Abstract
Semantic web technologies (SWT) represent data in a format that is easier for machines to understand. Validating the knowledge in data graphs created using SWT is critical to ensure that the axioms accurately represent the so-called “real” world. However, data graph validation is a significant challenge in the semantic web domain. The Shapes Constraint Language (SHACL) is the latest W3C standard developed with the goal of validating data-graphs. SHACL (pronounced as shackle) is a relatively new standard and hitherto has predominantly been employed to validate generic data graphs like WikiData and DBPedia. In generic data graphs, the name of a class does not affect the shape of a class, but this is not the case with biomedical ontology data graphs. The shapes of classes in biomedical ontology data graphs are highly influenced by the names of the classes, and the SHACL shape creation methods developed for generic data graphs fail to consider this characteristic difference. Thus, the existing SHACL shape creation methods do not perform well for domain-specific biomedical ontology data graphs. Maintaining the quality of biomedical ontology data graphs is crucial to ensure accurate analysis in safety-critical applications like Electronic Health Record (EHR) systems referencing such data graphs. Thus, in this work, we present a novel method to create enhanced SHACL shapes that consider the aforementioned characteristic difference to better validate biomedical ontology data graphs. We leverage the knowledge available from lexical auditing techniques for biomedical ontologies and incorporate this knowledge to create smart SHACL shapes. We also create SHACL shapes (baseline SHACL graph) without incorporating the lexical knowledge of the class names, as is performed by existing methods, and compare the performance of our enhanced SHACL shapes with the baseline SHACL shapes. The results demonstrate that the enhanced SHACL shapes augmented with lexical knowledge of the class names identified 176 violations which the baseline SHACL shapes, void of this lexical knowledge, failed to detect. Thus, the enhanced SHACL shapes presented in this work significantly improve the validation performance of biomedical ontology data graphs, thereby reducing the errors present in such data graphs and ensuring safe use in the life-critical applications referencing them.
1. Introduction
Semantic web technologies (SWT), like the Web Ontology Language (OWL) and Resource Description Framework (RDF) [1], represent data in a format that is easy for machines to understand and interpret. This allows machines to process data intelligently and derive new information. Given these benefits, information systems are increasingly representing their data in SWT-compliant RDF and OWL data graphs. The healthcare domain has also adopted the use of SWT for efficient information exchange and improved semantic interoperability. For example, biomedical ontologies that represent clinical information about diseases, procedures, diagnoses, etc. have found SWT to be quite beneficial. The Systematized Nomenclature Of Medicine–Clinical Terms (SNOMED-CT) [2], which is one of the most widely adopted biomedical ontologies in the world and consists of more than 300,000 biomedical concepts, has also released an SWT-complaint version: SNOMED-CT OWL [3].
Validating the knowledge represented in these data graphs is critical, ensuring that the RDF and OWL statements are accurate and correspond to the so-called “real” world. However, data graph validation is a significant challenge in the semantic web domain [4]. SWT (like OWL and RDF) were developed with the goal of inferring new data rather than validating existing ones, and therefore using them as validation technologies is not a straightforward task. Using OWL as a validation technology requires employing many counterintuitive mechanisms, which were developed originally for inferring and modifying them to perform the task of validation, which is not very straightforward [5]. For example, OWL restrictions are not used to constrain data but rather used to make inferences from the existing data. This leads to many unexpected results when employing OWL restrictions as a validation mechanism. For example, if we assume that the restriction owl:maxCardinality 1 states that a person can only have one value for the property hasFather, then the assumption would be incorrect. Instead if a person is assigned two values for the property hasFather, then the OWL processor would assume that both these values represent the same real world entity. Figure 1 illustrates this with the help of an example. In the figure, if an OWL ontology states that the rdfs:range of the property hasFather is the class Person, and a data graph only contains a triple stating John hasFather Bob, then the OWL processor will not assume that Bob is not an instance of class Person. In fact, the processor will assume the opposite and automatically infer the triple Bob rdf:type Person instead of reporting this as a violation. This happens because OWL was developed with the goal of inferring rather than validating. This is the reason why OWL cannot be directly employed in the task of validation, as this may lead to unexpected outcomes. OWL is based on the open-world assumption (OWA) and does not assume the absence of a statement as a violation or a false statement. Instead, it infers the missing statement from the existing statement. This can be counterintuitive and lead to unwanted outcomes in validation tasks.
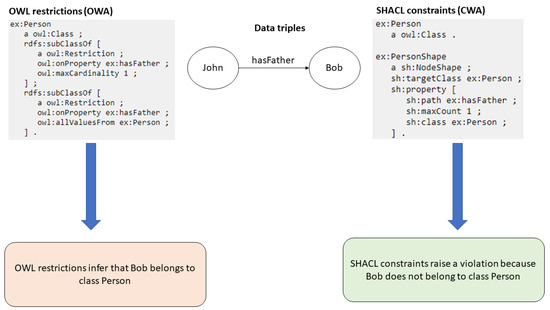
Figure 1.
OWL (OWA) vs. SHACL (CWA).
In the absence of a dedicated validation standard for SWT, the counterintuitive mechanisms of OWL, developed for inferring, were employed in the validation of data graphs [5]. For example, OWL and RDF data graphs were validated by writing SPARQL queries that tested for the presence or absence of certain triples by expressing relevant conditions in the WHERE clause of the SPARQL query. However, this process was ad hoc and lacked structure. Given these pitfalls, W3C developed the Shapes and Constraint Language (SHACL) [1], a dedicated standard with the predominant goal of validation. SHACL (pronounced as shackle) is based on the closed-world assumption (CWA), and whenever a statement is absent, it assumes the statement to be false instead of inferring a new triple (as is performed by OWL), and it reports this as a violation in the data graph (see Figure 1). This makes SHACL highly suitable for validation. A SHACL processor takes a data graph and a shapes graph as the input and outputs a validation report, which reports violations if any of the constraints mentioned in the shapes graph are not followed by the data graph. Figure 2 illustrates the process of SHACL validation with the help of an example. As can be seen from the figure, a SHACL shapes graph (on the left side) provides a shape for the class Person by listing all properties along with the restrictions for that class. In the data graph (on the right side), it can be seen that the birthdate of Robert, who belongs to the class Person, violates the SHACL constraint for the property birth date, sh:lessThan schema:deathDate, and therefore is reported as a violation in the validation report (at the bottom).
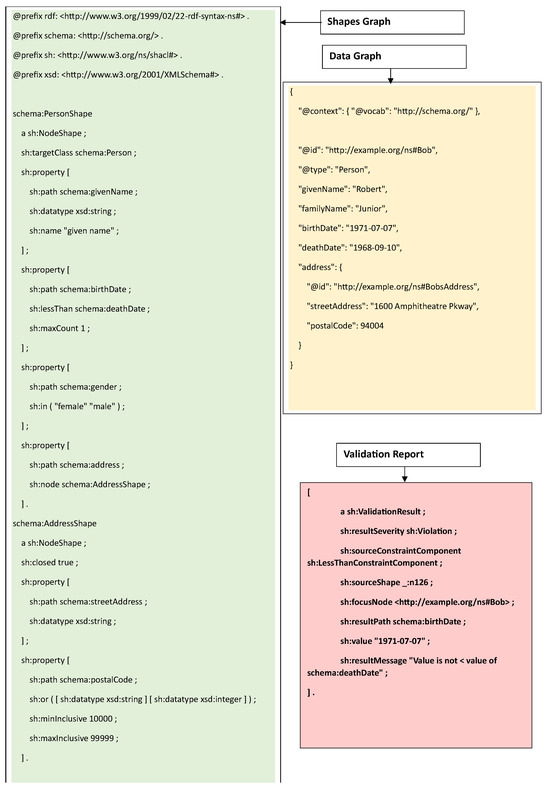
Figure 2.
Validation using Shapes and Constraint Language (SHACL).
Despite the availability of a dedicated validation standard for data graphs, given the volume and variety of existing data graphs, generating effective SHACL shapes that identify inaccurate or missing statements in a data graph is a challenging task [6]. Furthermore, SHACL is a relatively new standard and has hitherto been predominantly applied to generic data graphs. There is a need to develop effective SHACL shape creation methods and test the technology for domain-specific data graphs representing specialized knowledge. Domain-specific data graphs, like biomedical ontology data graphs, are inherently different from generic data graphs (WikiData, DBpedia, etc.) and have specific requirements which need to be satisfied in order to successfully validate their knowledge. Furthermore, maintaining the quality of biomedical ontology data graphs to the highest standard is crucial as they are referenced by safety-critical applications like Hospital Information Systems (HISs), automated decision-making systems, and EHR systems. Erroneous representation of clinical concepts in biomedical ontology data graphs can lead to inaccurate analysis and serious consequences in such safety-critical applications. Given this indispensability, in this paper, we present a novel SHACL shape creation method explicitly catering to the characteristic needs of domain-specific data graphs representing biomedical ontologies. The presented method aids SHACL shape creation for a biomedical ontology data graph by leveraging the lexical features and hidden semantics present in biomedical ontology data graph class names. Incorporating this lexical knowledge in SHACL shapes helps to create enhanced SHACL shapes which are better suited to validating domain-specific biomedical ontology data graphs. We also compare the enhanced shapes created by our method with the shapes created using existing methods that do not incorporate the lexical knowledge into SHACL shapes. The results of the comparison demonstrate that the proposed approach significantly improved the validation performance for domain-specific biomedical ontology data graphs by identifying 176 violations which could not be identified by a baseline SHACL shape graph created using existing methods for generic data graphs. Thus, our enhanced SHACL shapes helped reduce the errors present in biomedical ontology data graphs, thereby ensuring safe use in life-critical applications referencing them.
2. Related Work
Many techniques have been developed to create good-quality SHACL shape graphs which are effective at catching violations in a data graph. The majority of the existing methods employed in the creation of SHACL shapes either convert ontology constraints (OWL axioms) into SHACL constraints [7,8,9] or use data-driven approaches that use machine learning (ML) algorithms to predict the cardinalities of properties and then convert these cardinalities into SHACL (sh:minCount/sh:maxCount) constraints [9]. Some methods employ semantic profiling tools that combine data-driven and statistical approaches [10]. Finally, in cases where SHACL shapes cannot be predicted a priori, the “data graph first, shapes graph later” approach is used, which creates a shapes graph from an existing RDF data graph [11,12,13]. To address the challenge of automation in SHACL shape creation, Cimmino et al. [6] presented a tool (named Astrea) that refers to a knowledge base of ontology constraint patterns mapped to SHACL constraints in order to automatically create SHACL shapes for a data graph. Detailed summaries of the existing SHACL shape creation methods and their comparison based on parameters like whether the shapes are extracted from data (data-driven or semantic profiling approach) or extracted from ontology (using ontology axioms) and whether or not the method creates SHACL shapes automatically is available in the literature [6,14]. The majority of the aforementioned papers as well as the works studied in these literature reviews involve creating SHACL shapes for generic data graphs. As a result, the techniques mentioned above follow a data-driven or statistical approach to predicting cardinalities, and therefore, the SHACL constraints are solely created using the external features of a class in a data graph (In this work, we refer to OWL and RDF axioms representing the properties of a class as external features and the lexical features of a class name as internal features.). Despite these differences, one of the limitations identified by Rabbani et al. [14] states that complete SHACL shapes are not created by any of the existing methods, and the semantics of the object properties are often not reflected in the SHACL constraints unless they are explicitly present as OWL axioms in the ontology (e.g., to indicate that objects for “takes course” should be of the literal type “Course”). This supports our hypothesis that the semantics of class names and property names contain valuable information that can be used to create richer and more effective SHACL shapes.
As stated earlier, SHACL is a relatively new standard [1] (developed in 2017) and has mainly been tested on generic data graphs like DBpedia and WikiData [11,13]. Very few initiatives exist in creating SHACL shapes for biomedical data graphs, and they mainly focus on data graphs representing Electronic Health Record (EHR) models [15] and patient information [16]. Other clinical use cases for SHACL shapes include validating medical guidelines to integrate Fast Healthcare Interoperability Resources (FHIR) into decision-making systems [17], validating medical reports to identify missing data [18], and validating clinical trial study data to detect missing values, wrong cardinalities, and incorrect values that do not adhere to a predefined set [16]. However, these biomedical data graphs are not as sensitive to class name semantics as biomedical ontology data graphs. Thus, the nature of SHACL shape creation methods employed in these studies is similar to generic data graphs, which include Ontology Design Patterns (ODPs) and existing clinical reference model constraints being converted to SHACL shape constraints. For example, the authors of [16] derived SHACL shapes to regulate the values for fields like gender, study ID, and study type in a clinical trial report. Developing SHACL shapes for such fields is mainly focused on constraining the data types and values for such fields rather than ensuring the presence of missing properties in a class based on the semantics of the class name, which is the case with biomedical ontology data graphs.
Limited works exist where SHACL shapes were created for biomedical ontologies in particular. For example, the Swiss Personalized Health Network (SPHN) [19,20] provides a tool, named SHACLer, that caters toward validation of biomedical ontologies including SNOMED-CT. SNOMED-CT shapes [21] are in another study that creates SHACL shapes for SNOMED-CT, but the goal of this study was not validation but rather the representation of SNOMED-CT in an SWT-compliant format, which is easier to integrate with healthcare applications and other biomedical ontologies. The existing OWL representation of SNOMED-CT depicts properties as a combination of OWL intersections and restrictions, which makes the ontology (data graph representation) too complex to query or integrate with other biomedical ontologies [3]. The persistent demand of representing SNOMED-CT in a simplified SWT-compliant format led to the development of SNOMED-CT shapes [21], which creates a simplified representation of SNOMED-CT OWL and makes the ontology easily integrable with other biomedical ontologies as well as healthcare applications that refer to SNOMED-CT.
SNOMED-CT shapes [21] provide a feature for converting the simplified SNOMED-CT RDF representation into SHACL shapes. However, since the main goal of developing SNOMED-CT shapes was ease of interoperability with other biomedical ontologies and ease of querying, the SHACL shapes created by [21] simply convert existing ontology axioms into SHACL constraints. Similarly, SHACLer, the module responsible for creating SHACL shape graphs in SPHN [19], again generates all shape constraints based on the existing axioms of the class in RDF schema. Thus, all restrictions and cardinality constraints are created based on the existing RDF schema. This leaves us with SHACL shapes that are completely dependent on the richness of ontology axioms.
It has been demonstrated that biomedical ontologies are richer in natural language content than OWL axioms and logical definitions [22]. Although auditing techniques are working toward enriching OWL axioms with the lexical knowledge available in concept names [23], this is a gradual process. Thus, the existing OWL axioms do not sufficiently represent the lexical richness of concept names. As a result, the SHACL shapes created by simply converting existing ontology axioms into SHACL constraints fail to capture the lexical richness available in the class names (i.e., concept names). Furthermore, as stated earlier in this section, SHACL is a relatively new technology and has not been sufficiently tested on domain-specific data graphs representing biomedical ontology knowledge. While the exclusive conversion of ontology axioms into SHACL constraints may work for generic data graphs like WikiData and DBpedia [11,13,24], data graphs representing biomedical ontological knowledge are inherently different in nature.
For instance, consider the previous example of the class Person (see Figure 2). The name of a person (i.e., the number of words or other lexical features appearing in a person’s name) will not affect the shape of the Person class. Robert as well as any other name containing any number of words will still have the same properties (name, age, gender, birth date, and address). Now, consider the clinical concepts Burn of skin (disorder) and Burn caused by fire (disorder) belonging to the Disorder class in a biomedical ontology. As one can see in Figure 3, depending on the lexical features of the name of the disorder, the properties (attribute relationships) appearing in the concept definition change (i.e., the shape of the class) are dependent on the lexical features of the concept name. While the concept Burn caused by fire (disorder) has the properties Associated morphology, Due to, and Causative agent (owing to the stop word caused by in its name), the concept Burn of skin (disorder) has the properties Associated morphology, Due to, and Finding site (given the presence of a body structure in its name). This example illustrates the influence that class names have on the properties and thereby the shape of a class in a biomedical ontology data graph.
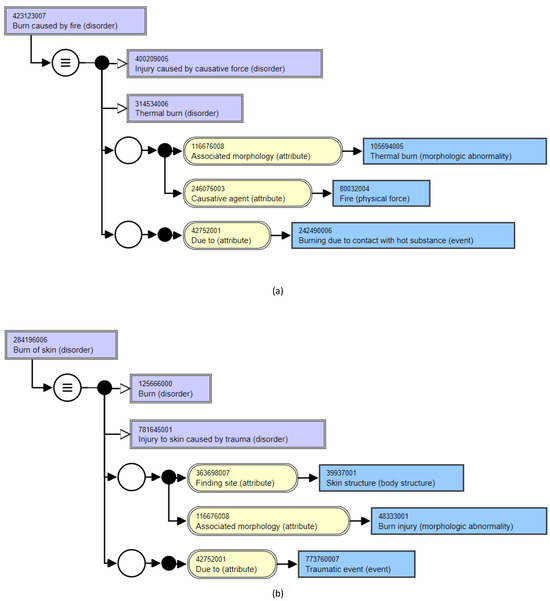
Figure 3.
Example to illustrate the characteristic difference of a biomedical ontology data graph. The shape of a disorder is dependent on the lexical features in its name. (a) Burn caused by fire (disorder). (b) Burn of skin (disorder).
Owing to the novelty of the technology, SHACL has not been widely tested on biomedical ontology data graphs. The limited biomedical data graphs validated using SHACL mainly represent clinical reports, EHR models, and FHIR resources, which (despite belonging to the same domain) are not as heavily influenced by the semantics of a class name as biomedical ontology data graphs. There is a need to test the new technology for validation of biomedical ontologies, which form the backbone of the aforementioned clinical applications. Biomedical ontologies provide data to EHR systems and clinical reports. Thus, validating biomedical ontology data graphs is crucial. The existing SHACL shape creation methods created for generic data graphs, which do not consider the lexical features of a class name while creating SHACL shapes, may not be sufficient for such domain-specific biomedical ontology data graphs. We hypothesize that there is a need to devise a method that creates enhanced SHACL shapes and is better suited for validation of biomedical ontology data graphs, which take into consideration the lexical richness of class names and effectively capture violations in biomedical ontologies.
3. Materials and Methods
In this work, we propose a novel approach for creating enhanced SHACL shapes that reflect the internal features (i.e., hidden semantics) of a biomedical ontology data graph. Our method is based on the hypothesis that, unlike generic data graphs, the shape of a class in a biomedical ontology data graph is dependent on the lexical features of the class name. To this end, our method creates SHACL shapes by augmenting the hidden semantics of class names (referred to as internal features) along with OWL and RDF axioms (referred to as external features) in the shape creation process. We refer to this graph as a SHACLex graph (and the corresponding data graph as an RDFLex graph). To demonstrate the benefits of our method, we also create a SHACL shape graph by exclusively converting OWL and RDF axioms (external features) into SHACL constraints (without considering the lexical features of the class names), as can be performed by existing methods [19,25]. We refer to this graph as a SHACL graph (and the corresponding data graph as an RDF graph). We then compare the validation performance of the SHACLex graph with the baseline SHACL graph.
The manner in which hidden semantics affect the shape of a class in a biomedical ontology data graph can be studied by analyzing the knowledge and insights gained from decades of lexical auditing techniques available in the literature [26]. To demonstrate the extensive nature of our proposed approach, we chose the knowledge gained from two lexical auditing techniques [27,28]. These two methods are quite different in nature and target different types of inconsistencies. More specifically, the method in [27] captures violations in class shapes related to attribute relationships, whereas the one in [28] captures violations related to hierarchical relationships (please refer to Appendix A for further details about the two types of relationships). These diverse methods were chosen to demonstrate the extensive nature of our approach and how it can be tailored to any of the existing auditing techniques available in the literature to create more efficient SHACL shapes. In particular, the authors of [27] showed that stop words in biomedical concept names carry significant semantic information which can be used to identify missing attribute relationships. Their work analyzed the semantic and lexical patterns of around 11 stop words and provided a list of mandatory attribute relationships that should be present in the concept definition, based on the presence of a stop word, to complete its definition. To demonstrate this, the authors of [27] created 26 sample sets for 11 stop words (see Table 1) which consisted of SNOMED-CT concepts that exhibited the same semantic and lexical pattern and therefore demanded the presence of a mandatory attribute in its logical definition. Table 1 lists all the sample sets along with the exhibited mandatory relationships, which are used to build a knowledge base in this work. Likewise, the authors of [28] stated that if a biomedical concept name consisted of an adjective followed by a noun (ADJ NOUN), then that concept gave more information about the NOUN concept and, as a result, demanded the presence of a hierarchical relationship between the two concepts (please refer to Appendix A for the detailed steps involved in the two studies [27,28]).

Table 1.
Sample-sets and the corresponding mandatory attribute relationships from [27].
In our method, we use the insights summarized from [27,28] to create enhanced SHACL shapes for concepts conforming to the lexical patterns. As a part of this, we follow the methodology presented in these two studies and obtain results that demonstrate the influence of the lexical features of a concept name on the relationships present in its definition (see Appendix A). The obtained results are then organized into a knowledge base (KB) which is used by our method while constructing enhanced (SHACLex) shapes for the SNOMED-CT concepts (see Figure 4b). The information from this KB is also considered while creating RDFLex data graphs (see Figure 5), which will be explained in detail in the next section. As stated earlier, we also create baseline (SHACL) shapes by exclusively converting the existing RDF and OWL axioms into SHACL constraints without augmenting the information gained from the created KB, as performed by the existing methods [19,25] (see Figure 4a). We then compare the validation performance of the SHACLex graph constructed using our method with the baseline SHACL graph. Figure 4 provides an overview of the proposed approach, where (a) represents the baseline method for SHACL and RDF graph creation and validation and (b) represents the proposed method for SHACLex and RDFLex graph creation and validation, incorporating additional lexical knowledge from the auditing techniques. The remainder of this section explains the experimentation and evaluation.
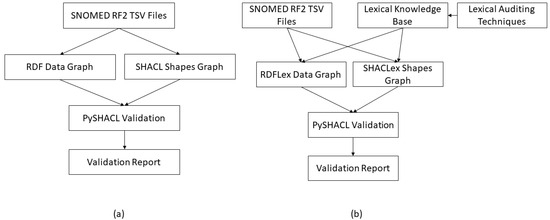
Figure 4.
(a) SHACL validation process without considering internal lexical features of class names (baseline). (b) SHACL validation process after augmenting internal lexical features of class names in the SHACL shape construction.
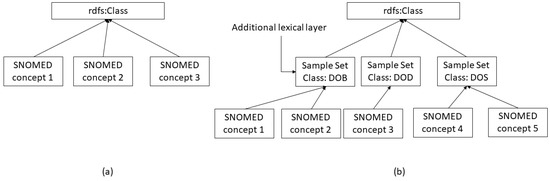
Figure 5.
(a) RDF data graph structure representing SNOMED-CT concepts targeted by non-conjunctive stop word method. (b) RDFLex data graph structure representing SNOMED-CT concepts targeted by non-conjunctive stop word method.
To assess the utility of our method, we used the Release Format 2 (RF2) files of the January 2021 international version of SNOMED-CT [2], one of the world’s most widely used biomedical ontologies. As per the requirements of [27,28], the Disorder hierarchy of SNOMED-CT was used during this experimentation. To create a data graph, the records in the RF2 files (tab-delimited) were converted into simple RDF triples. We used the inferred relationship RF2 files while creating the data graphs. The rules from the SNOMED-CT OWL guide were followed during this conversion. For example, each SNOMED-CT concept was represented as an RDFS class. The name of the class consisted of the prefix snomed followed by the SNOMED-CT identifier (SCTID) that uniquely identifies a SNOMED-CT concept. Two RDF properties, attribute and hierarchical, were created. All IS-A relationships (SCTID:116680003) were added as a subPropertyOf hierarchical, and all other relationships, including roleGroup (SCTID:609096000), were added as a subPropertyOf attribute. Within the concept definition, IS-A relationships were represented using the rdfs:subClassOf property as stated in the SNOMED-CT OWL guide. Furthermore, to simplify the data graph, only fully specified names (FSNs) of SNOMED-CT concepts were added to the data graph (using rdfs:label), and synonyms were not included. Role group information was represented in the RDF data graphs using the rdf:statement as a reification approach [29]. The following subsections discuss in detail the three major steps (Figure 4b) constituting our method.
3.1. Lexical Knowledge Base Creation
Knowledge bases (KBs) were created that reflected the insights gained from both studies [27,28]. The KB creation process representing the insights gained from [27] consisted of designing two types of files:
- A (tab-delimited) file containing information about which SNOMED-CT concept belongs to which sample set. There were 11 such files, with one for each stop word and columns organized as follows: sample-set, atomically annotated concept FSN, and SCTID.
- A (tab-delimited) file containing information about the mandatory attributes for each sample set, organized as the stop word, semantic pattern, sample set, SCTID of the mandatory attribute, and FSN of the mandatory attribute.
For example, Figure 6a illustrates the results of [27], with an example for the stop word due to organized in the two KB files ((i) and (ii)) designed by us. To incorporate the knowledge of this KB into graphs, 26 additional classes (one for each sample-set, as discussed in Section 3) were created and included in the RDFLex and SHACLex graphs, as explained later in this section.

Figure 6.
(a) Knowledge base files representing the insights from [27]. (b) Knowledge base files representing the insights from [28].
To create a KB representing the insights from [28], we applied a part of speech (POS) tagger to all one-word and two-word concept FSNs in the Disorder hierarchy of SNOMED-CT. We used Spacy’s “en_core_sci_sm” model trained on biomedical datasets [30], which is available in Python, to implement the POS tagger. The result of the POS tagger was filtered to extract all two-word “ADJ NOUN” (Adjective Noun) concepts and one-word “NOUN” concepts, which were segregated into two separate files. A randomly chosen set of 200 “ADJ NOUN” concepts was selected to test the hypothesis. For each “ADJ NOUN” concept, a recursive traversal was applied to record all Is-A relationships (all ancestors) reachable from the “ADJ NOUN” concept, which constituted the KB representing the insights from [28]. The KB was organized as follows:
- A (tab-delimited) file which contained information about all Is-A relationships (SCTID:116680003) reachable from each “ADJ NOUN” concept. It consisted of the following columns: the source SCTID, destination SCTID, source FSN, and destination FSN of all Is-A relationships (SCTID:116680003) reachable from each “ADJ NOUN” concept.
- A (tab-delimited) file which consisted of one-word Disorder concepts tagged as “NOUN” by the POS tagger, which were segregated earlier into a separate file.
For example, Figure 6b displays the results demonstrating the insights gained from [28] organized in the two KB files ((i) and (ii)) designed by us. Both of these files were used in combination to check if there were any potential “ADJ NOUN” concepts which had corresponding “NOUN” concepts but were currently not linked to them via an Is-A (hierarchical) relationship (see Algorithm 1).
| Algorithm 1 SHACLex graph Generation algorithm for [28] |
|
3.2. Graph Generation
To test the hypothesis that enhanced SHACL shapes can be created by taking into consideration the internal features of biomedical class names, we generated two types of RDF graphs (RDF and RDFLex) and two kinds of SHACL graphs (SHACL and SHACLex) representing the SNOMED-CT concepts examined by each of the studies [27,28]. To represent the results of [27], 22 data graphs (11 RDF and 11 RDFLex) and 22 shape graphs (11 SHACL and 11 SHACLex) for each stop word were constructed. The 11 RDF graphs were created as described in Section 3. To demonstrate SHACLex creation representing the results of [27], the lexical features of the class names, available from the constructed KB, were incorporated into the RDFLex data graph using an additional lexical layer. Again, 11 such RDFLex graphs were created, with 1 for each stopword. The extra lexical layer consisted of RDFS classes representing the sample sets extracted in the results of [27], which consisted of the mandatory attributes (see Table 1) identified for that sample-set as properties. All SNOMED-CT concept classes conforming to a particular semantic and lexical pattern were made into instances (using rdf:type) of the respective sample set classes by referencing the constructed KB. Figure 5 displays the structure of the RDF and RDFLex data graphs representing the SNOMED-CT concepts targeted by [27]. The SHACL and SHACLex graphs were created using Python programming.
Algorithms 2 and 3 illustrate the steps involved in the creation of RDFLex and SHACLex graphs, respectively, representing the results of [27]. The SHACL shape graph simply converted the existing RDF axioms into SHACL constraints. By contrast, SHACLex included additional information available from the developed KB and created shapes for each of the sample set classes, which ensured the presence of the mandatory attributes in all their instances [27]. Of note, in this method, for the purpose of demonstration, and to increase the efficiency, we implemented additional classes representing lexical and semantic patterns (i.e., sample sets) in the RDFLex graph. Creating sample set classes and making all adhering SNOMED-CT concept classes instances of the sample set class allowed us to apply the SHACL constraint to a single parent class and thereby all its children, thus avoiding redundant constraints in each SNOMED-CT concept class. However, we understand that this may not always be feasible when working with an actual ontology. In such cases, simply creating additional constraints based on the KB information in the shape of each SNOMED-CT concept class instead of applying the constraints through a parent sample set class would also work.
| Algorithm 2 RDFLex graph generation algorithm for [27] |
|
| Algorithm 3 SHACLex graph generation algorithm for [27] |
|
Similarly, to test the hypothesis by using the insights gained from [28], an RDF and an RDFLex graph were created and validated against a SHACL and a SHACLex graph. In this case, both the RDF graph and RDFLex graph were created normally, as mentioned in Section 3. However, only hierarchical Is-A relationships (SCTID:116680003) were converted into RDF triples to keep the data graph relevant to the experiment [28]. The SHACL and SHACLex graphs were created using Python programming. Algorithm 1 illustrates the steps followed to generate the SHACLex shape graph for the SNOMED-CT concepts targeted by [28]. The SHACL shape graph simply converted the existing RDF axioms into SHACL constraints, whereas SHACLex considered the information available from the developed KB and included additional SHACL constraints for the property “subClassOf”. Thus, the SHACLex graph ensured that missing hierarchical relationships were flagged and “ADJ NOUN” concepts were represented as children of the respective “NOUN” concepts in the biomedical ontology data graph (see Algorithm 1).
During a manual inspection of the experimentation results, several cases were identified as violations where an “ADJ NOUN” concept was linked to another “ADJ NOUN” concept but not linked to a “NOUN” concept. For example, the concept Gonococcal pericarditis (SCTID:90428001) (ADJ NOUN) was flagged as a violation due to missing an Is-A (hierarchical) relationship with the concept Pericarditis (SCTID:3238004) (NOUN). However, a more specific hierarchical relationship already existed with Bacterial pericarditis (SCTID:233883000) (ADJ NOUN). In such cases, we believed that the existing relationship between the two “ADJ NOUN” concepts superseded the proposed relationship between the “ADJ NOUN” and “NOUN” concepts. As a result, the algorithm was modified, and such cases were not flagged as violations by the final SHACLex graph (see Algorithm 1).
3.3. Graph Validation
The 11 RDF and RDFLex graphs, with 1 for each stop word [27], were validated against the 11 SHACL and SHACLex graphs, respectively, using the validate() method of the pySHACL library in Python. Similarly, the RDF and RDFLex graphs representing SNOMED-CT concepts targeted by [28] were also validated against the respective SHACL and SHACLex graphs. The validation performance was compared by examining the number of violations raised by the SHACL and SHACLex graphs for the same set of SNOMED-CT concepts in the RDF and RDFLex graphs. The violations raised by the SHACLex graphs were cross-verified with the results of the lexical auditing techniques [27,28], which additionally substantiated the validation performance of the SHACLex shapes.
4. Results
We tested the hypothesis by comparing the validation performance of the SHACL and SHACLex graphs to identify potentially inconsistent concepts in SNOMED-CT. As stated earlier, the validation performance was compared on the basis of the number of violations raised by each shape graph (SHACL and SHACLex) for the same set of SNOMED-CT concepts represented in the data graphs. The raised violations were manually inspected to ensure that they were congruous with the results of each study. The total number of violations identified by the SHACLex graphs for the two studies combined was 176 (171 for [27] and 5 for [28]), whereas the total number of violations raised by the SHACL graphs for the two studies combined was 0. This was expected since, in the SHACL shape graphs, the existing OWL axioms were exclusively converted into SHACL constraints, and therefore the shape graphs did not contain pertinent lexical information. As discussed earlier, biomedical ontologies are richer in natural language content than OWL axioms or logical definitions [22], and thus the SHACL shape graphs could not incorporate the semantic knowledge available from the lexical features of class names into SHACL constraints (which was essential to identify the violations) by exclusively using OWL axioms for shape creation. Contrarily, the SHACLex shape graphs were augmented with the lexical knowledge of class names in the biomedical ontologies by exploiting the insights gained from lexical auditing techniques employed to enrich OWL axioms, and thus the SHACLex shape graphs were able to identify all the missing properties in a class that could be derived from the lexical features of the class name. This proves the hypothesis that the methods employed in SHACL shape graph creation for generic data-graphs, in which class names do not influence the shape of a class, are not sufficient for domain-specific biomedical ontology data graphs, in which class names contain pertinent lexical information that influences the shape of a class. The results clearly indicate that the SHACLex graphs outperformed the SHACL graphs by raising 176 more violations corresponding to inconsistencies such as missing relationships in biomedical concepts.
Figure 7 illustrates the validation performance of the SHACL and SHACLex shape graphs using the SNOMED-CT concept Injury due to sword (SCTID:243051008) as an example. As one can see from the FSN, the concept belongs to the lexical pattern “A due to B” and the semantic pattern “Disorder due to Object (DdtO)” and therefore demands the presence of the mandatory attributes due to (SCTID:42752001) and causative agent (SCTID:246075003) in its definition, according to the template created in [27] (see Table 1). On the right side of the figure, one can see how the RDFLex data graph and SHACLex shapes-graph incorporate this lexical information in the form of sample set classes, whereas on the left side of the figure, the RDF data graph and SHACL shape graph fail to consider the internal lexical features of the class name and therefore do not convert it into SHACL constraints. Finally, from the validation results, we can see that SHACLex flagged the concept as a violation due to missing the mandatory attribute relationships due to (SCTID:42752001) and causative agent (SCTID:246075003) in its definition. In the figure, the raised violation is depicted using a red color and the SCTID of Injury due to sword (SCTID:243051008) is highlighted in the violation message using yellow and orange colors. By contrast, the SHACL simply validated the concept as true.
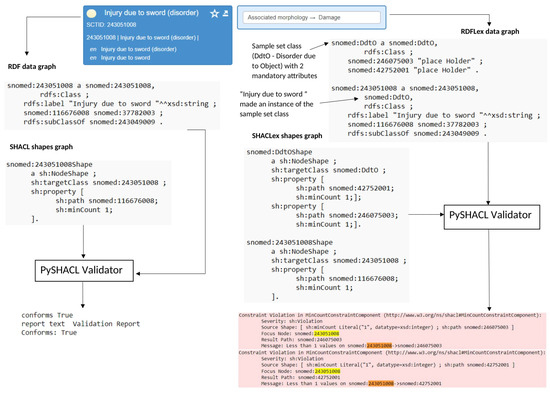
Figure 7.
SHACL vs. SHACLex validation report for the concept Injury due to sword (SCTID:243051008) belonging to the sample set “Disorder due to Object” (DdtO). Missing mandatory attributes due to (SCTID:42752001) and causative agent (SCTID:246075003) were caught as violations only by SHACLex.
Figure 8 illustrates the validation performance of the SHACL and SHACLex shape graphs for the SNOMED-CT concepts targeted by [28], using Pulmonary hypostasis (SCTID:196116008) as an example. As one can see from the FSN, the concept belongs to the lexical pattern “ADJ NOUN” and therefore demands the presence of a hierarchical relationship with its parent concept Hypostasis (SCTID:72127003), having the POS tag “NOUN”, according to the insight gained from [28]. On the right side of the figure, one can see how the SHACLex shape graph incorporated this lexical information in the form of an additional property, rdfs:subClassOf, with the designated “NOUN” concept in its sh:hasValue field along with the sh:minCount constraint. On the left side of the figure, the SHACL shape graph failed to consider the internal lexical features of the class name and therefore did not convert it into SHACL constraints. Finally, the validation results show that SHACLex flagged the concept as a violation due to the missing hierarchical relationship between the concept Pulmonary hypostasis (SCTID:196116008) and Hypostasis (SCTID:72127003) in its definition. In the figure, the raised violation is depicted using a red color and the FSN of Pulmonary hypostasis (SCTID:196116008) is highlighted in the violation message using yellow and orange colors. By contrast, the SHACL simply validated the concept as true.
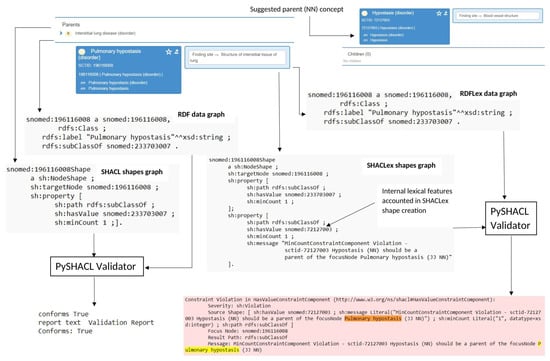
Figure 8.
SHACL vs. SHACLex validation report for the concept Pulmonary hypostasis (SCTID:196116008) conforming to the POS pattern “ADJ NOUN”. Missing hierarchical relationship with Hypostasis (SCTID:72127003) was caught as a violation only by SHACLex.
To demonstrate the validation performance of the SHACLex graphs against the SHACL graphs, for cases where a sample set did not have any SNOMED-CT concepts with missing mandatory attributes (as per the results of [27]), we introduced deliberate errors by removing the mandatory attributes from their definitions and creating new concepts with fictitious SCTIDs in the data graph, which we refer to as negative test cases. For every negative test case, a concept containing the mandatory attribute with a fictitious SCTID was introduced in the data graph, which we refer to as a positive test case. The positive test cases were added to ensure that SHACLex only flagged the negative test cases as violations. Five such positive and negative test cases were added for each sample set that did not exhibit any SNOMED-CT concepts with missing mandatory attributes in the biomedical ontology. We then created RDF and RDFLex data graphs, including these newly added fictitious correct and incorrect concepts (positive and negative test cases) and validated them against the SHACL and SHACLex shape graphs, respectively. Table 2 shows a comparative evaluation of the violations caught by both the shape graphs in SNOMED-CT concepts targeted by [27]. Zero violations indicate that the data graph conforms (conforms true) to the shape graph. Violations >0 indicate that the data graph does not conform (conforms false) to the shape graph and harbors inconsistencies, and N/A indicates that the sample sets did not have any mandatory attributes whose presence or absence needed to be tested.

Table 2.
Comparative evaluation of SHACL vs. SHACLex validation performance for SNOMED-CT concepts targeted by [27].
Similarly, for validating the created RDF and RDFLex data graphs representing SNOMED-CT concepts targeted by [28] against the SHACL and SHACLex shape graphs, we observed five missing hierarchical relationships in the definitions of the “ADJ NOUN” concepts as caught by SHACLex. Table 3 shows a comparative evaluation of the violations caught by both the shape graphs in the SNOMED-CT concepts targeted by [28]. The results indicate that the SHACLex graphs generated by considering additional lexical information of class names in a data graph created better shapes that were able to capture violations in a biomedical ontology data graph that could not be captured by the baseline SHACL graph. Please note that the violations captured by the SHACLex graphs and the missing attribute relationships thereby suggested requiring manual evaluation by a domain expert. The inconsistencies in the modeling of SNOMED-CT concepts were raised as violations by the SHACLex graphs because they were congruous with the results of the auditing methods. Thus, in Table 3, the concepts were flagged as violations because they were not consistent with the principles mentioned in [28], and in Table 2, the concepts flagged as violations because they were not consistent with other lexically and semantically similar FD concepts [27].

Table 3.
Comparative evaluation of SHACL vs. SHACLex validation performance for SNOMED-CT concepts targeted by [28].
5. Discussion
Biomedical ontologies provide resources to be used in all downstream applications like EHR systems, clinical reports, and discharge summaries. Thus, it is crucial to ensure that the highest quality information is represented in biomedical ontology data graphs. Failing to do so would have adverse consequences not only in the aforementioned downstream applications but also the tertiary, safety-critical, data analysis, and automated decision making systems relying those applications. While a handful of SHACL shape creation methods exist to validate these downstream applications, due to the novelty of the technology, greatly limited research has been conducted on applying SHACL to validate biomedical ontology data graphs.
Furthermore, the existing SHACL shape creation techniques created for generic data graphs have been adopted in the biomedical domain without any modifications or consideration of characteristic differences in the nature of the domain-specific data graphs and the knowledge represented by them. As a result, research on the creation of SHACL shapes for biomedical ontologies has also focused on exclusively converting TSV files or OWL restrictions into SHACL shape constraints [19,25] as performed for the majority of generic data graphs like WikiData and DBpedia [11,13,24]. SHACL shape graphs created using such methods fail to consider the characteristic differences of biomedical ontology data graphs. As stated earlier, in biomedical ontologies, the class names (internal features) contain semantic information that can be exploited to create better SHACL shapes [25]. SHACL shape graphs, created using generic methods, convert OWL axioms devoid of lexical richness of concept names into SHACL constraints. Therefore, they do not raise any potential violations linked to the lexical structure of concept names in the data graph. Table 2 and Table 3 show the difference in the number of violations caught with and without incorporating the internal features (i.e., the lexical information available in class names for each of the studies) [27,28]. The SHACLex graphs caught 176 violations for the same set of SNOMED-CT concepts by exploiting the hidden semantics of class names, whereas the SHACL graphs failed to raise a single violation due to the lack of this knowledge in SHACL constraints. The work presented here has addressed a crucial research gap that will improve the validation performance of biomedical ontologies represented using SWT, like OWL and RDF. In the absence of such a method, the existing methods (adopted from generic data graphs) that rely solely on the quality of existing ontology axioms would continue to create substandard SHACL shapes incapable of highlighting missing information in biomedical ontology data graphs, which can be derived from the lexical structure of the class names and impede the QA of biomedical ontology data graphs.
With the increase in the amount of data being produced every day, there is a need to represent these data in intelligent machine-processable formats, such as those provided by SWTs, and with that arises the need to improve the newly developed SWTs, like the SHACL, which are focused on validation. The results of this research show the importance of bespoke solutions in KG validation. They emphasize that all data graphs cannot be treated alike, and creative solutions need to be developed for effective data validation pertaining to each domain. Domain-specific data graphs should be scrutinized individually to identify characteristic differences which can be exploited to create enhanced SHACL shapes for improved data validation in each domain. The method presented in this chapter can be adapted to other domains where the lexical knowledge of class names influences the properties and, as a result, the shape of a class. The presented work can also be used as an inspiration to identify other characteristic differences which can be exploited and included in SHACL constraints.
SHACL shapes created simply by adopting methods for generic data graphs without any consideration for the domain-specific characteristics of biomedical ontology data graphs also fail to utilize the knowledge available from decades of evolution of lexical auditing techniques in the validation process [26]. The SHACL is a relatively new standard developed in 2017 [1], whereas the auditing techniques employed in the quality assurance (QA) of biomedical ontologies have existed and been perfected since the early 1980s [26]. There is a large gap in incorporating the knowledge of existing auditing techniques employed in the QA of biomedical ontologies into SHACL shape graph creation. The obtained results demonstrate that the methods that work for generic data graphs are not sufficient for biomedical ontology data graphs, and SHACL shape creation for domain-specific data graphs like biomedical ontologies require additional knowledge augmentation to create efficient SHACL shapes.
The methodology presented here can also be used as a framework to design lexical knowledge bases for a variety of lexical auditing techniques existing in the literature [26] and then convert the lexical knowledge of biomedical class names into SHACL constraints for improved validation of biomedical ontology data graphs. One of the major challenges in accomplishing this is the variety of lexical auditing techniques and therefore the variation in the structure of the created knowledge bases. Each auditing technique will require the development of a unique approach to organize the hidden semantical knowledge into a KB which can later be incorporated into RDFLex and SHACLex graphs, implying that each method would demand bespoke solutions and programming only applicable to that particular auditing technique. Thus, making seamless automation a challenge. However, given the poor performance of the SHACL for biomedical ontology data graphs, implementing SHACLex using bespoke solutions is the most feasible approach at the moment to bridge the gap between SHACL shape creation methods and the existing knowledge in the QA of biomedical ontologies. Although the variety in the results of lexical auditing techniques makes it difficult to standardize the method, the proposed method can be incorporated officially in the SNOMED-CT validation process by creating SHACLex shapes for the description modeling templates provided by IHTSDO [31].
6. Conclusions
The SHACL is a relatively new technology, and the existing SHACL shape creation methods are predominantly tested on generic data graphs, in which the name of a class does not affect the shape of a class. In this work, we highlighted how domain-specific data graphs can have different characteristics due to the nature of the information stored in them and therefore may require additional resources to create efficient SHACL shapes for accurate knowledge validation. We identified and addressed the requirement of testing the relatively new validation technology, SHACL, for domain-specific data graphs. To this end, we presented a novel method to create enhanced SHACL shapes better suited for biomedical ontology data graphs by incorporating the internal features (i.e., lexical knowledge of class names) into SHACL constraints. We also presented a comparative evaluation of the shapes created by our method (SHACLex shapes) which incorporated lexical knowledge of class names into SHACL constraints, with the shapes created using existing techniques (baseline SHACL shapes) that exclusively convert existing ontology axioms into SHACL constraints. The comparative evaluation demonstrated that the enhanced SHACLex shapes identified 176 violations which the baseline SHACL shapes, void of this lexical knowledge, failed to detect. Thus, the validation performance for domain-specific biomedical ontology data graphs significantly improved by creating enhanced SHACL shapes as proposed in our novel approach. Thus, the work presented in this paper proved the hypothesis that, unlike generic data graphs, biomedical ontology data graphs require additional knowledge augmentation to create effective SHACL shapes. A promising direction for future work would be devising methods that normalize the structure of KBs while incorporating the knowledge of unique auditing techniques into SHACL shape creation to reduce the amount of bespoke programming and increase the rate of automation. The results of this research also encourage scrutinizing other domain-specific data graphs to identify characteristic differences which can be exploited to create enhanced SHACL shapes catered to the data validation requirements of specific domains.
Author Contributions
Conceptualization, R.B.; methodology, R.B.; writing—original draft preparation, R.B.; writing—review and editing, M.B. and G.M.; supervision, M.B. and G.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A. SNOMED-CT Structure and Lexical Auditing Technique Details
This appendix provides further details about the structure of SNOMED-CT to gain a deeper understanding of the data graphs constructed in this study. We also provide a detailed overview of the methodologies for the two lexical auditing studies [27,28] which were used to create the knowledge bases referenced by our algorithms to develop the enhanced SHACL shapes.
SNOMED-CT [2], due to its comprehensive nature, is the world’s most widely adopted biomedical ontology. SNOMED-CT [2] categorizes biomedical information into 19 major hierarchies. Each hierarchy contains concepts belonging to a particular semantic type. This semantic category is mentioned as a hierarchy tag or semantic tag in parenthesis after the name of the concept. Figure A1 displays a screenshot of the SNOMED-CT browser, listing the 19 hierarchies of SNOMED-CT. The SNOMED-CT logical model consists of three components: the concept, description, and relationship. Figure A2 illustrates the logical model followed by SNOMED-CT. As illustrated in Figure A2, concepts in SNOMED are represented using a unique identifier (SCTID) and a unique name called a Fully Specified Name (FSN). Concepts are linked to each other using two types of relationships:
- Is-A relationships, which are hierarchical in nature and represent the subsumption relationships between two concepts that belong to the same hierarchy.
- Attribute (lateral) relationships, which give more information about a concept by linking it to concepts from other hierarchies based on the domain and range constraints defined in the logical model of SNOMED-CT.

Figure A1.
SNOMED-CT hierarchies [32].
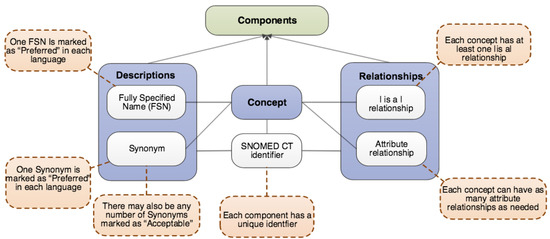
Figure A2.
SNOMED-CT logical nodel [2].
Furthermore, SNOMED-CT contains two types of concepts: (1) Fully Defined (FD) concepts [2], which are sufficiently defined to distinguish them from other concepts, and (2) primitive or Partially Defined (PD) concepts, which are not sufficiently defined. There are multiple reasons as to why a concept may be a PD concept [33]. One of the reasons is that the attribute relationships that distinguish the concept from other concepts may not be present in its definition. FD concepts are more suitable for machine processing because for one, they are sufficiently differential (i.e., the concept has at least one sufficient definition that distinguishes it from any concepts or expressions that are neither equivalent to nor sub-types of the defined concept) [2], and secondly, FD concepts are manually inspected by SNOMED authors [33] to attain the FD status and therefore are more reliable than PD concepts. The higher the number of FD concepts in an ontology, the more reliable the biomedical ontology is [34]. Thus, auditing techniques employed in the QA of biomedical ontologies strive to identify missing relationships which can be used to fully define a concept.
As stated earlier, biomedical ontologies are richer in natural language content than OWL axioms and logical definitions, and the existing lexical auditing techniques strive to enrich the logical definitions of biomedical concepts with the hidden semantics [23] available in the concept names. Despite this effort, the process is gradual, and the existing OWL axioms do not represent all the necessary information available in the lexical structure of concept names [22]. Figure A3 and Figure A4 give an overview of the steps followed to obtain the results of the two lexical auditing techniques ([27] and [28], respectively) used to create knowledge bases in this study. In Figure A3, we used the output templates for mandatory attribute relationships to create enhanced SHACL shapes (see Table 1). The templates check for the presence of a stop word in a concept name and, based on that, provide information about which attribute relationship (property in RDF terminology) should be present in the class. This knowledge was incorporated as a constraint to create enhanced SHACL shapes. In Figure A4, we mainly used the insight gained from [28] that a “NOUN” concept should ideally be linked to an “ADJ NOUN” concept via a hierarchical (Is-A) relationship. Using this insight, we checked for the presence of a hierarchical relationship (subclass property in RDF terminology) between an existing “ADJ NOUN” concept and the corresponding “NOUN” concept, and this knowledge was incorporated as a constraint to create enhanced SHACL shapes.
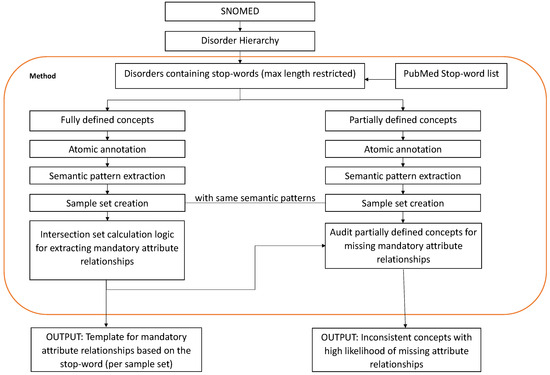
Figure A3.
Overview of methodology in [27].
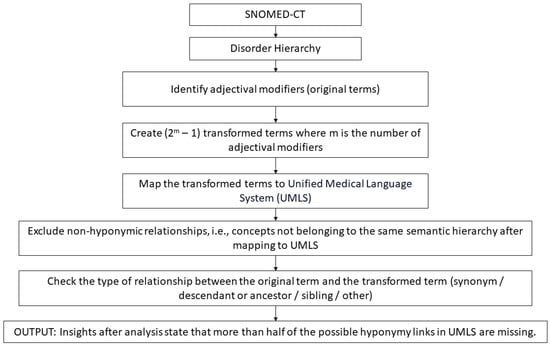
Figure A4.
Overview of methodology in [28].
References
- W3C. OWL 2 Web Ontology Language (2012). 2012. Available online: https://www.w3.org/TR/ (accessed on 31 May 2023).
- IHTSDO. IHTSDO SNOMED International Confluence. 2002. Available online: https://confluence.ihtsdotools.org (accessed on 2 July 2023).
- IHTSDO. SCT Template Syntax Specification. 2018. Available online: https://github.com/IHTSDO/snomed-owl-toolkit (accessed on 31 May 2023).
- Tiwari, S.M.; Abraham, A. Semantic assessment of smart healthcare ontology. Int. J. Web Inf. Syst. 2020, 16, 475–491. [Google Scholar] [CrossRef]
- Knublauch, H. SHACL and OWL Compared. 2017. Available online: https://spinrdf.org/shacl-and-owl.html (accessed on 31 May 2023).
- Cimmino, A.; Fernández-Izquierdo, A.; García-Castro, R. Astrea: Automatic Generation of SHACL Shapes from Ontologies. Semant. Web 2020, 12123, 497–513. [Google Scholar]
- Pandit, H.J.; O’Sullivan, D.; Lewis, D. Using Ontology Design Patterns To Define SHACL Shapes. In Proceedings of the WOP@ISWC, Monterey, CA, USA, 9 October 2018. [Google Scholar]
- Boneva, I.; Dusart, J.; Fernández-Álvarez, D.; Gayo, J.E.L. Shape Designer for ShEx and SHACL constraints. In Proceedings of the International Workshop on the Semantic Web, Auckland, New Zealand, 26–30 October 2019. [Google Scholar]
- Mihindukulasooriya, N.; Rashid, M.R.A.; Rizzo, G.; García-Castro, R.; Corcho, Ó.; Torchiano, M. RDF shape induction using knowledge base profiling. In Proceedings of the 33rd Annual ACM Symposium on Applied Computing, Pau, France, 9–13 April 2018. [Google Scholar]
- Spahiu, B.; Maurino, A.; Palmonari, M. Towards Improving the Quality of Knowledge Graphs with Data-driven Ontology Patterns and SHACL. In Proceedings of the WOP@ISWC, Monterey, CA, USA, 9 October 2018. [Google Scholar]
- Fernández-Álvarez, D.; García-González, H.; Frey, J.; Hellmann, S.; Gayo, J.E.L. Inference of Latent Shape Expressions Associated to DBpedia Ontology. In Proceedings of the International Workshop on the Semantic Web, Monterey, CA, USA, 8–12 October 2018. [Google Scholar]
- Fernández-Álvarez, D.; Labra-Gayo, J.E.; García-González, H. Inference and Serialization of Latent Graph Schemata Using ShEx. In Proceedings of the SEMAPRO 2016: The Tenth International Conference on Advances in Semantic Processing, Venice, Italy, 9–13 October 2016. [Google Scholar]
- González, L.; Hogan, A. Modelling Dynamics in Semantic Web Knowledge Graphs with Formal Concept Analysis. In Proceedings of the 2018 World Wide Web Conference, Lyon, France, 23–27 April 2018. [Google Scholar]
- Rabbani, K.; Lissandrini, M.; Hose, K. SHACL and ShEx in the Wild: A Community Survey on Validating Shapes Generation and Adoption. In Proceedings of the Companion Proceedings of the Web Conference 2022, Lyon, France, 25–29 April 2022. [Google Scholar]
- Martínez-Costa, C.; Schulz, S. Validating EHR clinical models using ontology patterns. J. Biomed. Inform. 2017, 76, 124–137. [Google Scholar] [CrossRef] [PubMed]
- Keuchel, D.; Spicher, N. Automatic Detection of Metadata Errors in a Registry of Clinical Studies Using Shapes Constraint Language (SHACL) Graphs. Stud. Health Technol. Inform. 2021, 281, 372–376. [Google Scholar] [PubMed]
- Kober, G.; Robaldo, L.; Paschke, A. Modeling Medical Guidelines by Prova and SHACL Accessing FHIR/RDF. Use Case: The Medical ABCDE Approach. Stud. Health Technol. Inform. 2022, 293, 59–66. [Google Scholar] [PubMed]
- Keuchel, D.; Spicher, N.; Wang, J.; Völcker, M.; Gong, Y.; Deserno, T.M. SHACL-Based Report Quality Evaluation for Health IT-Induced Medication Errors. Stud. Health Technol. Inform. 2022, 290, 414–418. [Google Scholar] [PubMed]
- Gaudet-Blavignac, C.; Raisaro, J.L.; Touré, V.; Österle, S.; Crameri, K.; Lovis, C. SPHN Semantic Framework. 2020. Available online: https://sphn-semantic-framework.readthedocs.io/en/latest/index.html (accessed on 2 June 2023).
- Gaudet-Blavignac, C.; Raisaro, J.L.; Touré, V.; Österle, S.; Crameri, K.; Lovis, C. A National, Semantic-Driven, Three-Pillar Strategy to Enable Health Data Secondary Usage Interoperability for Research within the Swiss Personalized Health Network: Methodological Study. JMIR Med. Inform. 2021, 9, e27591. [Google Scholar] [CrossRef] [PubMed]
- Hodgson, R.; Polikoff, I. SNOMED-CT Expo 2020—SNOMED-CT-SHAPES: A Simpler Approach to Working with SNOMED in RDF. 2020. Available online: https://www.youtube.com/watch?v=mrlNn3oYH3k (accessed on 2 June 2023).
- van Damme, P.; Quesada-Martínez, M.; Cornet, R.; Fernández-breis, J.T. From lexical regularities to axiomatic patterns for the quality assurance of biomedical terminologies and ontologies. J. Biomed. Inform. 2018, 84, 59–74. [Google Scholar] [CrossRef] [PubMed]
- Third, A. “Hidden semantics”: What can we learn from the names in an ontology? In Proceedings of the International Conference on Natural Language Generation, Utica, IL, USA, 30 May–1 June 2012.
- Omran, P.G.; Taylor, K.L.; Méndez, S.J.R.; Haller, A. Learning SHACL shapes from knowledge graphs. Semant. Web 2022, 14, 101–121. [Google Scholar] [CrossRef]
- TopQuadrant. SNOMED-CT Shapes. 2020. Available online: https://www.topquadrant.com/wp-content/uploads/2020/11/SNOMED_CT_Expo2020-TopQuadrant-sFINAL2.pdf (accessed on 27 May 2023).
- Amith, M.T.; He, Z.; Bian, J.; Lossio-Ventura, J.A.; Tao, C. Assessing the practice of biomedical ontology evaluation: Gaps and opportunities. J. Biomed. Inform. 2018, 80, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Burse, R.; Mcardle, G.; Bertolotto, M. Targeting stopwords for quality assurance of SNOMED-CT. Int. J. Med. Inform. 2022, 167, 104870. [Google Scholar] [CrossRef] [PubMed]
- Bodenreider, O.; Burgun, A.; Rindflesch, T. Lexically-suggested hyponymic relations among medical terms and their representation in the UMLS. In Proceedings of the TIA 2001: Terminologie et Intelligence Artificielle, Nancy, France, 3–4 May 2001. [Google Scholar]
- Hartig, O. RDF* and SPARQL*: An Alternative Approach to Annotate Statements in RDF. In Proceedings of the International Workshop on the Semantic Web, Vienna, Austria, 21–15 October 2017. [Google Scholar]
- Neumann, M.; King, D.; Beltagy, I.; Ammar, W. ScispaCy: Fast and Robust Models for Biomedical Natural Language Processing. arXiv 2019, arXiv:1902.07669. [Google Scholar]
- IHTSDO. SCT Modeling Templates and Description Patterns. 2018. Available online: https://confluence.ihtsdotools.org/display/SCTEMPLATES/SCT+Modeling+Templates+and+description+patterns (accessed on 11 July 2023).
- IHTSDO. SNOMED International SNOMED CT Browser. 2002. Available online: https://browser.ihtsdotools.org/ (accessed on 10 August 2023).
- IHTSDO. What Does It Mean If a Concept Is Fully-Defined or Primitive and How Do I Tell the Difference? 2002. Available online: https://ihtsdo.freshdesk.com/support/solutions/articles/4000050378-what-does-it-mean-if-a-concept-is-fully-defined-or-primitive-and-how-do-i-tell-the-difference- (accessed on 15 December 2021).
- Schulz, S.; Suntisrivaraporn, B.; Baader, F.; Boeker, M. SNOMED reaching its adolescence: Ontologists’ and logicians’ health check. Int. J. Med. Inform. 2009, 78 (Suppl. S1), S86–S94. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).