Explainable Machine Learning (XAI) for Survival in Bone Marrow Transplantation Trials: A Technical Report
Abstract
:1. Introduction
2. Materials and Methods
Statistical Analysis
3. Results
| 1—Model performance for the whole cohort |
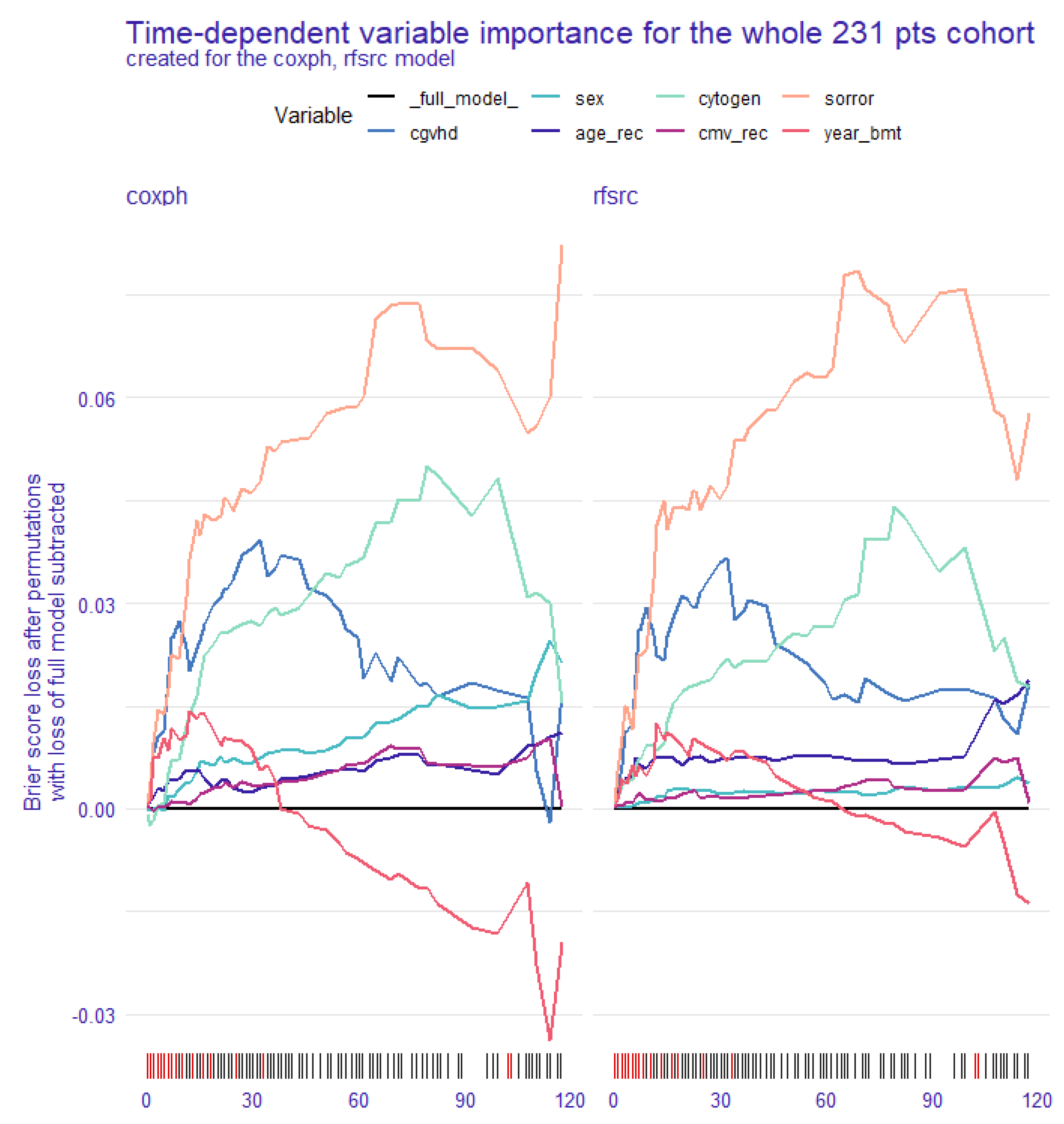
| 2a—Global explanation: time-dependent feature importance for the whole cohort |
| 2b—Global explanation: partial dependence survival profile (PDP) for the whole cohort |
| 3a—Local explanation: SurvSHAP(t) plot per single patient |
| 3b—Local explanation: SurvLIME plot per single patient |
| 3c—Local explanation: ceteris paribus survival profile per single patient |
3.1. Model Performance for the Whole Cohort
3.2. Global Explanation: Time-Dependent Feature Importance for the Whole Cohort
3.3. Global Explanation: Partial Dependence Survival Profile for the Whole Cohort
3.4. Local Explanation: SurvSHAP(t) Plot per Single Patient
3.5. Local Explanation: SurvLIME Plot per Single Patient
3.6. Local Explanation: Ceteris Paribus Survival Profile per Single Patient
4. Discussion
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Joshi, G.; Jain, A.; Adhikari, S.; Garg, H.; Bhandari, M. FDA approved Artificial Intelligence and Machine Learning (AI/ML)-Enabled Medical Devices: An. updated 2022 landscape. MedRxiv Prepr. Serv. Health Sci. 2022. [Google Scholar] [CrossRef]
- Salah, H.T.; Muhsen, I.N.; Salama, M.E.; Owaidah, T.; Hashmi, S.K. Machine learning applications in the diagnosis of leukemia: Current trends and future directions. Int. J. Lab. Hematol. 2019, 41, 717–725. [Google Scholar] [CrossRef] [PubMed]
- Shouval, R.; Fein, J.A.; Savani, B.; Mohty, M.; Nagler, A. Machine learning and artificial intelligence in haematology. Br. J. Haematol. 2021, 192, 239–250. [Google Scholar] [CrossRef] [PubMed]
- Muhsen, I.N.; Hashmi, S.K. Utilizing machine learning in predictive modeling: What’s next? Bone Marrow Transpl. 2022, 57, 699–700. [Google Scholar] [CrossRef] [PubMed]
- Barredo Arrieta, A.; Díaz-Rodríguez, N.; Del Ser, J.; Bennetot, A.; Tabik, S.; Barbado, A.; Garcia, S.; Gil-Lopez, S.; Molina, D.; Benjamins, R.; et al. Explainable Artificial Intelligence (XAI): Concepts, taxonomies, opportunities and challenges toward responsible AI. Inf. Fusion. 2020, 58, 82–115. [Google Scholar]
- Knapic, S.; Malhi, A.; Saluja, R.; Främling, K. Explainable Artificial Intelligence for Human Decision Support System in the Medical Domain. Mach. Learn. Knowl. Extr. 2021, 3, 740–770. [Google Scholar] [CrossRef]
- Di Martino, F.; Delmastro, F. Explainable AI for clinical and remote health applications: A survey on tabular and time series data. Artif. Intell. Rev. 2023, 56, 5261–5315. [Google Scholar] [PubMed]
- Biecek, P.; Burzykowski, T. Explanatory Model Analysis; Chapman and Hall/CRC: New York, NY, USA, 2021. [Google Scholar]
- Baniecki, H.; Parzych, D.; Biecek, P. The grammar of interactive explanatory model analysis. Data Min. Knowl. Disc. 2023, 14, 1–37. [Google Scholar]
- Spytek, M.; Krzyziński, M.; Baniecki, H.; Biecek, P. Survex: Explainable Machine Learning in Survival Analysis; R Package Version 1.0.0.9000. Available online: https://modeloriented.github.io/survex/ (accessed on 15 August 2023).
- Kovalev, M.S.; Utkin, L.V.; Kasimov, E.M. SurvLIME: A method for explaining machine learning survival models. Knowl.-Based Syst. 2020, 203, 106164. [Google Scholar]
- Krzyziński, M.; Spytek, M.; Baniecki, H.; Biecek, P. SurvSHAP(t): Time-dependent explanations of machine learning survival models. Knowl.-Based Syst. 2023, 262, 110234. [Google Scholar]
- Cox, D.R. Regression models and life-tables. J. Royal. Stat. Soc. Ser. B 1972, 34, 187–220. [Google Scholar] [CrossRef]
- Ishwaran, H.; Kogalur, U.B.; Blackstone, E.H.; Lauer, M.S. Random survival forests. Ann. Appl. Stat. 2008, 2, 841–860. [Google Scholar]
- Lo Schirico, M.; Passera, R.; Gill, J.; Dellacasa, C.; Dogliotti, I.; Giaccone, L.; Zompi, S.; Busca, A. Graft-versus-host disease prophylaxis with anti-thymocyte globulin in patients receiving stem cell transplantation from unrelated donors: An observational retrospective single-center study. Cancers 2023, 15, 2761. [Google Scholar]










| Characteristics | All Patients |
|---|---|
| Age at transplant, median (IQR), years | 51 (43–60) |
| Gender | |
| Male | 126 (54.5%) |
| Female | 105 (45.5%) |
| HCT-CI | |
| Low/intermediate (0–2) | 119 (68.4%) |
| High (>3) | 55 (31.6%) |
| Cytogenetics | |
| Low risk | 53 (35.6%) |
| Intermediate/High risk | 96 (64.4%) |
| CMV Recipient status | |
| negative | 67 (30.3%) |
| positive | 154 (69.7%) |
| Conditioning regimen | |
| MAC | 199 (86.1%) |
| RIC | 32 (13.9%) |
| Year of transplant | |
| 2008–2012 | 47 (20.3%) |
| 2013–2017 | 98 (42.4%) |
| 2018–2021 | 86 (37.3%) |
| Chronic GvHD | |
| absent/mild | 131 (68.6%) |
| moderate/severe | 60 (31.4%) |
| Overall Survival | |
| alive | 133 (57.6%) |
| dead | 98 (42.4%) |
| Univariate Models | Multivariate Model | |||
|---|---|---|---|---|
| HR (95% CI) | p | HR (95%CI) | p | |
| Age | 0.080 | 0.262 | ||
| (40–60 vs. <40 years) | 1.30 (0.73–2.31) | 0.369 | 0.56 (0.23–1.38) | 0.210 |
| (>60 vs. <40 years) | 1.93 (1.04–3.59) | 0.038 | 0.91 (0.37–2.25) | 0.839 |
| Gender (male vs. female) | 1.30 (0.87–1.95) | 0.201 | 2.25 (1.18–4.29) | 0.014 |
| HCT-CI (≥3 vs. 0–2) | 3.11 (1.86–5.21) | <0.001 | 3.10 (1.70–5.65) | <0.001 |
| Cytogenetics (interm/high vs. low risk) | 1.80 (1.02–3.20) | 0.044 | 1.54 (0.81–2.92) | 0.191 |
| CMV recipient status (positive vs. negative) | 1.69 (1.04–2.76) | 0.035 | 1.04 (0.47–2.32) | 0.925 |
| Conditioning (RIC vs. MAC) | 1.71 (1.02–2.90) | 0.044 | 1.04 (0.44–2.47) | 0.933 |
| Year of transplant | 0.007 | 0.382 | ||
| (2013–2017 vs. 2008–2012) | 0.51 (0.32–0.82) | 0.005 | 1.64 (0.38–6.99) | 0.506 |
| (2018–2021 vs. 2008–2012) | 0.49 (0.29–0.82) | 0.007 | 1.04 (0.24–4.56) | 0.959 |
| Chronic GvHD moderate-severe (yes vs. no) * | 0.19 (0.09–0.40) | <0.001 | 0.22 (0.09–0.53) | <0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Passera, R.; Zompi, S.; Gill, J.; Busca, A. Explainable Machine Learning (XAI) for Survival in Bone Marrow Transplantation Trials: A Technical Report. BioMedInformatics 2023, 3, 752-768. https://doi.org/10.3390/biomedinformatics3030048
Passera R, Zompi S, Gill J, Busca A. Explainable Machine Learning (XAI) for Survival in Bone Marrow Transplantation Trials: A Technical Report. BioMedInformatics. 2023; 3(3):752-768. https://doi.org/10.3390/biomedinformatics3030048
Chicago/Turabian StylePassera, Roberto, Sofia Zompi, Jessica Gill, and Alessandro Busca. 2023. "Explainable Machine Learning (XAI) for Survival in Bone Marrow Transplantation Trials: A Technical Report" BioMedInformatics 3, no. 3: 752-768. https://doi.org/10.3390/biomedinformatics3030048
APA StylePassera, R., Zompi, S., Gill, J., & Busca, A. (2023). Explainable Machine Learning (XAI) for Survival in Bone Marrow Transplantation Trials: A Technical Report. BioMedInformatics, 3(3), 752-768. https://doi.org/10.3390/biomedinformatics3030048









