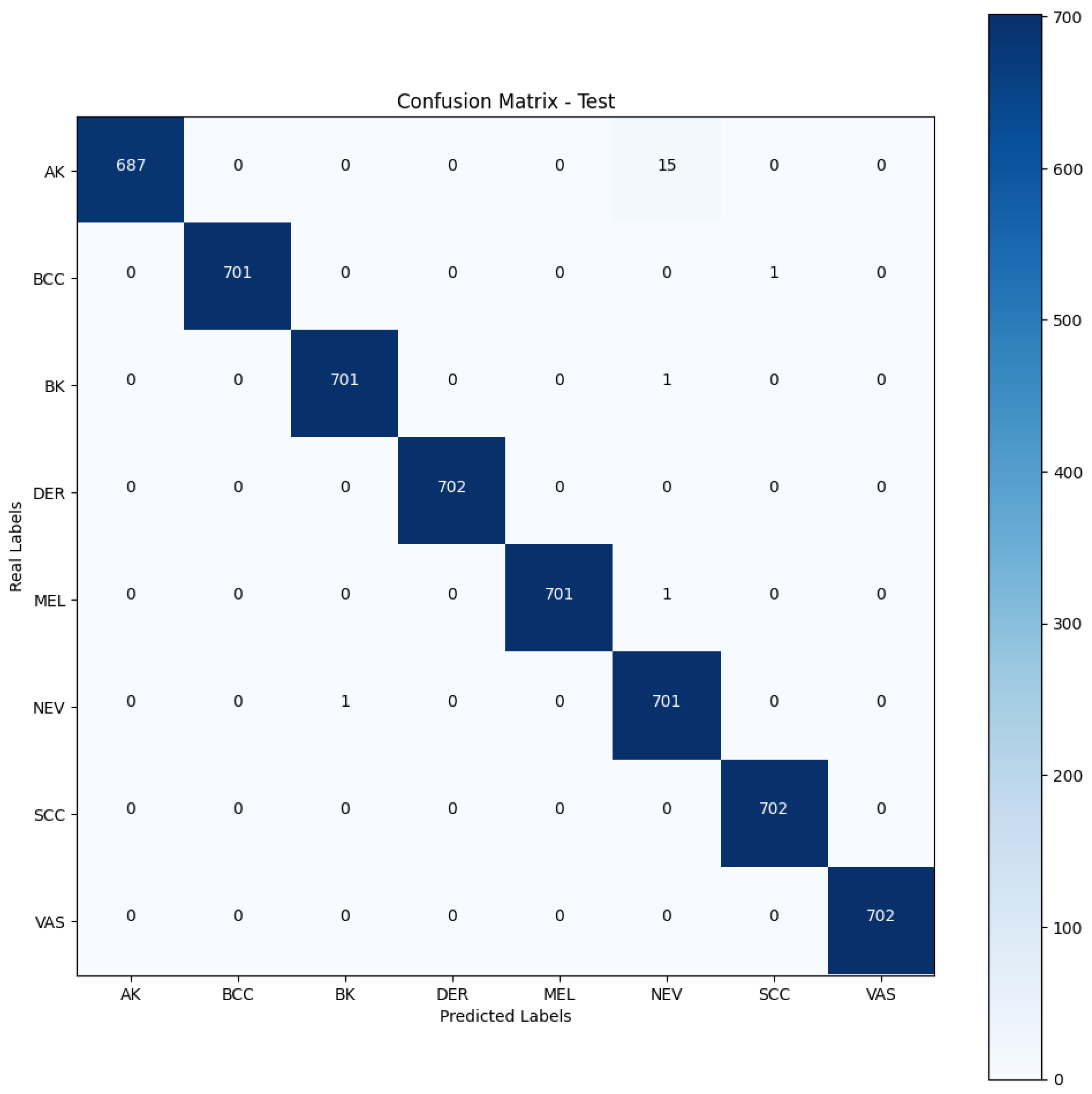
Confusion matrix—fine-tuned DenseNet201 with the Naturalized 7.2K dataset.
Figure 10.
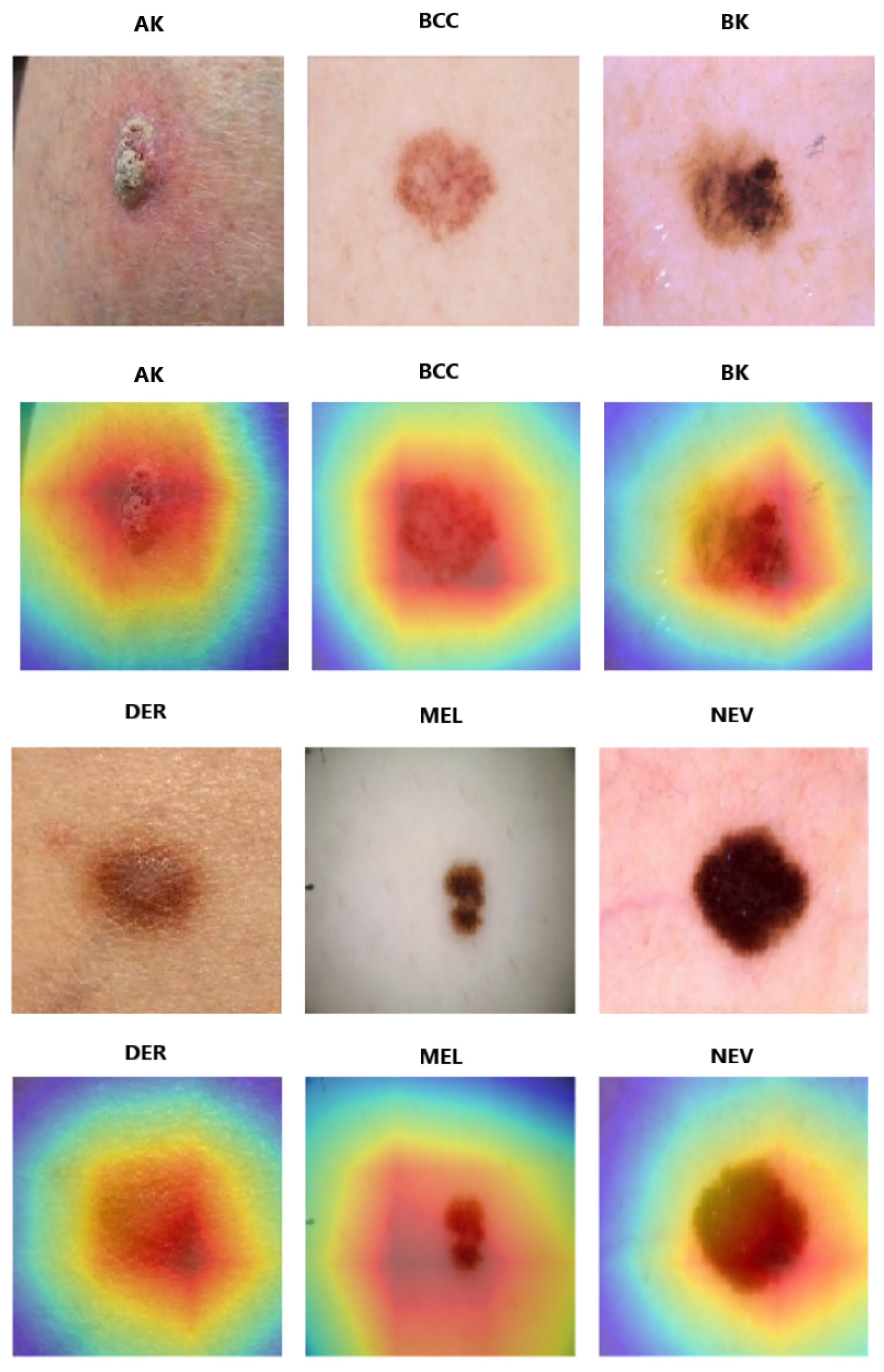
Score-CAM for fine-tuned DenseNet-201.
Table 1.
Overview of related work.
| Ref. | Model and Approach | Dataset | Split Ratio | Accuracy | Recall | Precision | F1-Score |
|---|
| [11] | GoogleNet (Inception V1) and Transfer Learning | ISIC2019 | 80/10/10 | 94.92 | 79.80 | 80.36 | 80.07 |
| [12] | Ensemble CNN-EfficientNet | ISIC2019 | 75/25 | 89.5 | 89.5 | 89.5 | 89.5 |
| [13] | Ensemble Inception-ResNet | ISIC2019 | 60/20/20 | 96.72 | 95.47 | 84.70 | 89.76 |
| [14] | Quantum Inception-ResNet-V1 | ISIC2019 | 80/10/10 | 98.76 | 98.26 | 98.40 | 98.33 |
| [15] | MobileNet and Transfer Learning | ISIC2019 | 80/10/10 | 83 | 83 | 83 | 82 |
| [16] | Wide ShuffleNet and Segmentation | ISIC2019 | 90/10 | 84.80 | 70.71 | 75.15 | 72.61 |
| [17] | Four-layer DCNN | ISIC2019 | 60/10/30 | 84.80 | 83.80 | 80.50 | 81.60 |
| [18] | Residual Deep CNN Model | ISIC2019 | 70/15/15 | 94.65 | 70.78 | 72.56 | 71.33 |
| [19] | Modified EfficientNetV2 | ISIC2019 | 80/20 | 95.49 | 95 | 96 | 95 |
| [20] | DSCC-Net with SMOTE Tomek | ISIC2019 | 80/10/10 | 94.17 | 94.28 | 93.76 | 93.93 |
| [21] | MSCDNet Model | ISIC2019 | 70/20/10 | 98.77 | 98.42 | 98.56 | 98.76 |
Table 2.
Summary of the ISIC-2019 dataset.
| Number | Cell Type | Total of Images by Type | Percent |
|---|
| 1 | Actinic Keratosis | 867 | 3.322 |
| 2 | Basal Cell Carcinoma | 3323 | 13.11 |
| 3 | Benign Keratosis | 2624 | 10.35 |
| 4 | Dermatofibroma | 239 | 0.94 |
| 5 | Melanocytic Nevi | 12,875 | 50.82 |
| 6 | Melanoma | 4522 | 17.85 |
| 7 | Vascular Skin Lesion | 253 | 1.138 |
| 8 | Squamous Cell Carcinoma | 628 | 2.47 |
| | Total | 25,331 | 100 |
Table 3.
Summary of the Pruned 2.4K ISIC-2019 dataset.
| Number | Cell Class | Symbol | Images by Class | (%) |
|---|
| 1 | Actinic Keratosis | AK | 867 | 7.5 |
| 2 | Basal Cell Carcinoma | BCC | 2400 | 20.7 |
| 3 | Benign Keratosis | BK | 2400 | 20.7 |
| 4 | Dermatofibroma | DER | 239 | 2 |
| 5 | Melanocytic Nevi | NEV | 2400 | 20.7 |
| 6 | Melanoma | MEL | 2400 | 20.7 |
| 7 | Vascular Skin Lesion | VAS | 253 | 2.1 |
| 8 | Squamous Cell Carcinoma | SCC | 628 | 5.6 |
| | Total | | 11,587 | 100 |
Table 4.
Summary of the Naturalized 2.4K ISIC-2019 dataset.
| Number | Cell Class | Symbol | Images by Class | (%) |
|---|
| 1 | Actinic Keratosis | AK | 2400 | 12.5 |
| 2 | Basal Cell Carcinoma | BCC | 2400 | 12.5 |
| 3 | Benign Keratosis | BK | 2400 | 12.5 |
| 4 | Dermatofibroma | DER | 2400 | 12.5 |
| 5 | Melanocytic Nevi | NEV | 2400 | 12.5 |
| 6 | Melanoma | MEL | 2400 | 12.5 |
| 7 | Vascular Skin Lesion | VAS | 2400 | 12.5 |
| 8 | Squamous Cell Carcinoma | SCC | 2400 | 12.5 |
| | Total | | 19,200 | 100 |
Table 5.
Summary of the Naturalized 7.2K ISIC-2019 dataset.
| Number | Cell Class | Symbol | Images by Class | (%) |
|---|
| 1 | Actinic Keratosis | AK | 7200 | 12.5 |
| 2 | Basal Cell Carcinoma | BCC | 7200 | 12.5 |
| 3 | Benign Keratosis | BK | 7200 | 12.5 |
| 4 | Dermatofibroma | DER | 7200 | 12.5 |
| 5 | Melanocytic Nevi | NEV | 7200 | 12.5 |
| 6 | Melanoma | MEL | 7200 | 12.5 |
| 7 | Vascular Skin Lesion | VAS | 7200 | 12.5 |
| 8 | Squamous Cell Carcinoma | SCC | 7200 | 12.5 |
| | Total | | 57,600 | 100 |
Table 6.
Naturalized 2.4K ISIC 2019—summary of models’ training, validation, and testing accuracies.
| Model | Accuracy |
|---|
| Training | Validation | Testing |
|---|
| ConvNexTBase | 0.99 | 0.95 | 0.92 |
| ConvNeXtLarge | 0.87 | 0.84 | 0.84 |
| DenseNet-201 | 0.97 | 0.95 | 0.95 |
| EfficientNetV2 B0 | 0.88 | 0.85 | 0.82 |
| InceptionResNetV2 | 0.94 | 0.90 | 0.89 |
| VGG16 | 0.97 | 0.93 | 0.94 |
| VGG-19 | 0.96 | 0.89 | 0.90 |
| ViT | 0.89 | 0.87 | 0.90 |
| Xception | 0.94 | 0.91 | 0.82 |
Table 7.
Naturalized 2.4K ISIC 2019—summary of models’ macro-average precision, recall, and F1-scores.
| Model | Macro Average |
|---|
| Precision | Recall | F1-Score |
|---|
| ConvNexTBase | 0.93 | 0.92 | 0.91 |
| ConvNeXtLarge | 0.87 | 0.86 | 0.87 |
| DenseNet-201 | 0.96 | 0.95 | 0.95 |
| EfficientNetV2 B0 | 0.86 | 0.82 | 0.80 |
| InceptionResNetV2 | 0.90 | 0.89 | 0.88 |
| VGG16 | 0.94 | 0.94 | 0.94 |
| VGG-19 | 0.90 | 0.90 | 0.89 |
| ViT | 0.91 | 0.90 | 0.90 |
| Xception | 0.86 | 0.87 | 0.86 |
Table 8.
DenseNet201—classification report for the original ISIC 2019.
| Class | Precision | Recall | F1-Score | Support |
|---|
| AK | 0.61 | 0.67 | 0.60 | 66 |
| BCC | 0.74 | 0.69 | 0.79 | 333 |
| BK | 0.58 | 0.88 | 0.79 | 263 |
| DER | 0.56 | 0.75 | 0.69 | 24 |
| NEV | 0.88 | 0.93 | 0.90 | 1287 |
| MEL | 0.65 | 0.36 | 0.46 | 452 |
| VAS | 0.85 | 0.87 | 0.89 | 63 |
| SCC | 0.75 | 0.94 | 0.86 | 25 |
| Accuracy | 0.78 | 2513 |
| Macro Avg. | 0.76 | 0.68 | 0.70 | 2513 |
| Weighted Avg. | 0.85 | 0.81 | 0.81 | 2513 |
Table 9.
DenseNet201—classification report for the Pruned 2.4K ISIC 2019.
| Class | Precision | Recall | F1-Score | Support |
|---|
| AK | 0.55 | 0.67 | 0.60 | 66 |
| BCC | 0.92 | 0.69 | 0.79 | 240 |
| BK | 0.71 | 0.88 | 0.79 | 240 |
| DER | 0.64 | 0.75 | 0.69 | 24 |
| NEV | 0.75 | 0.25 | 0.38 | 240 |
| MEL | 0.57 | 0.82 | 0.67 | 240 |
| VAS | 0.57 | 0.87 | 0.69 | 63 |
| SCC | 0.77 | 0.96 | 0.86 | 25 |
| Accuracy | 0.68 | 1138 |
| Macro Avg. | 0.69 | 0.74 | 0.68 | 1138 |
| Weighted Avg. | 0.72 | 0.68 | 0.66 | 1138 |
Table 10.
DenseNet201—classification report for the Naturalized 2.4K ISIC 2019 with the testing dataset sourced from the Naturalized 2.4K ISIC 2019.
| Class | Precision | Recall | F1-Score | Support |
|---|
| AK | 0.98 | 0.99 | 0.98 | 240 |
| BCC | 0.99 | 0.95 | 0.97 | 240 |
| BK | 0.93 | 0.97 | 0.95 | 240 |
| DER | 0.98 | 1.00 | 0.99 | 240 |
| NEV | 0.98 | 0.75 | 0.85 | 240 |
| MEL | 0.81 | 0.99 | 0.89 | 240 |
| VAS | 0.99 | 0.97 | 0.98 | 240 |
| SCC | 1.00 | 1.00 | 1.00 | 240 |
| Accuracy | 0.95 | 1920 |
| Macro Avg. | 0.96 | 0.95 | 0.95 | 1920 |
| Weighted Avg. | 0.96 | 0.95 | 0.95 | 1920 |
Table 11.
DenseNet201—classification report for the Naturalized 2.4K ISIC 2019 with the testing dataset sourced from the Original ISIC 2019.
| Class | Precision | Recall | F1-Score | Support |
|---|
| AK | 0.98 | 0.98 | 0.98 | 240 |
| BCC | 0.99 | 0.98 | 0.99 | 240 |
| BK | 0.95 | 1.00 | 0.97 | 240 |
| DER | 1.00 | 1.00 | 1.00 | 240 |
| NEV | 0.85 | 0.98 | 0.91 | 240 |
| MEL | 0.99 | 0.80 | 0.89 | 240 |
| VAS | 1.00 | 1.00 | 1.00 | 240 |
| SCC | 1.00 | 0.99 | 0.99 | 240 |
| Accuracy | 0.97 | 1920 |
| Macro Avg. | 0.97 | 0.97 | 0.97 | 1920 |
| Weighted Avg. | 0.97 | 0.97 | 0.97 | 1920 |
Table 12.
DenseNet201—classification report for the Naturalized 7.2K ISIC 2019.
| Class | Precision | Recall | F1-Score | Support |
|---|
| AK | 1.00 | 0.98 | 0.99 | 760 |
| BCC | 1.00 | 1.00 | 1.00 | 760 |
| BK | 1.00 | 1.00 | 1.00 | 760 |
| DER | 1.00 | 1.00 | 1.00 | 760 |
| NEV | 0.98 | 1.00 | 0.99 | 760 |
| MEL | 1.00 | 1.00 | 1.00 | 760 |
| VAS | 1.00 | 1.00 | 1.00 | 760 |
| SCC | 1.00 | 1.00 | 1.00 | 760 |
| Accuracy | 1.00 | 5760 |
| Macro Avg. | 1.00 | 1.00 | 1.00 | 5760 |
| Weighted Avg. | 1.00 | 1.00 | 1.00 | 5760 |
Table 13.
DenseNet201—classification reports’ summaries for all ISIC 2019 datasets (original, Pruned 2.4K, Naturalized 2.4K and 7.2K).
| PBC Datasets | Macro Average |
|---|
| Precision | Recall | F1-Score | Accuracy |
|---|
| Imbalanced ISIC 2019 Datasets |
| Original | 0.76 | 0.68 | 0.70 | 0.93 |
| Pruned | 0.69 | 0.74 | 0.68 | 0.82 |
| Naturalized Balanced ISIC 2019 Datasets |
| 2.4K (Testing dataset from Naturalized 2.4K ISIC 2019) | 0.96 | 0.95 | 0.95 | 0.96 |
| 2.4K (Testing dataset sourced from Original ISIC 2019) | 0.97 | 0.97 | 0.97 | 0.97 |
| 7.2K | 1.00 | 1.00 | 1.00 | 1.00 |
Table 14.
Comparison with previous works.
| Ref. | Model and Approach | Dataset | Split Ratio | Accuracy | Recall | Precision | F1-Score |
|---|
| [11] | GoogleNet (Inception V1) and Transfer Learning | ISIC2019 | 80/10/10 | 94.92 | 79.80 | 80.36 | 80.07 |
| [12] | Ensemble CNN-EfficientNet | ISIC2019 | 75/25 | 89.5 | 89.5 | 89.5 | 89.5 |
| [13] | Ensemble Inception-ResNet | ISIC2019 | 60/20/20 | 96.72 | 95.47 | 84.70 | 89.76 |
| [14] | Quantum Inception-ResNet-V1 | ISIC2019 | 80/10/10 | 98.76 | 98.26 | 98.40 | 98.33 |
| [15] | MobileNet and Transfer Learning | ISIC2019 | 80/10/10 | 83 | 83 | 83 | 82 |
| [16] | Wide ShuffleNet and Segmentation | ISIC2019 | 90/10 | 84.80 | 70.71 | 75.15 | 72.61 |
| [17] | Four-layer DCNN | ISIC2019 | 60/10/30 | 84.80 | 83.80 | 80.50 | 81.60 |
| [18] | Residual Deep CNN Model | ISIC2019 | 70/15/15 | 94.65 | 70.78 | 72.56 | 71.33 |
| [19] | Modified EfficientNetV2 | ISIC2019 | 80/20 | 95.49 | 95 | 96 | 95 |
| [20] | DSCC-Net with SMOTE Tomek | ISIC2019 | 80/10/10 | 94.17 | 94.28 | 93.76 | 93.93 |
| [21] | MSCDNet Model | ISIC2019 | 70/20/10 | 98.77 | 98.42 | 98.56 | 98.76 |
| Ours | FT DenseNet201 | Naturalized 7.2K | 80/10/10 | 100 | 100 | 100 | 100 |
| Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).