Optimized FIR Filter Using Genetic Algorithms: A Case Study of ECG Signals Filter Optimization
Abstract
:1. Introduction
1.1. Problem Statement
1.2. Contribution Statement
2. Related Work
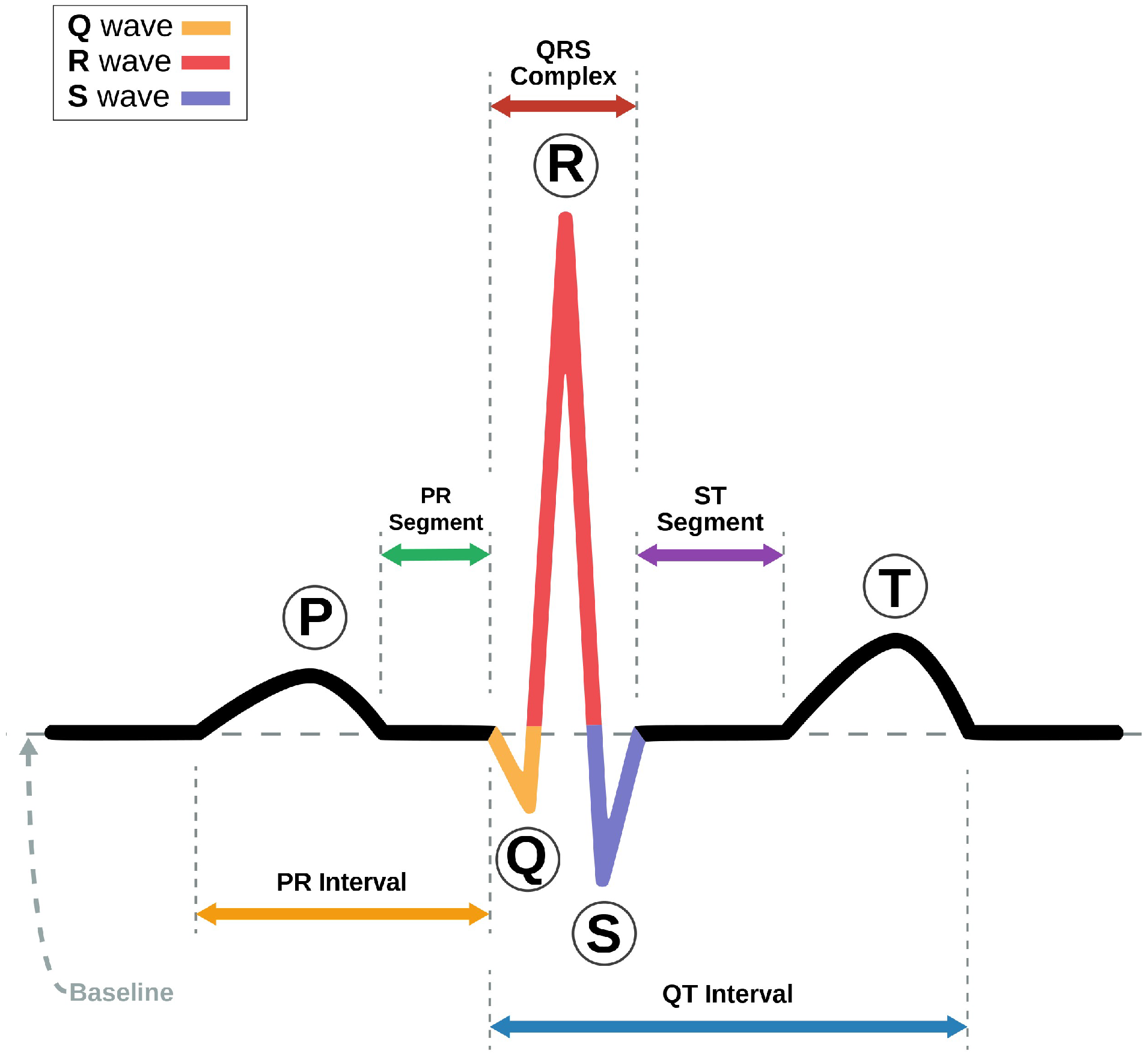
3. Methodology
4. Comparative Analysis of the GA Algorithm Options for FIR Filter Optimization
4.1. Algorithm Settings Results and Analysis
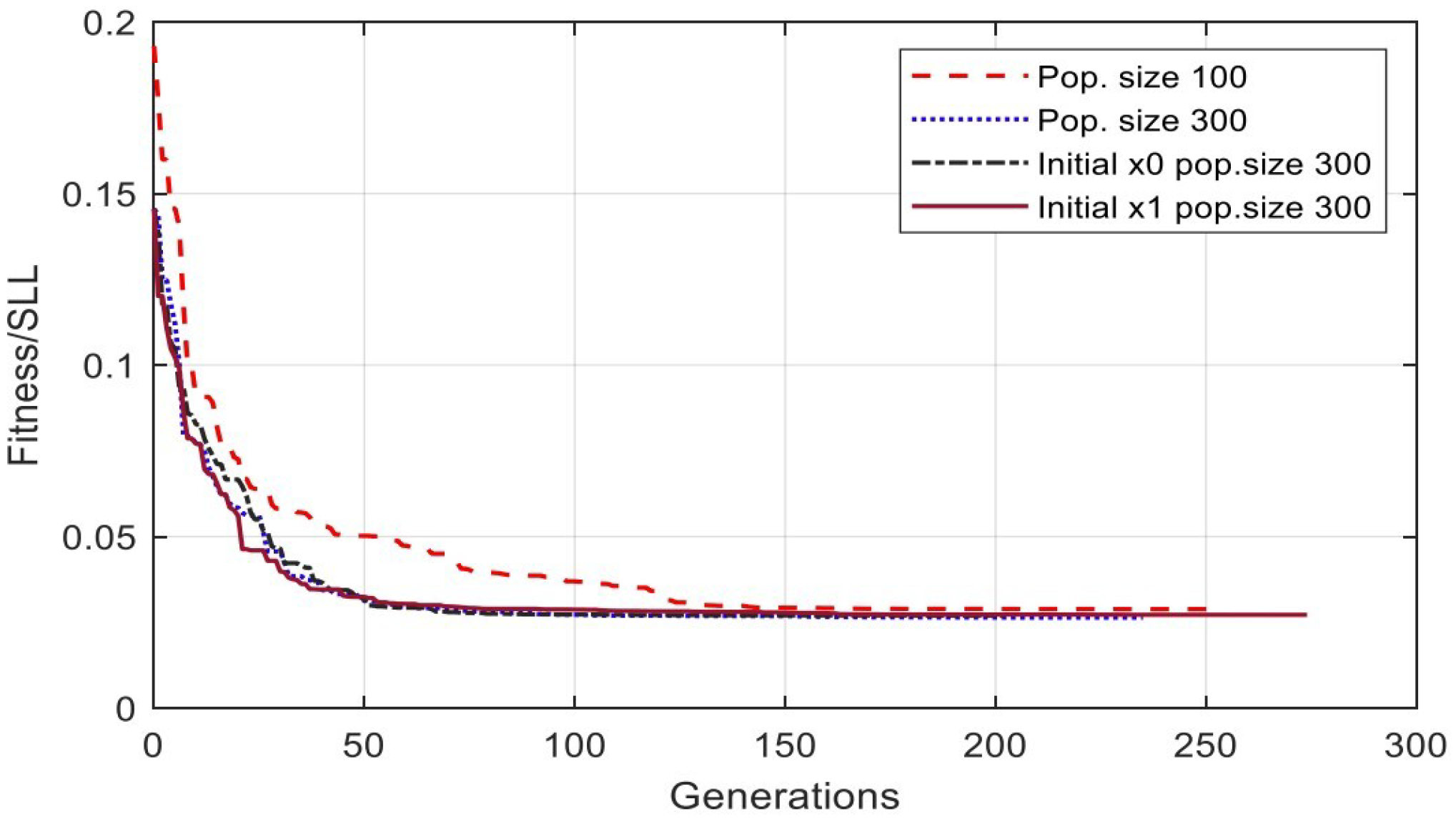
4.2. Population Settings Results and Analysis
4.3. Runtime Limits Results and Analysis
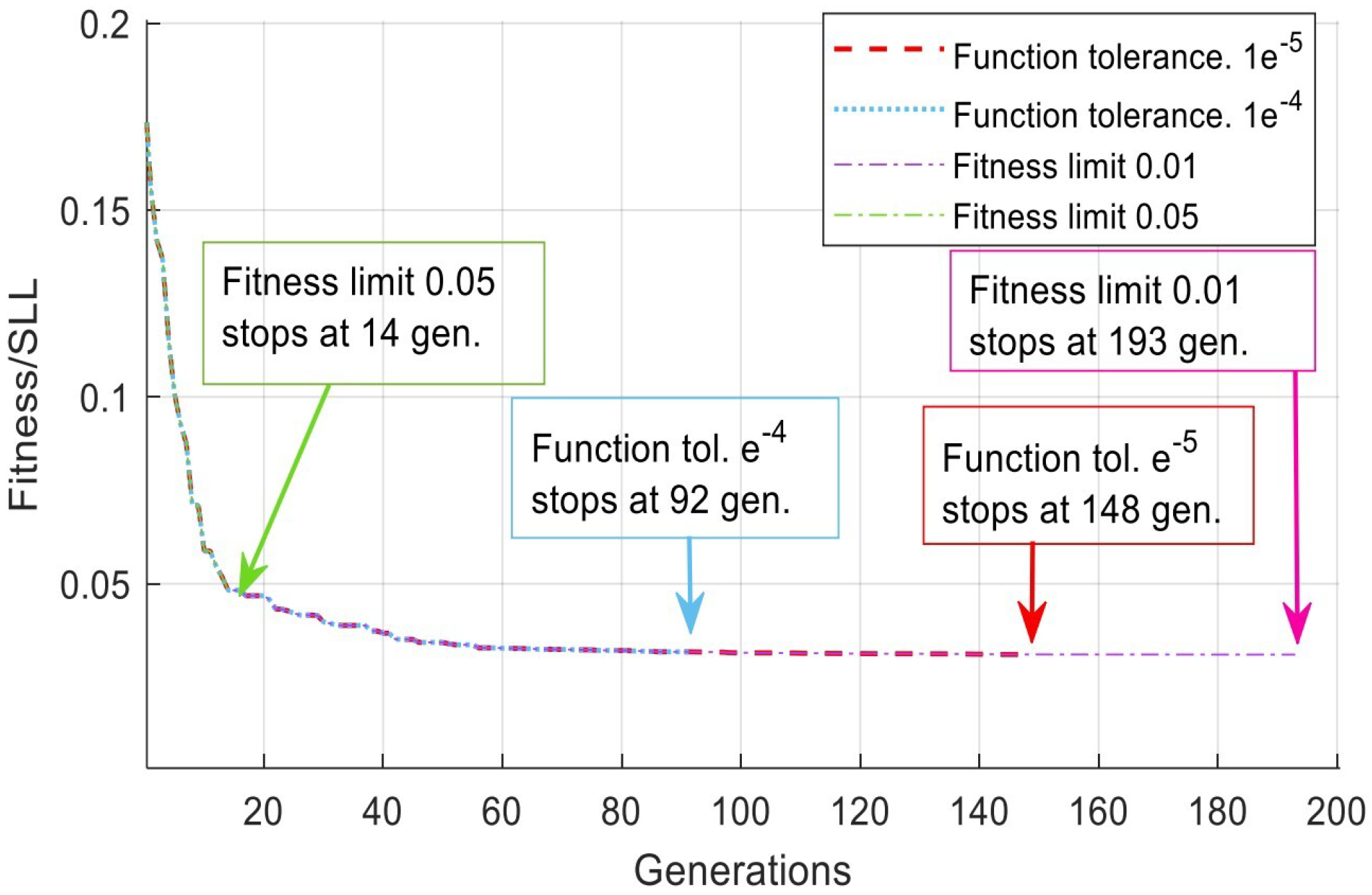
4.4. Tolerance Results and Analysis
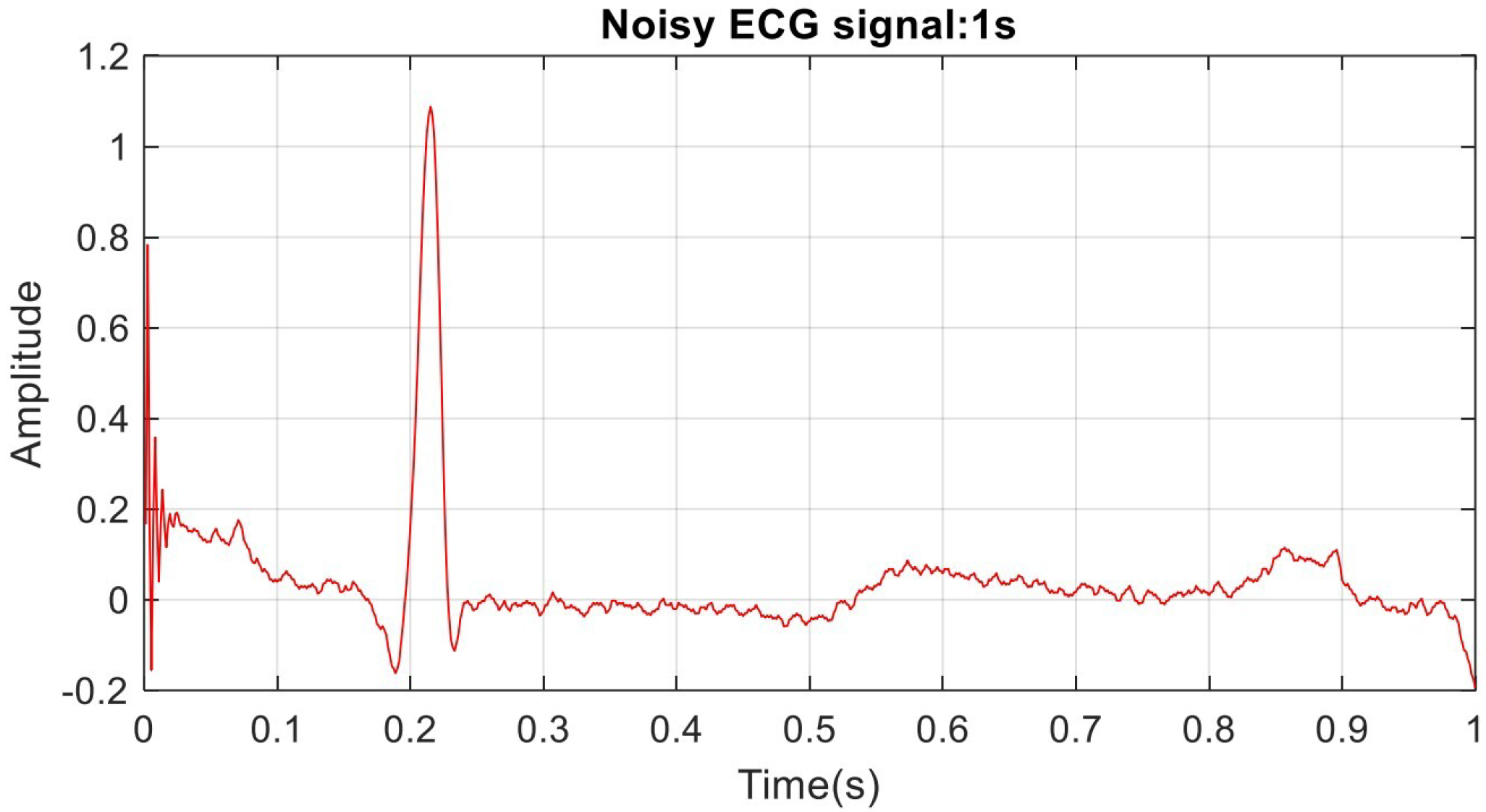
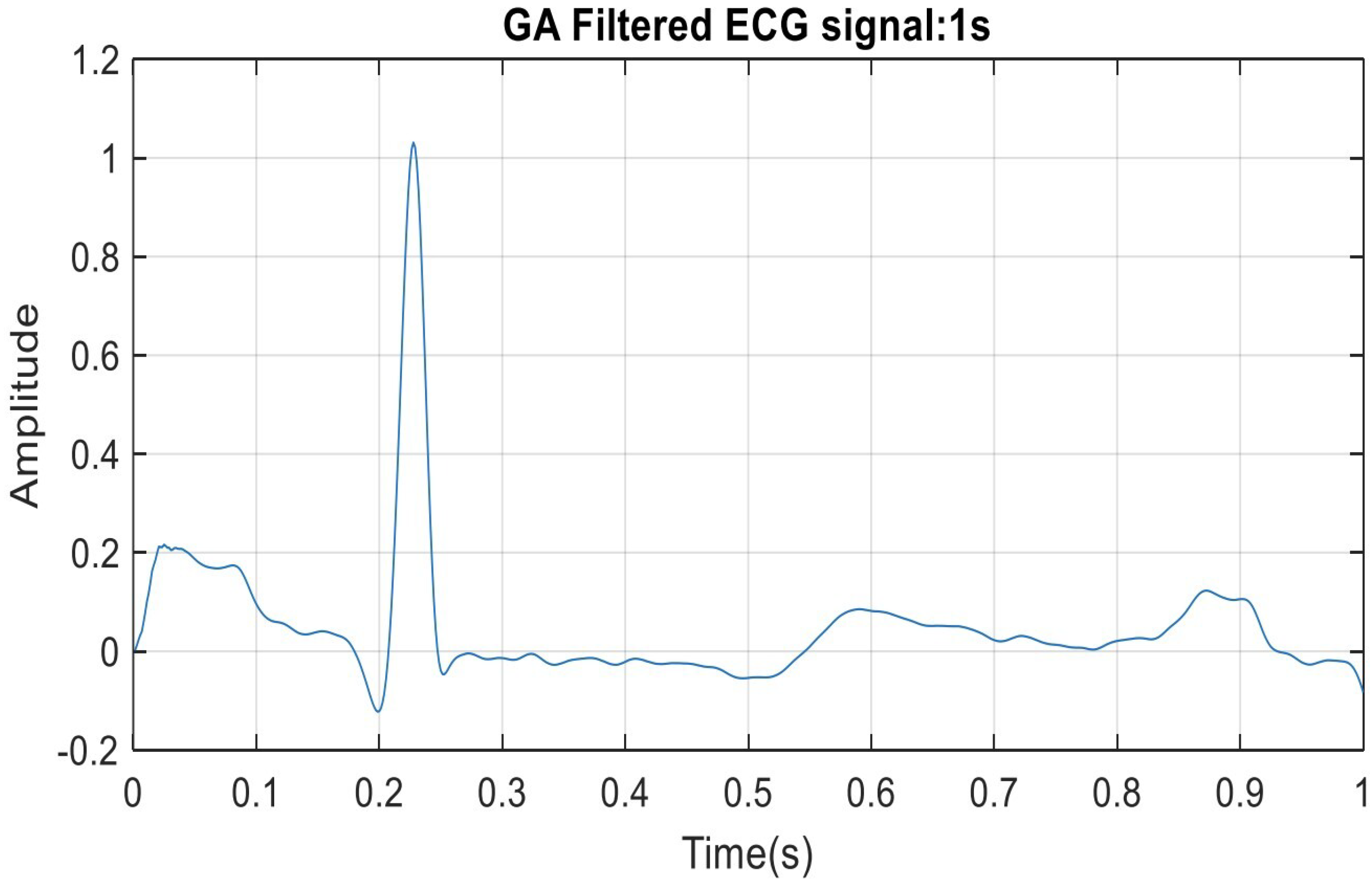
5. ECG Filtering Results Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ACO | Ant colony optimization |
| Alg. | Algorithm |
| CRO | Chemical reaction optimization |
| DAPSO | Dynamic adjustable particle swarm optimization |
| DSP | Digital signal processing |
| dB | Decibel unit |
| ECG | Electrocardiogram |
| EEG | Electroencephalogram |
| FIR | Finite impulse response |
| GA | Genetic algorithm |
| Gen. | Generation |
| IIR | Infinite impulse response |
| PSO | Particle swarm optimization |
| SLL | Side lobe level |
Appendix A. Comparative Analysis of Different GA Categories and Options
| Case No. | GA Algorithm Option | Category | Sub-Function Solver Setting | SSL (dB) | Filter Coefficients |
|---|---|---|---|---|---|
| 1 | Algorithm Setting | Crossover function | Heuristic | −19.0941 | [0.1728 0.5336 0.5986 0.2363 0.4960 0.7534 0.6947 0.5135 0.4703 0.6803 0.6439 0.6650 0.7603 0.2939 0.4812 0.3335 0.4335 0.3923 0.3140 0.3530] |
| 2 | Algorithm Setting | Crossover function | Crossover two-point | −30.1599 | [0.1119 0.1180 0.1937 0.3267 0.3476 0.4794 0.5589 0.6952 0.7050 0.7677 0.8614 0.7511 0.7524 0.7646 0.6211 0.5923 0.4037 0.4056 0.3399 0.2514] |
| 3 | Algorithm Setting | Mutation Function | Gaussian mutation | −30.8327 | [0.1359 0.2439 0.2813 0.3658 0.4837 0.6359 0.6791 0.8025 0.8434 0.8723 0.9373 0.9739 0.8859 0.8013 0.6949 0.5921 0.5364 0.4193 0.2180 0.3143] |
| 4 | Algorithm Setting | Mutation Function | Adapt-feasible mutation | −30.4694 | [0.2063 0.2968 0.3638 0.5322 0.6796 0.7954 0.8321 0.9273 1.0000 1.0000 0.9863 0.9579 0.9579 0.8361 0.6728 0.4515 0.5123 0.2934 0.2521 0.2428] |
| 5 | Population Setting | Population Size | Size = 100 | −30.7976 | [0.1575 0.2661 0.2467 0.4623 0.5886 0.5737 0.7992 0.9097 0.9834 0.9999 0.9954 1.0000 0.9933 0.8660 0.7521 0.6720 0.5120 0.4924 0.2738 0.2611] |
| 6 | Population Setting | Population Size | Size = 300 | −31.5810 | [0.1916 0.2769 0.3671 0.3364 0.6227 0.6609 0.7728 0.8591 0.9155 0.9637 0.9567 0.9225 0.8709 0.7921 0.6803 0.5446 0.5021 0.3453 0.2449 0.2105] |
| 7 | Population Setting | Initial Population | value = [0.5425 0.1422 0.3733 0.6741 0.4418 0.4340 0.6178 0.5131 0.6504 0.6010 0.8052 0.5216 0.9086 0.3192 0.0905 0.3007 0.1140 0.8287 0.0469 0.6263] | −31.3811 | [0.3034 0.2449 0.3330 0.4338 0.5682 0.7429 0.8100 0.8999 0.9730 0.9947 0.9979 0.9346 0.8853 0.8487 0.6816 0.5995 0.5000 0.3849 0.2243 0.1835] |
| 8 | Population Setting | Initial Population | value = [0.5476 0.8193 0.1989 0.8569 0.3517 0.7546 0.2960 0.8839 0.3255 0.1650 0.3925 0.0935 0.8211 0.1512 0.3841 0.9443 0.9876 0.4563 0.8261 0.2514] | −31.3141 | [0.2438 0.2903 0.4232 0.5260 0.5379 0.7947 0.8925 0.8748 0.9824 1.0000 0.9999 0.9441 0.8710 0.7934 0.6194 0.5883 0.4087 0.3561 0.2031 0.1954] |
| 9 | Runtime Limits | Max Generations | value = 100 | −30.0219 | [0.1087 0.1176 0.1907 0.3214 0.3442 0.4764 0.5565 0.6933 0.7032 0.7667 0.8613 0.7482 0.7511 0.7637 0.6208 0.5923 0.4035 0.4112 0.3396 0.2504] |
| 10 | Runtime Limits | Max Generations | value = 200 | −30.1599 | [0.1119 0.1180 0.1937 0.3267 0.3476 0.4794 0.5589 0.6952 0.7050 0.7677 0.8614 0.7511 0.7524 0.7646 0.6211 0.5923 0.4037 0.4056 0.3399 0.2514] |
| 11 | Runtime Limits | Stall Time Limit | value (T) = 2 s | −30.1599 | [0.1119 0.1180 0.1937 0.3267 0.3476 0.4794 0.5589 0.6952 0.7050 0.7677 0.8614 0.7511 0.7524 0.7646 0.6211 0.5923 0.4037 0.4056 0.3399 0.2514] |
| 12 | Runtime Limits | Stall Time Limit | value (T) = 4 s | −30.1599 | [0.1119 0.1180 0.1937 0.3267 0.3476 0.4794 0.5589 0.6952 0.7050 0.7677 0.8614 0.7511 0.7524 0.7646 0.6211 0.5923 0.4037 0.4056 0.3399 0.2514] |
| 13 | Tolerances | Function Tolerances | value = | −30.1490 | [0.1114 0.1179 0.1935 0.3265 0.3472 0.4791 0.5588 0.6947 0.7047 0.7677 0.8613 0.7507 0.7521 0.7644 0.6208 0.5923 0.4037 0.4054 0.3399 0.2510] |
| 14 | Tolerances | Function Tolerances | value = | −29.9617 | [0.1085 0.1163 0.1896 0.3215 0.3422 0.4723 0.5575 0.6922 0.7006 0.7659 0.8626 0.7454 0.7475 0.7630 0.6218 0.5917 0.4032 0.4120 0.3399 0.2487] |
| 15 | Tolerances | Fitness Limit | Fitness Limit Value = 0.01 | −30.1599 | [0.1119 0.1180 0.1937 0.3267 0.3476 0.4794 0.5589 0.6952 0.7050 0.7677 0.8614 0.7511 0.7524 0.7646 0.6211 0.5923 0.4037 0.4056 0.3399 0.2514] |
| 16 | Tolerances | Fitness Limit | Fitness Limit Value = 0.05 | −26.3296 | [0.0376 0.0092 0.1892 0.2362 0.3246 0.4347 0.4748 0.6437 0.6288 0.7338 0.8300 0.7065 0.7334 0.7504 0.6110 0.5910 0.3996 0.3229 0.2517 0.3651] |
Appendix B. Analysis of Random Solutions for Crossover and Mutation
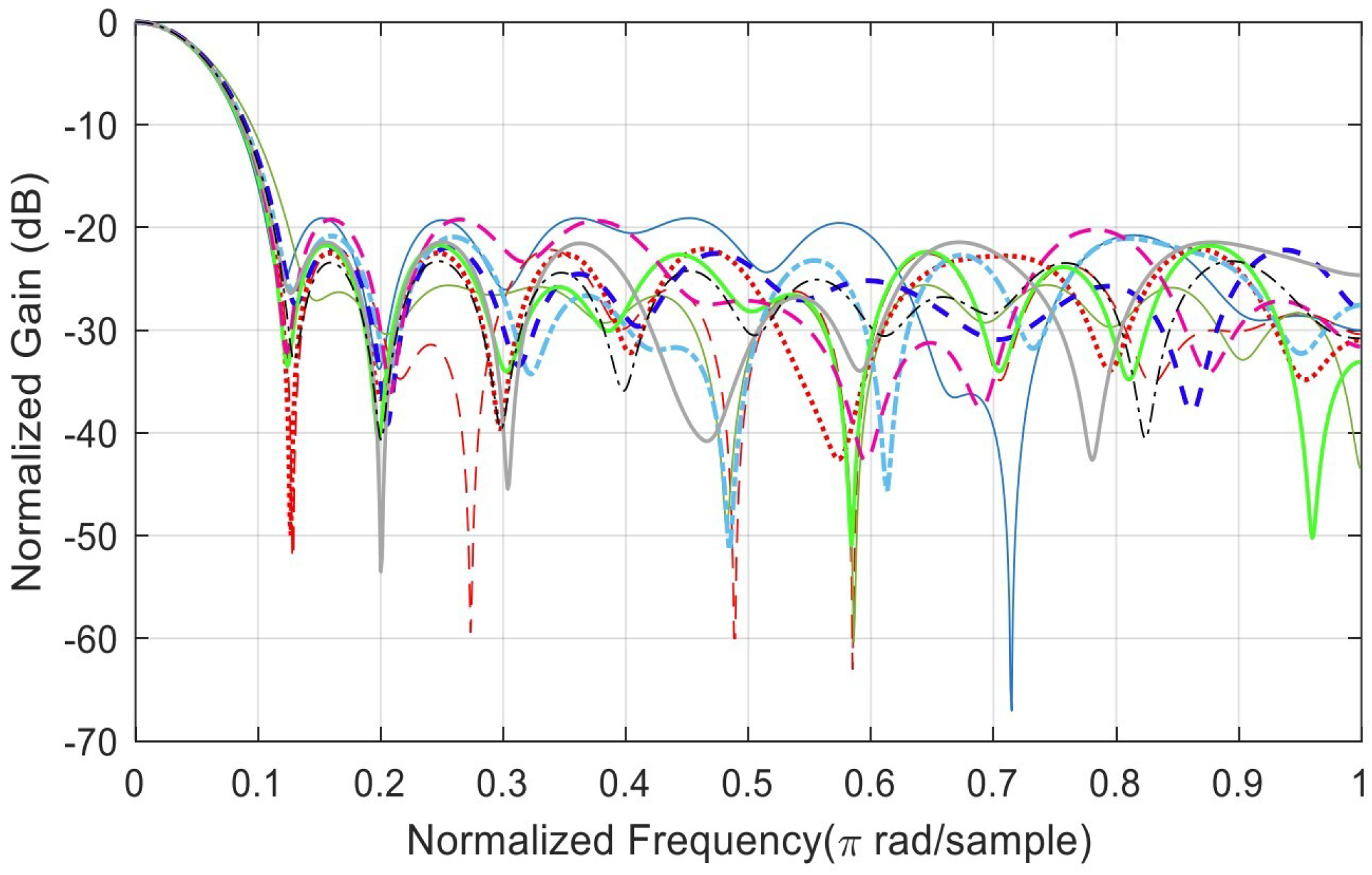
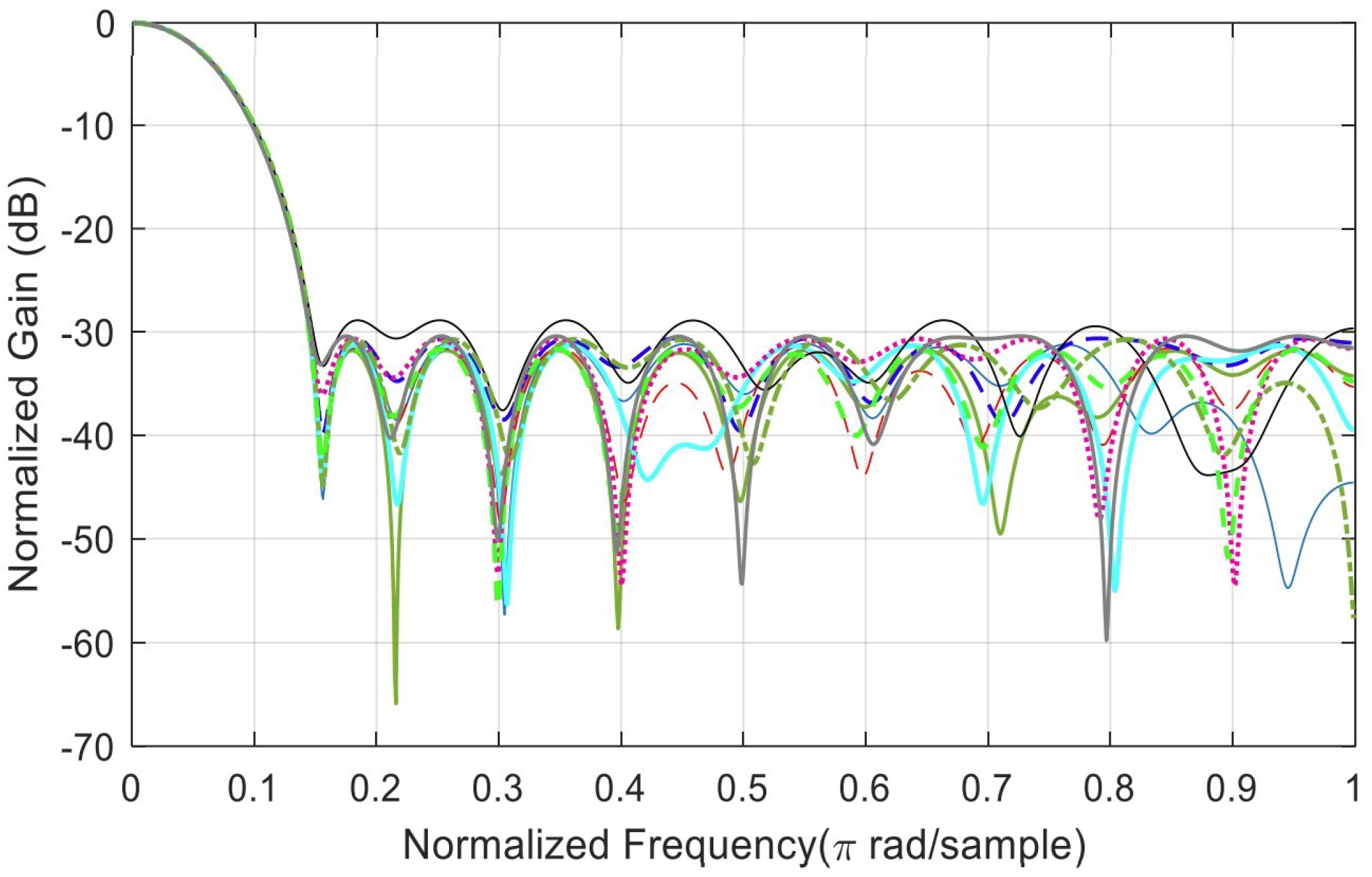
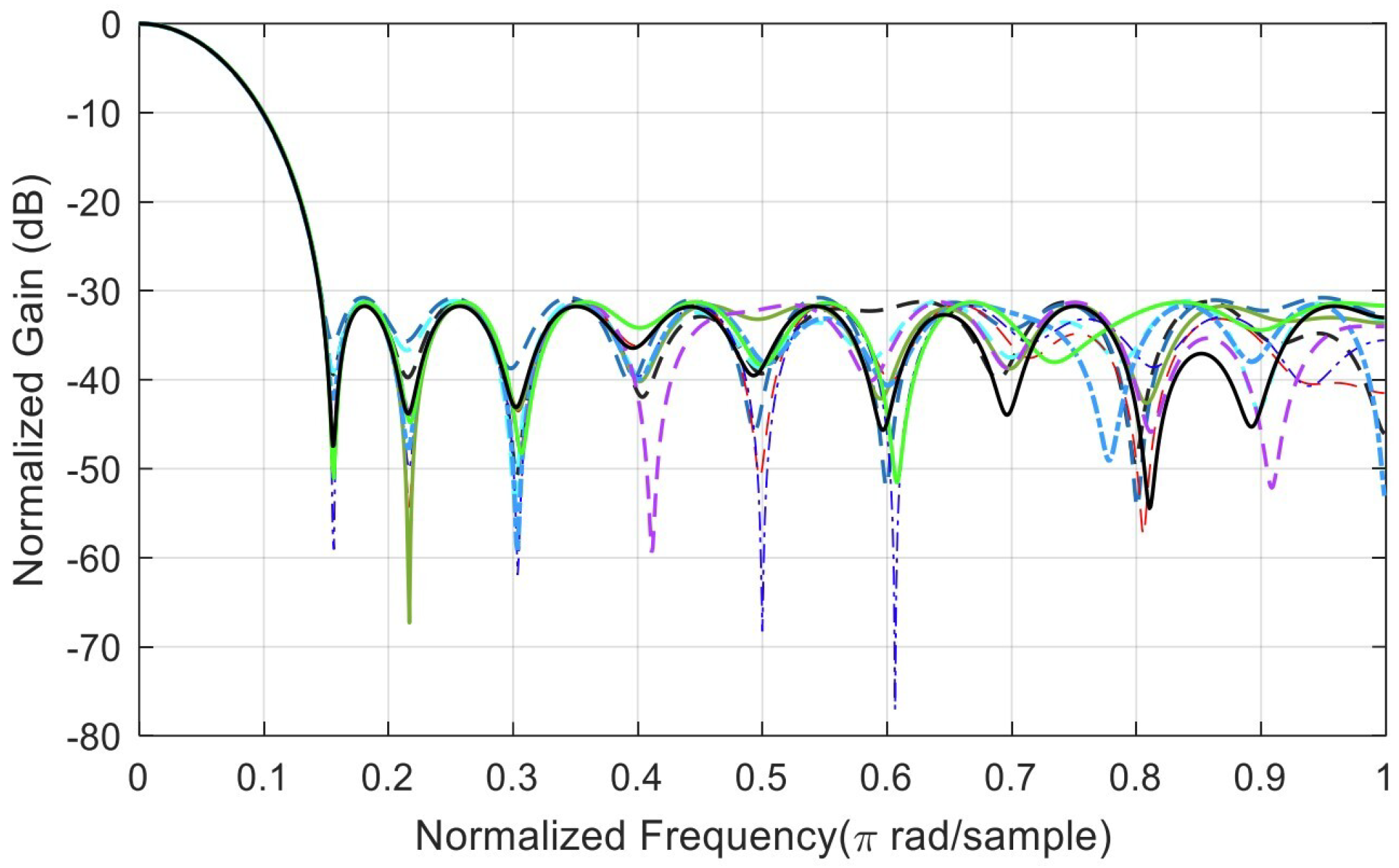

References
- Ansari, S.; Alnajjar, K.A.; Mahmoud, S.; Alabdan, R.; Alzaabi, H.; Alkaabi, M.; Hussain, A. Blind Source Separation Based on Genetic Algorithm-Optimized Multiuser Kurtosis. In Proceedings of the 2023 46th International Conference on Telecommunications and Signal Processing (TSP), Prague, Czech Republic, 12–14 July 2023; pp. 164–171. [Google Scholar] [CrossRef]
- Figueroa-García, J.C.; Neruda, R.; Hernandez-Pérez, G. A genetic algorithm for multivariate missing data imputation. Inf. Sci. 2023, 619, 947–967. [Google Scholar] [CrossRef]
- Li, C. Digital Filters. In Time Series Data Analysis in Oceanography; Cambridge University Press: Cambridge, UK, 2022; pp. 374–398. [Google Scholar] [CrossRef]
- Kaur, H.; Saini, S.; Raut, Y. The Application of Hybrid Optimization For FIR Filter Design. In Proceedings of the 2023 1st International Conference on Innovations in High Speed Communication and Signal Processing (IHCSP), Bhopal, India, 4–5 March 2023; IEEE: Piscataway, NJ, USA, 2023; pp. 386–391. [Google Scholar]
- Gen, M.; Lin, L. Genetic algorithms and their applications. In Springer Handbook of Engineering Statistics; Springer: Berlin/Heidelberg, Germany, 2023; pp. 635–674. [Google Scholar]
- Cosoli, G.; Spinsante, S.; Scardulla, F.; D’Acquisto, L.; Scalise, L. Wireless ECG and cardiac monitoring systems: State of the art, available commercial devices and useful electronic components. Measurement 2021, 177, 109243. [Google Scholar] [CrossRef]
- Chauhan, S.; Singh, M.; Aggarwal, A.K. Designing of optimal digital IIR filter in the multi-objective framework using an evolutionary algorithm. Eng. Appl. Artif. Intell. 2023, 119, 105803. [Google Scholar] [CrossRef]
- Kockanat, S.; Karaboga, N. The design approaches of two-dimensional digital filters based on metaheuristic optimization algorithms: A review of the literature. Artif. Intell. Rev. 2015, 44, 265–287. [Google Scholar] [CrossRef]
- Kolupaev, A.Y.; Tyapkin, V.N.; Dmitriev, D.D.; Ratushniak, V.N.; Tyapkin, I.V.; Smolev, I.A. Synthesis of a Filter of Phase-Domanipulated Signals with a Minimum Side-Lobe Level. J. Sib. Fed. Univ. Eng. Technol. 2023, 16, 497–508. [Google Scholar]
- Alzahrani, A.; Kanan, A. Machine Learning Approaches for Developing Land Cover Mapping. Appl. Bionics Biomech. 2022, 2022, 5190193. [Google Scholar] [CrossRef] [PubMed]
- Eldos, T.; Kanan, A.; Aljumah, A. Adapting the Ant Colony Optimization Algorithm to the Printed Circuit Board Drilling Problem. World Comput. Sci. Inf. Technol. J. 2013, 3, 100–104. [Google Scholar]
- Eldos, T.; Kanan, A.; Nazih, W.; Khatatbih, A. Adapting the Chemical Reaction Optimization Algorithm to the Printed Circuit Board Drilling Problem. Int. J. Comput. Inf. Eng. 2015, 9, 247–252. [Google Scholar]
- Prakash, R.; Yadav, S.; Kumar Saha, S. A Metaheuristic Approach for the Design of Linear Phase FIR Differentiator. In Proceedings of the 2023 International Conference on Computational Intelligence and Sustainable Engineering Solutions (CISES), Uttar Pradesh, India, 28–30 April 2023; pp. 744–748. [Google Scholar] [CrossRef]
- Maghsoudi, Y.; Kamandar, M. Low delay digital IIR filter design using metaheuristic algorithms. In Proceedings of the 2017 2nd Conference on Swarm Intelligence and Evolutionary Computation (CSIEC), Kerman, Iran, 7–9 March 2017; IEEE: Piscataway, NJ, USA, 2017. [Google Scholar] [CrossRef]
- Bhowmick, C.; Dutta, P.K.; Mahadevappa, M. Computer Aided Classification of Benign and Malignant Breast Lesions using Maximum Response 8 Filter Bank and Genetic Algorithm. In Proceedings of the 2020 IEEE Region 10 Conference (TENCON), Virtual, 16–19 November 2020; IEEE: Piscataway, NJ, USA, 2020. [Google Scholar] [CrossRef]
- Romsai, W.; Nawikavatan, A.; Hlangnamthip, S.; Puangdownreong, D. Design of Optimal Active Low-Pass Filter by Hybrid Intensified Current Search. In Proceedings of the 2022 Joint International Conference on Digital Arts, Media and Technology with ECTI Northern Section Conference on Electrical, Electronics, Computer and Telecommunications Engineering (ECTI DAMT & NCON), Chiang Rai, Thailand, 26–28 January 2022; IEEE: Piscataway, NJ, USA, 2022. [Google Scholar] [CrossRef]
- Stubberud, P. Digital IIR Filter Design Using a Differential Evolution Algorithm with Polar Coordinates. In Proceedings of the 2022 IEEE 12th Annual Computing and Communication Workshop and Conference (CCWC), Virtual, 26–29 January 2022; IEEE: Piscataway, NJ, USA, 2022. [Google Scholar] [CrossRef]
- Tripathi, K.K.; Kidwai, M.S.; Khan, I.U. Performance Analysis of Low Pass FIR Filter Design using Dynamic and Adjustable Particle Swarm Optimization Techniques. In Proceedings of the 2021 10th International Conference on System Modeling & Advancement in Research Trends (SMART), Moradabad, India, 10–11 December 2021; IEEE: Piscataway, NJ, USA, 2021. [Google Scholar] [CrossRef]
- Miyata, T.; Asou, H.; Aikawa, N. Design Method for FIR Filter with Variable Multiple Elements of Stopband Using Genetic Algorithm. In Proceedings of the 2018 IEEE 23rd International Conference on Digital Signal Processing (DSP), Shanghai, China, 19–21 November 2018; IEEE: Piscataway, NJ, USA, 2018. [Google Scholar] [CrossRef]
- Guliashki, V.; Marinova, G. An Approach for Coefficients optimization in Adaptive Filter Signal Equalization. In Proceedings of the 2022 29th International Conference on Systems, Signals and Image Processing (IWSSIP), Sofia, Bulgaria, 1–3 June 2022; IEEE: Piscataway, NJ, USA, 2022. [Google Scholar] [CrossRef]
- Debnath, K.; Dhabal, S.; Venkateswaran, P. Design of High-Pass FIR Filter using Arithmetic Optimization Algorithm and its FPGA Implementation. In Proceedings of the 2022 IEEE Region 10 Symposium (TENSYMP), Mumbai, India, 1–3 July 2022; IEEE: Piscataway, NJ, USA, 2022. [Google Scholar] [CrossRef]
- Chauhan, R.S.; Arya, S.K. An Optimal Design of FIR Digital Filter Using Genetic Algorithm. In Communications in Computer and Information Science; Springer: Berlin/Heidelberg, Germany, 2011; pp. 51–56. [Google Scholar] [CrossRef]
- Chandra, A.; Kumar, A.; Roy, S. Design of FIR filter ISOTA with the aid of genetic algorithm. Integration 2021, 79, 107–115. [Google Scholar] [CrossRef]
- Dliou, A.; Elouaham, S.; Laaboubi, M.; Zougagh, H.; Saddik, A. Denoising Ventricular tachyarrhythmia Signal. In Proceedings of the 2018 9th International Symposium on Signal, Image, Video and Communications (ISIVC), Rabbat, Morocco, 27–30 November 2018; IEEE: Piscataway, NJ, USA, 2022; pp. 124–128. [Google Scholar]
- Elouaham, S.; Latif, R.; Dliou, A.; Maoulainine, F.; Laaboubi, M. Biomedical signals analysis using time-frequency. In Proceedings of the 2012 IEEE International Conference on Complex Systems (ICCS), Agadir, Morocco, 5–6 November 2012; IEEE: Piscataway, NJ, USA, 2012; pp. 1–6. [Google Scholar]
- Elouaham, S.; Dliou, A.; Laaboubi, M.; Latif, R.; Elkamoun, N.; Zougagh, H. Filtering and analyzing normal and abnormal electromyogram signals. Indones. J. Electr. Eng. Comput. Sci. 2020, 20, 176–184. [Google Scholar] [CrossRef]
- Strohmenger, H.U.; Lindner, K.H.; Brown, C.G. Analysis of the ventricular fibrillation ECG signal amplitude and frequency parameters as predictors of countershock success in humans. Chest 1997, 111, 584–589. [Google Scholar] [CrossRef] [PubMed]





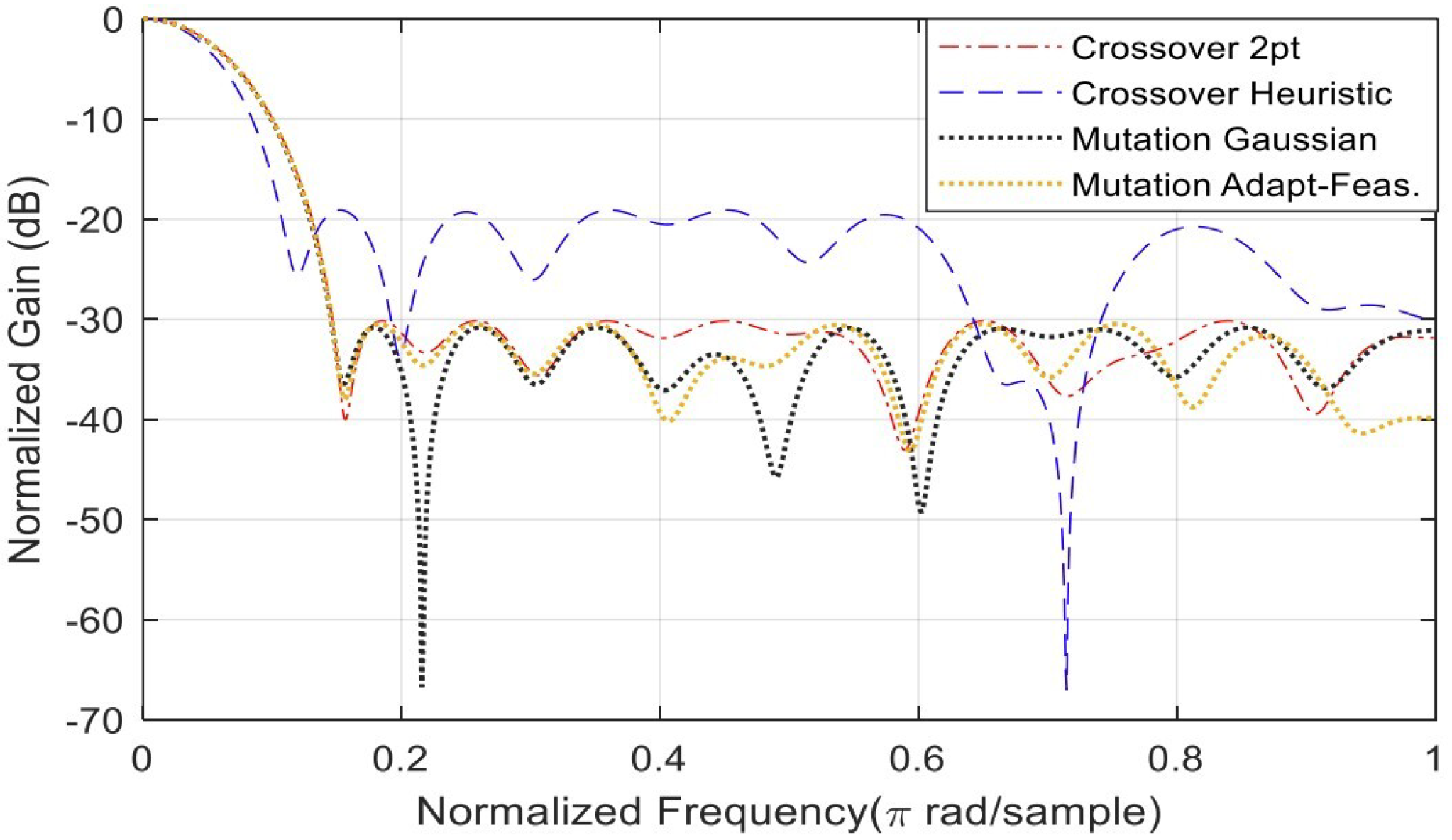
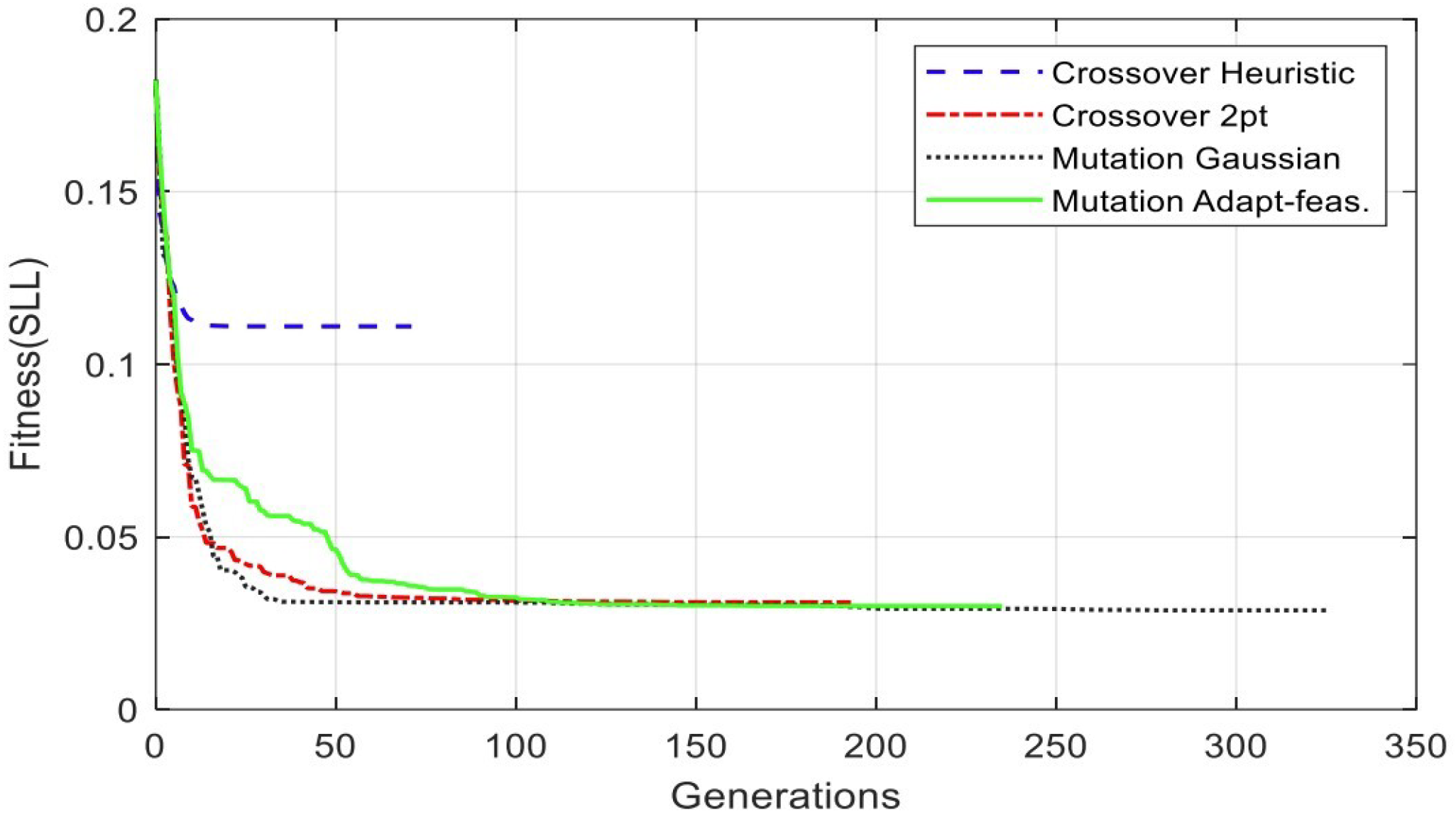
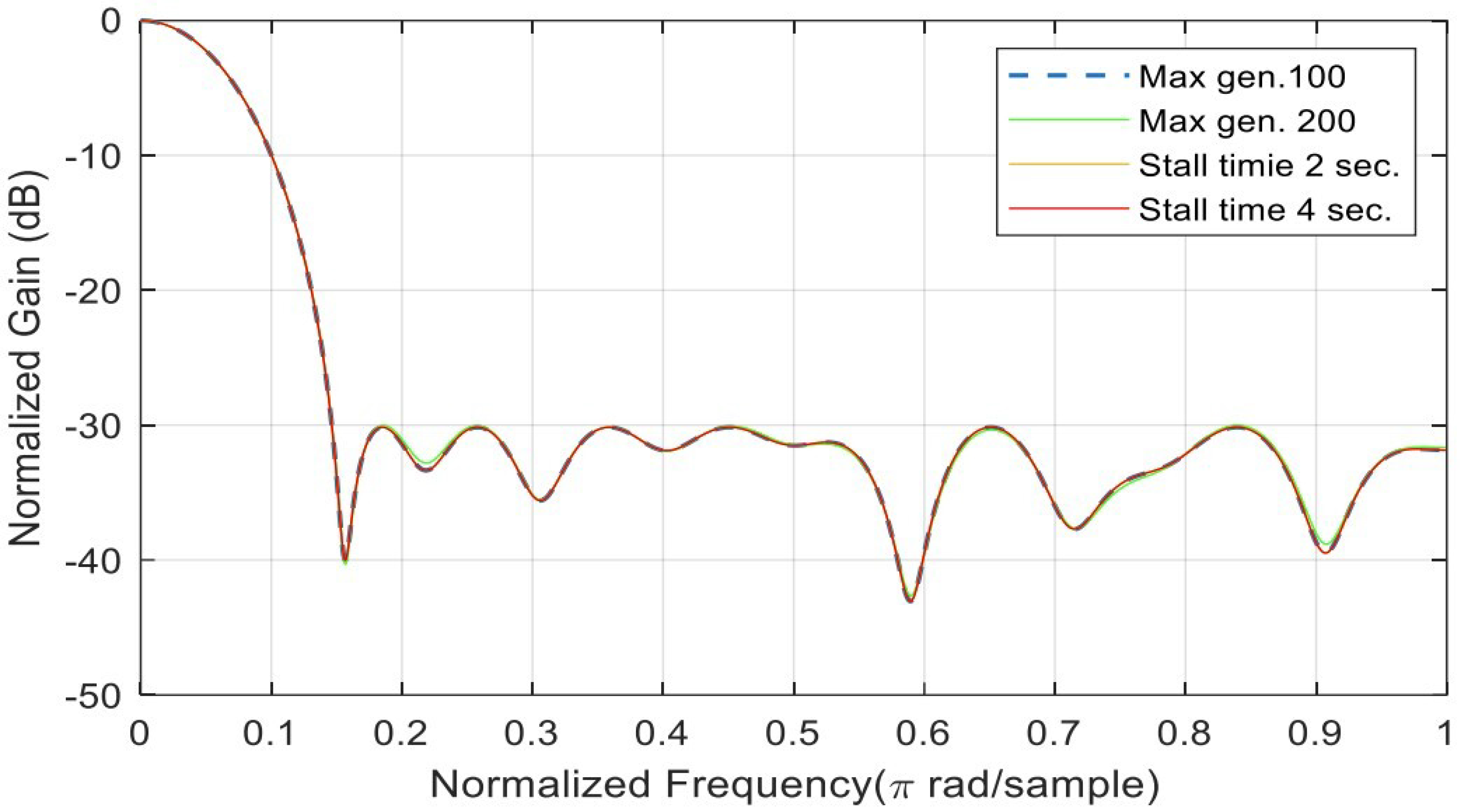
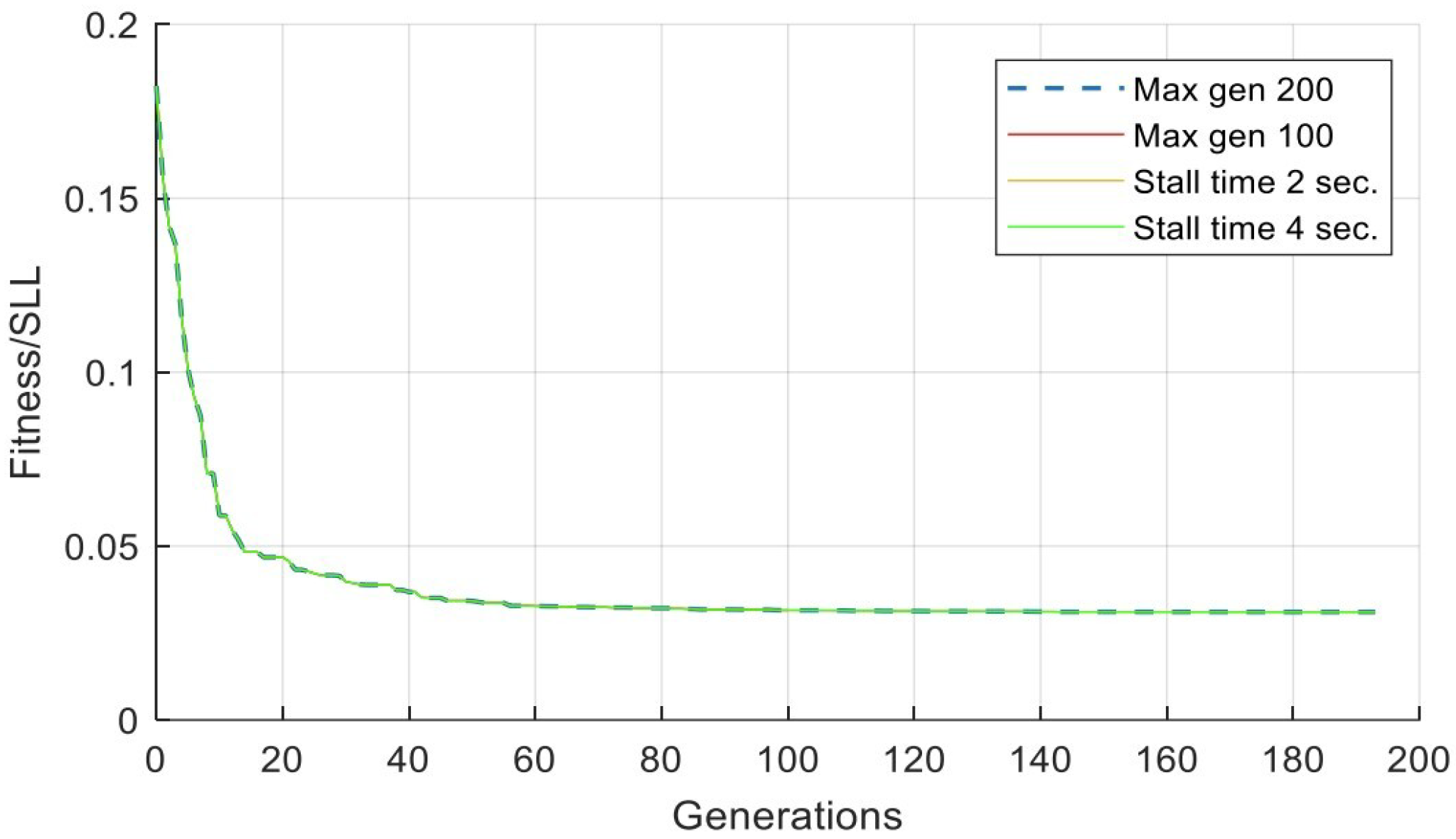
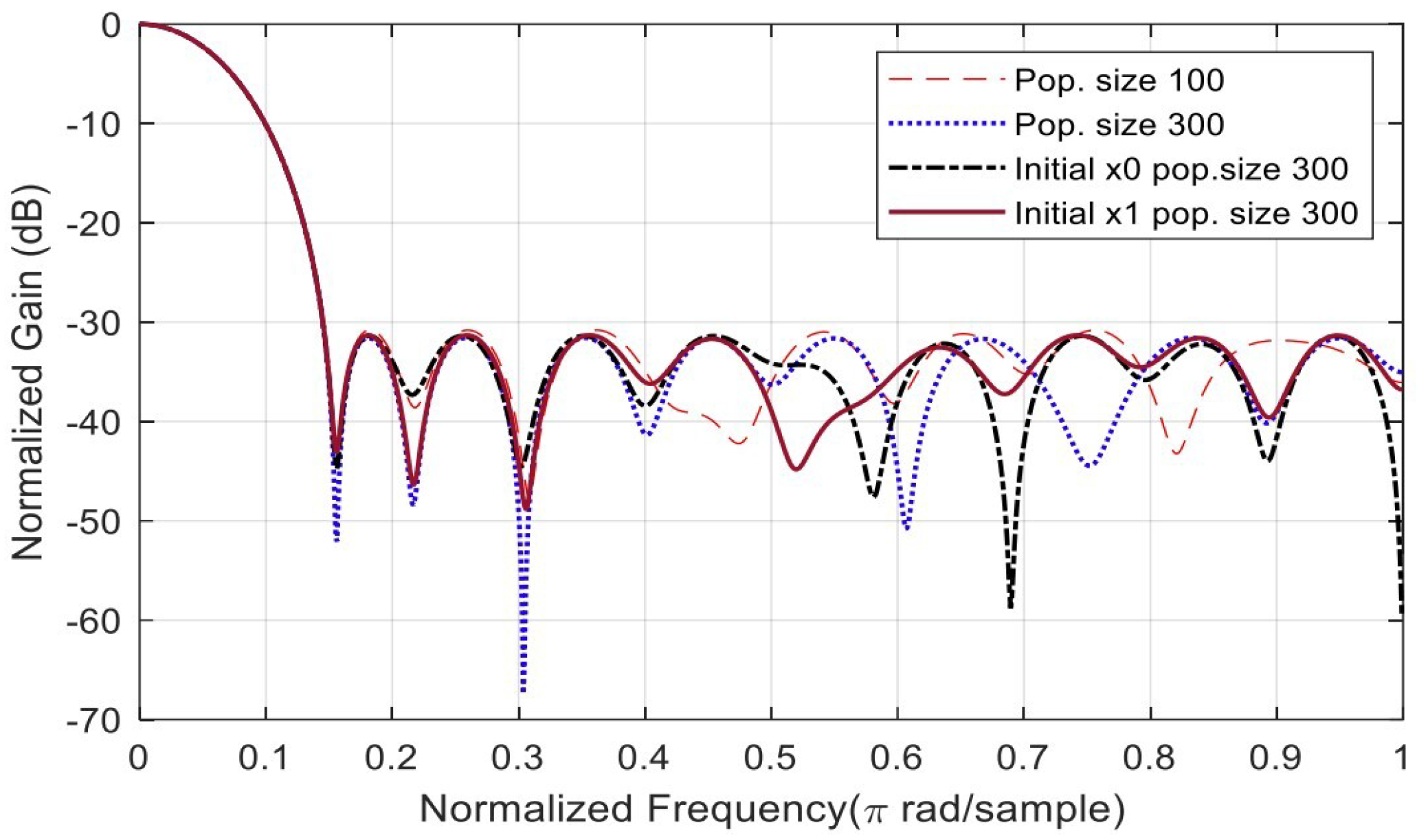




| Alg. Setting | No. | Sub-Function Solver Setting | SSL (dB) | Gen. No. Convergence |
|---|---|---|---|---|
| Crossover | 1 | Heuristic | −19.0941 | 71 |
| Crossover | 2 | Two-point | −30.1599 | 193 |
| Mutation | 1 | Adapt-Feasible | −30.4694 | 235 |
| Mutation | 2 | Gaussian | −30.8327 | 236 |
| Solver Option | No. | Sub-Function Solver Setting | SSL (dB) | Gen. No. Convergence |
|---|---|---|---|---|
| Max gens | 1 | 100 | −30.0219 | 100 |
| Max gens | 2 | 200 | −30.1599 | 193 |
| Stall time limit | 1 | 2 s | −30.1599 | 193 |
| Stall time limit | 2 | 4 s | −30.1599 | 193 |
| Pop. Setting | No. | Sub-Function Solver Setting | SSL (dB) | Gen. No. Convergence |
|---|---|---|---|---|
| Pop. Size | 1 | 100 | −30.7976 | 253 |
| Pop. Size | 2 | 300 | −31.5810 | 235 |
| Initial. Pop | 1 | Initial Pop. () | −31.3811 | 274 |
| Initial. Pop | 2 | Initial Pop. () | −31.3141 | 202 |
| Solver. Options | No. | Sub-Function Solver Setting | SSL (dB) | Gen. No. Convergence |
|---|---|---|---|---|
| Fun. tolerance | 1 | −29.9617 | 92 | |
| Fun. tolerance | 2 | −30.1490 | 148 | |
| Fitness limit | 1 | 0.01 | −26.3296 | 14 |
| Fitness limit | 2 | 0.001 | −30.1599 | 193 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hamici, H.; Kanan, A.; Al-hammuri, K. Optimized FIR Filter Using Genetic Algorithms: A Case Study of ECG Signals Filter Optimization. BioMedInformatics 2023, 3, 1197-1215. https://doi.org/10.3390/biomedinformatics3040071
Hamici H, Kanan A, Al-hammuri K. Optimized FIR Filter Using Genetic Algorithms: A Case Study of ECG Signals Filter Optimization. BioMedInformatics. 2023; 3(4):1197-1215. https://doi.org/10.3390/biomedinformatics3040071
Chicago/Turabian StyleHamici, Houssam, Awos Kanan, and Khalid Al-hammuri. 2023. "Optimized FIR Filter Using Genetic Algorithms: A Case Study of ECG Signals Filter Optimization" BioMedInformatics 3, no. 4: 1197-1215. https://doi.org/10.3390/biomedinformatics3040071
APA StyleHamici, H., Kanan, A., & Al-hammuri, K. (2023). Optimized FIR Filter Using Genetic Algorithms: A Case Study of ECG Signals Filter Optimization. BioMedInformatics, 3(4), 1197-1215. https://doi.org/10.3390/biomedinformatics3040071







