Abstract
The retinal degenerative disease retinitis pigmentosa (RP) is a genetic disease that is the most common cause of blindness in adults. In 2016, Chow et. al. identified over 100 candidate modifier genes for RP through the genome-wide analysis of 173 inbred strains from the Drosophila Genetic Reference Panel (DGRP). However, this type of analysis may miss some modifiers lying in trans to the variation. In this paper, we propose an alternative approach to identify transcripts whose expression is significantly altered in strains demonstrating extreme phenotypes. The differences in the eye size phenotype will, therefore, be associated directly with changes in gene expression rather than indirectly through genetic variation that might then be linked to changes in gene expression. Gene expression data are obtained from the DGRP2 database, where each strain is represented by up to two replicates. The proposed algorithmic approach first chooses the strains’ replicate combination that best represents the relationship between gene expression level and eye size. The extensive correlation analysis identified several genes with known relationships to eye development, along with another set of genes with unknown functions in eye development. The modifiers identified in this analysis can be validated and characterized in biological systems.
Keywords:
retinitis pigmentosa; Drosophila melanogaster; DGRP; RNA-Seq; gene expression; correlation 1. Introduction
The goal of personalized medicine is to develop better ways to treat and diagnose disease on an individual level for the patient. This is particularly important because genetic diseases, even those with seemingly simple inheritance patterns, can be incredibly phenotypically heterogeneous. Much of this variability is due to differences in genetic background between patients, but the identity of those variants and the way they influence disease processes remain, in many cases, unknown [1,2]. A better understanding of this modifying variation could lead to new therapeutics that target modifier gene products, or better predictions of prognosis for patients.
One example of this can be seen in the retinal degenerative disease of retinitis pigmentosa (RP). RP is the most common cause of blindness in adults, at an incidence of ~1/4000 [3]. A common cause of dominantly inherited RP is the mutation of the light-sensing G-protein-coupled receptor rhodopsin (RHO). In many cases, the mutation of the rhodopsin protein sequence leads it to misfold and aggregate in the retinal cells, causing cell disfunction and eventually death [3,4]. Even in individuals with identical rhodopsin mutations, symptoms can vary dramatically in severity, ranging from night blindness to an almost complete loss of vision [5].
In a previous study [1], a genome-wide analysis (GWA) was developed using a well-characterized model of a collection of ~200 inbred, fully sequenced strains from the Drosophila Genetic Reference Panel (DGRP) [6,7]. A mutant form of Drosophila rhodopsin was overexpressed in the developing eye (Rh1G69D) and adult eye size was used as a quantitative proxy for the degree of degeneration. Because all environmental conditions were held stable and the genetic disease model was identical in all lines, any variation in phenotype could be attributed to differences in genetic background between the DGRP lines [1]. A genome-wide association analysis identified over 100 candidate modifier genes, including 84 with conserved human orthologues. Several of these have been validated through separate candidate modifier studies [1,8,9].
However, due to its design, this analysis missed any modifiers that lay more than one kilobase from a cis-acting variant in a regulatory region, as any variants farther than this from a gene were labeled as intergenic and not associated with a candidate modifier gene. This arbitrary cut-off reflects the fact that regulatory sequences that are distant from a coding sequence often regulate genes other than the nearest neighbor [10]. This is an issue for all GWA-style analyses as they will miss any modifiers that lie in trans to the variation. In other words, any modifier genes whose expression is altered as an indirect result of changes in the activation of downstream pathways will be missed in this type of genomic analysis because the genetic variation is linked to a different gene [11]. An alternative approach that could flag such modifiers would be to identify transcripts whose expression is significantly altered in strains demonstrating extreme phenotypes (either very large or very small eyes). This approach has been used successfully to identify causative and modifier genes in a variety of diseases and cancers [12,13], yet it has not been widely applied in the DGRP. Rather than identifying expression quantitative trait loci (QTLs) [14], wherein specific variants in the genome are linked to changes in gene expression as the phenotype [11,15], our approach focuses on associating differences in the eye size phenotype directly with expression. Such transcripts will be treated as candidate modifier genes to be validated later. A relevant study was published by the authors in [16].
In this paper, eye size data from a model of retinal degeneration in a fly and gene expression data from adult female flies are combined and analyzed through an algorithmic approach to identify genes whose expressions are correlated with eye size. Section 2 and Section 3 provide a description of input data and the proposed algorithmic approach, respectively. In Section 4, several candidate genes are identified that are related to eye development and degeneration. These potential modifiers are excellent candidates for characterization in a biological model of retinitis pigmentosa. However, the proposed approach has some limitations that are discussed in Section 5, considering future directions as well.
2. Materials and Methods
2.1. Input Data
This research uses two sets of data to investigate the candidate modifier’s retinal degeneration in Drosophila melanogaster. The first dataset consists of a list of the average eye sizes for each strain from the DGRP dataset, as shown in Table 1. According to Chow et al. [1], female flies expressing the Rh1G69D model of degeneration were at least three days old and flash-frozen prior to imaging. At least 10 flies were measured for each 173 DGRP lines. The second dataset includes the gene expression data (Table 2) that were collected as part of a previous independent study carried out by Huang et al. in [11]. RNA was isolated from adult females for the 184 DGRP strains and analyzed using next-generation RNA sequencing. Expression data were obtained for 18,140 genes with two samples for each strain. Each sample represents an independent biological replicate isolated from identical genetic backgrounds. Due to the limited space, Table 1 and Table 2 show a sample of the data showing the line replicates and their expression values for some genes. It is important to note that of the 184 strains, only 172 intersect with the first dataset and therefore were utilized in this analysis.

Table 1.
Average eye size data structure (sample).

Table 2.
Expression-level matrix data structure (sample).
Furthermore, it is worth noting that the gene expression levels for the two replicates are not the same for some genes. Figure 1 depicts the gene expression patterns observed for the 18,140 genes collected in Huang et al. [11] for the two biological replicates of strain RAL049. The standard deviation between the two replicates was measured to be 0.7085. This indicates that there might be some missing information if the expression profile for one replicate is chosen over the other, or even if an average expression level of the two is used. Therefore, in this study, we propose a method to find and select the most representative replicate combination to perform the correlation analysis between the genes’ expression levels and the eye size measurements. Further details will be discussed in the next section.
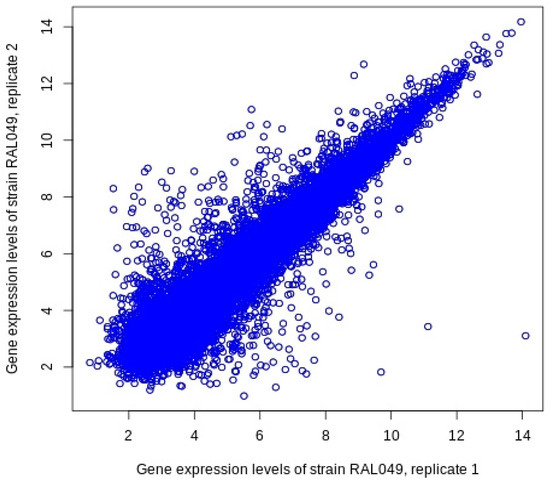
Figure 1.
Inconsistent gene expression levels between the two replicates of strain Ral049.
2.2. The Algorithmic Approach
The first step in the analysis is to identify the strains that exhibit extreme eye sizes. We identified two groups of stains in the first data set representing the largest eye sizes (N1) and the smallest eye sizes (N2), respectively. As shown in Algorithm 1, one of the input parameters specifies the top and bottom quantile thresholds for the eye size range. Those values can be optimized such that the number of selected strains in the two groups are balanced. After choosing strains to be analyzed, the next step is to select which replicate of each strain will be used in the analysis. As mentioned earlier, the disparity in gene expression levels between the two replicates of a strain may affect the accuracy of the analysis. Therefore, we generate a list of all possible combinations of replicates and select the one that best guides the correlation analysis. This task involves a number of steps that are listed in Algorithm 2.
| Algorithm 1: MainProcedure. |
|
| Algorithm 2: GenerateReplicateCombinations. |
|
The process starts by representing each replicate combination with a number. This encoding scheme is similar to the process of representing a number in the binary computer system. Since each strain has two replicates to choose from, a binary digit is used to indicate which replicate is selected, where 0 indicates the first replicate is selected and 1 indicates the second one is selected. Thus, for a given a set of strains, a series of binary digits will be generated representing all possibilities of such combinations. To illustrate this concept, suppose that there are two strains to be analyzed, one from the largest eye size group and another from the smallest eye size group. Because each strain has two replicates, there are four possible replicate combinations that can be represented in binary as: “00”, “01”, “10”, and “11”, which are equivalent to the decimal numbers 0, 1, 2, and 3, respectively. As we mentioned above, “00” means picking the first replicate of each strain, while “01” means picking the first replicate of strain 1 and the second replicate of strain 2, and so on. In general, for N number of strains, each replicate combination requires at least N binary digits to be represented. Furthermore, there are 2N combinations of such possibilities. This works in a similar way to the process of incrementing a digital counter from 0 to 2N−1, where each generated number uniquely identifies a replicate combination.
Therefore, the purpose of Algorithm 2 is to iterate every number from 0 to 2N−1, decode the number into its base-2 representation, and retrieve the choices of replicates based on the values of their binary digits. This will form a complete set of choices from which a replicate will be picked and passed to the next phase of analysis. After Algorithm 2 generates all possible replicate combinations, Algorithm 1 passes them to Algorithm 3 in order to find the best combination among them. That is, for each combination of replicates, the vector of expression levels that is associated with a gene is correlated with the average eye size vector for the extreme strains selected initially. The correlation coefficients are computed using Pearson’s method [17]. The Pearson correlation coefficient values lie between −1 and 1, where a value of 0 implies that there is no linear dependency between the variables under study. A + 1, on the other hand, implies that if one variable increases, the other variable increases and vice versa for −1.
The calculated correlation coefficient value for that combination is then checked against the given threshold value. Each gene whose correlation value passes the threshold condition counts as one point toward the score of that replicate combination. Replicate combinations then accumulate points through this process and eventually the combination possessing the highest score is selected as the best combination. Next, in Algorithm 1, the genes of the best combination are sorted in descending order based on the absolute values of their correlation coefficients, and then only those genes that satisfy the correlation condition will be printed.
| Algorithm 3: FindBestReplicateCombination. |
|
For example, let us consider two strains with eye sizes A and B corresponding to four replicates: namely, A0, A1, B0 and B1. Assume that, for a specific gene, the expression levels for each replicate are C0, C1, D0, and D1, respectively. Since correlation analysis is carried out on all replicate combinations: “00”, “01”, “10”, and “11”. In this case, “00” indicates C0 and D0 are correlated with A and B, “10” indicates C1 and D0 are correlated with A and B, and so forth. Each replicate combination is then awarded 1 point if the absolute value of its correlation coefficient is above the set threshold. This is repeated for all the genes in the expression dataset. Finally, the replicate combination with the top score is considered the best combination and hence is passed to the final step in the analysis.
3. Results and Discussion
As we mentioned above, this study uses two sets of data: the average eye sizes of 173 DGRP strains [1], and a subset of the gene expression data from Huang et al. [11] with 18140 genes representing 184 DGRP female strains. Notably, 172 strains were shared between the two datasets and used in this analysis. In this experiment, we defined the extreme eye sizes by setting quantile values at 20.9% and 87.2%. These bounds were set to ensure only a few strains with the most extreme phenotypes were analyzed and that an equivalent number of strains would be found in each group. This resulted in eight strains designated as the small eye size group (eye sizes represent the bottom 20% of all strains) and eight strains designated as the large eye size group (eye sizes represent the top 12% of all strains). The set of selected strains is listed in Table 3 and is visually highlighted in Figure 2 using the color blue. With each strain having two replicates from the gene expression dataset, this means that there will be 216 or 65,536 different combinations to consider. The algorithm was implemented using the R language and executed on a computer with a Fourth generation Intel CPU 1.7 GHz and 8GB RAM. The total running time was measured as approximately 70 min.

Table 3.
Replicates selected for each strain in the best replicate combination.
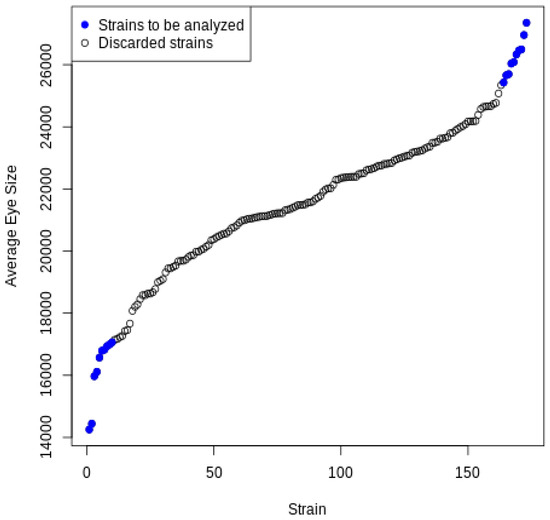
Figure 2.
Average eye sizes of all strains.
Out of the 216 replicate combinations generated, the identified best replicate combinations are indicated in the third column of Table 3. The strain numbers in Table 3 are indices applying to both datasets. By setting the correlation threshold at 0.6, this replicate combination yields 919 genes whose correlation coefficient values are either above 0.6 or below −0.6 (Table S1). We presume that, in most cases, the increased expression of the gene will correlate with the increased activity of the protein encoded by that gene, although we acknowledge that this may not always be the case and will need to be assessed in future studies. We treat these genes as candidate modifiers, as the expression of these genes appears to differentiate between positive and negative outcomes. Among the 919 genes passing the filtering conditions, we will focus our analysis on the top ten candidate genes. The genes were sorted based on their absolute correlation values.
Figure 3 shows the top ten candidate genes along with their correlation values. Some of these genes can be directly linked to eye development and degeneration. The increased expression of Pink is associated with reduced eye size (correlation of –0.8). This is particularly interesting because the human ortholog of Pink is associated with the eye degeneration syndrome human Hermansky–Pudlak syndrome 5 (HPS5) [18]. The most negative correlation value (−0.85) associates the increased expression of MRG15 with reduced eye size. While MRG15 does not have a known role in eye development, previous studies indicate that it interacts with the absent, small, and homeotic discs 1 (ash1) trithorax protein group. This complex, in turn, regulates eye development in Drosophila [19,20,21]. This suggests that MRG15 may have an indirect effect on eye degeneration and is a strong candidate for further validation and characterization in the Drosophila model and other biological systems. Finally, CG2004, an uncharacterized gene, was reported as a candidate modifier in the original screen paper associated with decreased eye size [1].
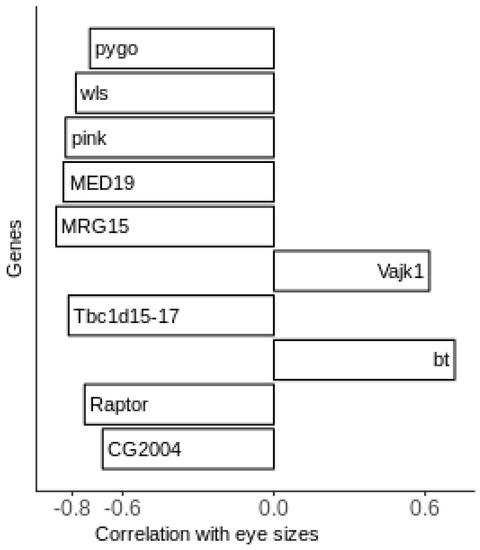
Figure 3.
Top candidate genes and their correlation values.
Effects on eye size phenotype in Drosophila can also come from two signaling pathways: wnt and TOR. Wnt signaling is associated with both pro-apoptopic and anti-apoptopic processes in retinitis pigmentosa [9]. The candidate genes of wntless (wls), whose product is involved in Wnt protein secretion, and pygopus (pygo), a nuclear transcription factor, are both involved in Wnt signaling [22,23]. The expression of both of these genes is associated with reduced eye size (correlation values of <−0.7). The Wnt pathway may, therefore, be a major contributor to the degenerative eye phenotype. To see how strong this influence may be, we examined the other 909 genes for possible roles in Wnt signaling using gene ontogeny analysis with the DAVID functional annotation tool (Supplementary Table S1) [24]. Eleven additional genes are involved in Wnt signaling, with the expression of all but one (fz2) negatively correlated with eye size (Supplementary Table S2). The TOR pathway is able to inhibit autophagy, which is one way a normal cell disposes of misfolded proteins. The increased expression of Raptor, a gene known to work with TOR complex 1 (TORC1), is associated with small eye size (correlation value < −0.7) [25]. To see how strong this influence may be, we examined the other 909 genes for possible roles in TOR signaling using gene ontogeny analysis with the DAVID functional annotation tool (Supplementary Table S1) [26]. Seven additional genes are involved in Wnt signaling, with the expression of all but one (fz2 once again) negatively correlated with eye size (Supplementary Table S3). Although there has been no direct evidence for the involvement of these two pathways in this retinitis pigmentosa model, there may be other genes in these pathways that influence the degenerative phenotype.
Other genes that also have high and consistent correlation values are Tbc1 domain family member 15/17 (Tbc1d15-17), Bent (bt), Mediator subunit 19 (Med19), and Vajk1. Tbc1d15-17, whose expression is correlated with reduced eye size (correlation value < −0.8), encodes a protein that activates Rab GTPases. Some of these, such as Rab11 and Rab1, are reported to be involved in the trafficking of rhodopsins to rhabdomeres [27]. Inability to transport rhodopsins is one pathogenic mechanism in retinitis pigmentosa. The increased expression of Med19 is also associated with reduced eye size, but a role for it in eye development has not been identified. However, Med19 has been associated with cancer cell proliferation, implying a negative effect on cellular function when overexpressed [28]. On the other side of the spectrum, the increased expression of bt is associated with increased eye size. It encodes a calmodulin-dependent protein kinase and is linked to eye development by calmodulin, which is essential for this function [27]. Finally, the increased expression of the chitin-binding protein Vajk1 is also associated with increased eye size, despite no previous evidence in the literature for its association with eye development [29]. These candidate modifier genes provide possible avenues of future research.
In order to further evaluate the analytical approach adopted in this study, the results are compared with the study conducted by Chow et al. [1]. As previously mentioned, a genome-wide association analysis was performed to identify genomic variants that were correlated with the eye degeneration phenotype. To validate their results, the authors reduced the expression of each of the top 14 candidates by driving RNAi in the developing eye. They then used changes in eye size and degeneration to determine if the candidate gene did in fact modify the phenotype [1]. As shown in Table 4, the first and the second columns show part of Chow’s study listing the top identified candidate genes and the measured impact of reduced candidate gene expression on eye size, respectively. The third column shows the correlation coefficients as calculated in this study for each of the candidate genes. Finally, the last column indicates whether this specific gene was identified as a candidate modifier by both methods.

Table 4.
Comparison with Chow’s study [1].
The results show some agreement between the two studies for three candidate genes: Cdk5, fred and CG16885. The listed correlation values indicate that a negative correlation coefficient for Cdk5 corresponds to larger eye sizes upon the reduced expression of that gene in Chow et al. [1]. The positive correlation values for fred and CG16885 correspond with a reduction in eye size upon the reduced expression of these genes in Chow et al. [1]. We also observe similarity in our results with hppy. While none of these genes meet our original correlation cut-off (−0.6/+0.6), the trend in the same direction is a promising result, nonetheless. Chow’s method showed a qualitative reduction in eye appearance, which corresponds to the very small negative correlation value [1]. It is important to note that this research uses different datasets and a different approach than the original study. It would not be unexpected that these approaches would identify different subsets of modifier genes. Because, in this study, transcriptional changes are monitored as opposed to genomic changes, we are more likely to capture candidate modifiers whose expression is linked to the phenotype in trans. In other words, the changes in the expression of these genes are due to genomic variation in regulators of gene expression, such as transcription factors. Alternatively, the genomic variation responsible for the changes in expression may indeed be found in a regulatory element for that gene, but that element may only be distantly linked to the gene of interest. In both cases, the gene would not have been identified in the original genomic variation study.
The results were also compared with the previous study [16]. Only two genes from the top candidates, as determined by the Pearson correlation coefficient, were shared between the two analyses: Ipk2 (−0.6165) and CG10657 (+0.7042). Ipk2 encodes an inositol kinase involved in the synthesis of the sugar inositol phosphate, but its link to retinitis pigmentosa is unclear [30]. CG10657, on the other hand, is orthologous to human RLBP1 that is mutated in Newfoundland rod–cone dystrophy and Bothnia retinal dystrophy [31,32]. The discrepancy in these two lists comes from independent study design. In Amstutz et al., more strains were included in the analysis and a lower absolute correlation coefficient cut-off was used [16]. Significance of the correlation was also factored into the analysis. Because of the use of samples with moderate phenotype and lower correlation, it is expected that the identified genes may be different.
4. Limitations and Future Extensions
Despite promising results, this study is limited by the temporal nature and the differences in genetic disease models utilized. Gene expression varies over time [33]. Because RNA was isolated at a single time point during adulthood, we only have a snapshot of gene expression at that point in time. While the phenotypes for the disease model are expressed during adulthood, the disease mechanisms are activated during larval development. The conclusions we can draw from this comparison are limited. In addition, gene expression was measured in wild-type DGRP strains, instead of in the presence of the degenerative RP model [1]. A better comparison could be made using RNA isolated from larval eye precursor cells in strains that are expressing the disease model. This work is already underway as part of a collaborative effort including the authors. Considering these limitations, it is extremely important to validate these candidates through individual characterization. Future studies will focus on this biological validation by examining the impact that the loss of function of each candidate has, in turn, on the phenotype exhibited by the disease model, in this case, eye degeneration and reduced eye area. Those passing the validation will be characterized in greater detail for their role in regulating the degenerative process, and possibly as candidates for therapeutic targeting or prognostic markers. Such characterization has proven extremely fruitful in previous studies [1,34,35,36,37].
From a computational point of view, there are several directions that further efforts could explore. One area relates to how the replicates are selected. The current approach is intensive in computing power. The computation increases on the order of the power of two as the total number of selected replicates increases. In our tests, a total of 16 strains leads to 216 replicate combinations. However, one should also be mindful that, as the strain count scales, the correlation range to filter meaningful genes may end up with fewer genes because of the noise in the gene expression dataset. In our tests, given the same selected window of correlation values, it has been observed that 7000 genes are returned for two strains from each group of eye size, while only approximately 1000 genes are returned for eight strains from each group. This tradeoff between increased data inclusion and increased data noise must be considered during quantile cut-off selection. Related to this, it could be interesting in the future to extend this method to deal with three or more replicates rather than just two. In this case, the number of possible combinations will increase from 2N to mN, where N refers to the number of selected stains and m represents the number of replicates. In addition, each combination will be represented by m digits instead of 2. This increase in replicate combinations will definitely require more computational time and power to complete the analysis, but should be possible when considering high-performance computing options which can provide more efficient solutions to the task of choosing the most accurate combination.
Another direction that one could explore is to eliminate the lines that have an unacceptable amount of noise between their replicates before diving into the main correlation computation. This approach has the advantage of avoiding expensive replicate selection because one can take a random replicate from a line without impacting the stability of the outcomes. However, the challenge with this approach is to define the condition needed to eliminate the lines that violate the noise threshold. Since this study does not explore this area, it could be the focus of future research.
5. Conclusions
Based on two datasets representing gene expression levels for individual DGRP strains and average eye sizes, a correlation study has been conducted to find out which genes are related to changes in eye size [1,11]. The correlation results reveal new genes and pathways that are likely to be associated with eye degeneration, and candidate genes for future experiments. Although a thorough understanding of these genes has not yet been reached, some of them—such as CG2004 and Pink—have been confirmed as modifiers of eye development and degenerative disease in previous studies. Others show promising potential with their indirect associations with the targeted phenotypes.
Given that each strain has two replicates to choose from, the approach used in this study employs a binary representation to encode replicate combinations for strains, picks the best replicate combination, and performs a correlation calculation to measure the relationship between each gene’s expression level and the eye size measurements. Due to its combinatorial nature, this approach faces challenges when the number of strains increases. This is one area that future efforts could improve, which could reveal better insights into the genotype–phenotype relationship.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biomedinformatics2040040/s1, Table S1: Candidate genes and correlations, Table S2: Candidate genes involved in Wnt signaling, Table S3: Candidate genes involved in TOR.
Author Contributions
T.N. was responsible for implementing the algorithm using R, creating the first draft of the paper, producing and visualizing the results, and editing the final manuscript. A.K. was involved in the conception of the project, was responsible for advising the computational aspects of the research, and was engaged in writing/revising/editing various drafts of the paper. R.P. was involved in the conception of the project, writing secondary drafts related to the biological functions of the genes in question, and editing the final manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Genomic sequence and gene expression data for the DGRP is available at http://dgrp.gnets.ncsu.edu/ (accessed on 24 October 2020). Gene expression data were initially published in Huang et al., 2015. Eye size data are available as a supplementary file from Chow et al., 2016.
Conflicts of Interest
The authors have no relevant financial or non-financial conflict of interest to disclose.
References
- Chow, C.Y.; Kelsey, K.J.P.; Wolfner, M.F.; Clark, A.G. Candidate Genetic Modifiers of Retinitis Pigmentosa Identified by Exploiting Natural Variation in Drosophila. Hum. Mol. Genet. 2016, 25, 651–659. [Google Scholar] [CrossRef] [PubMed]
- Queitsch, C.; Carlson, K.D.; Girirajan, S. Lessons from Model Organisms: Phenotypic Robustness and Missing Heritability in Complex Disease. PLoS Genet. 2012, 8, e1003041. [Google Scholar] [CrossRef] [PubMed]
- Hartong, D.T.; Berson, E.L.; Dryja, T.P. Retinitis Pigmentosa. Lancet 2006, 368, 1795–1809. [Google Scholar] [CrossRef]
- Sung, C.H.; Davenport, C.M.; Nathans, J. Rhodopsin Mutations Responsible for Autosomal Dominant Retinitis Pigmentosa. Clustering of Functional Classes along the Polypeptide Chain. J. Biol. Chem. 1993, 268, 26645–26649. [Google Scholar] [CrossRef]
- Chang, S.; Vaccarella, L.; Olatunji, S.; Cebulla, C.; Christoforidis, J. Diagnostic Challenges in Retinitis Pigmentosa: Genotypic Multiplicity and Phenotypic Variability. Curr. Genom. 2011, 12, 267–275. [Google Scholar] [CrossRef] [PubMed]
- Mackay, T.F.C.; Richards, S.; Stone, E.A.; Barbadilla, A.; Ayroles, J.F.; Zhu, D.; Casillas, S.; Han, Y.; Magwire, M.M.; Cridland, J.M.; et al. The Drosophila melanogaster Genetic Reference Panel. Nature 2012, 482, 173–178. [Google Scholar] [CrossRef]
- Huang, W.; Massouras, A.; Inoue, Y.; Peiffer, J.; Ramia, M.; Tarone, A.M.; Turlapati, L.; Zichner, T.; Zhu, D.; Lyman, R.F.; et al. Natural Variation in Genome Architecture among 205 Drosophila melanogaster Genetic Reference Panel Lines. Genome Res. 2014, 24, 1193–1208. [Google Scholar] [CrossRef]
- Palu, R.A.S.; Chow, C.Y. Baldspot/ELOVL6 is a Conserved Modifier of Disease and the ER Stress Response. PLoS Genet. 2018, 14, e1007557. [Google Scholar] [CrossRef]
- Palu, R.A.S.; Dalton, H.M.; Chow, C.Y. Decoupling of Apoptosis from Activation of the ER Stress Response by the Drosophila Metallopeptidase superdeath. Genetics 2020, 214, 913–925. [Google Scholar] [CrossRef]
- Visel, A.; Rubin, E.M.; Pennacchio, L.A. Genomic Views of Distant-Acting Enhancers. Nature 2009, 461, 199–205. [Google Scholar] [CrossRef]
- Huang, W.; Carbone, M.A.; Magwire, M.M.; Peiffer, J.A.; Lyman, R.F.; Stone, E.A.; Anholt, R.R.H.; Mackay, T.F.C. Genetic Basis of Transcriptome Diversity in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2015, 112, E6010–E6019. [Google Scholar] [CrossRef] [PubMed]
- Liang, P.; Pardee, A.B. Analysing Differential Gene Expression in Cancer. Nat. Rev. Cancer 2003, 3, 869–876. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Esteban, R.; Jiang, X. Differential Gene Expression in Disease: A Comparison between High-Throughput Studies and the Literature. BMC Med. Genom. 2017, 10, 59. [Google Scholar] [CrossRef] [PubMed]
- Members of the Complex Trait Consortium. The Nature and Identification of Quantitative Trait Loci: A Community’s View. Nat. Rev. Genet. 2003, 4, 911–916. [Google Scholar] [CrossRef]
- Everett, L.J.; Huang, W.; Zhou, S.; Carbone, M.A.; Lyman, R.F.; Arya, G.H.; Geisz, M.S.; Ma, J.; Morgante, F.; St. Armour, G.; et al. Gene Expression Networks in the Drosophila Genetic Reference Panel. Genome Res. 2020, 30, 485–496. [Google Scholar] [CrossRef]
- Amstutz, J.; Khalifa, A.; Palu, R.; Jahan, K. Cluster-Based Analysis of Retinitis Pigmentosa Modifiers Using Drosophila Eye Size and Gene Expression Data. Genes 2022, 13, 386. [Google Scholar] [CrossRef]
- Pearson, K. Notes on the history of correlation. Biometrika 1920, 13, 25–45. [Google Scholar] [CrossRef]
- Syrzycka, M.; McEachern, L.A.; Kinneard, J.; Prabhu, K.; Fitzpatrick, K.; Schulze, S.; Rawls, J.M.; Lloyd, V.K.; Sinclair, D.A.R.; Honda, B.M. The Pink Gene Encodes the Drosophila Orthologue of the Human Hermansky-Pudlak Syndrome 5 (HPS5) Gene. Genome 2007, 50, 548–556. [Google Scholar] [CrossRef]
- Huang, C.; Yang, F.; Zhang, Z.; Zhang, J.; Cai, G.; Li, L.; Zheng, Y.; Chen, S.; Xi, R.; Zhu, B. Mrg15 Stimulates Ash1 H3K36 Methyltransferase Activity and Facilitates Ash1 Trithorax Group Protein Function in Drosophila. Nat. Commun. 2017, 8, 1649. [Google Scholar] [CrossRef]
- Janody, F.; Lee, J.D.; Jahren, N.; Hazelett, D.J.; Benlali, A.; Miura, G.I.; Draskovic, I.; Treisman, J.E. A Mosaic Genetic Screen Reveals Distinct Roles for Trithorax and Polycomb Group Genes in Drosophila Eye Development. Genetics 2004, 166, 187–200. [Google Scholar] [CrossRef]
- Rozovskaia, T.; Tillib, S.; Smith, S.; Sedkov, Y.; Rozenblatt-Rosen, O.; Petruk, S.; Yano, T.; Nakamura, T.; Ben-Simchon, L.; Gildea, J.; et al. Trithorax and ASH1 Interact Directly and Associate with the Trithorax Group-Responsive Bxd Region of the Ultrabithorax Promoter. Mol. Cell. Biol. 1999, 19, 6441–6447. [Google Scholar] [CrossRef] [PubMed]
- Bänziger, C.; Soldini, D.; Schütt, C.; Zipperlen, P.; Hausmann, G.; Basler, K. Wntless, a Conserved Membrane Protein Dedicated to the Secretion of Wnt Proteins from Signaling Cells. Cell 2006, 125, 509–522. [Google Scholar] [CrossRef] [PubMed]
- Belenkaya, T.Y.; Han, C.; Standley, H.J.; Lin, X.; Houston, D.W.; Heasman, J.; Lin, X. Pygopus Encodes a Nuclear Protein Essential for Wingless/Wnt Signaling. Development 2002, 129, 4089–4101. [Google Scholar] [CrossRef] [PubMed]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef]
- Xu, T.; Nicolson, S.; Denton, D.; Kumar, S. Distinct Requirements of Autophagy-Related Genes in Programmed Cell Death. Cell Death Differ. 2015, 22, 1792–1802. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Xiong, B.; Bellen, H.J. Rhodopsin Homeostasis and Retinal Degeneration: Lessons from the Fly. Trends Neurosci. 2013, 36, 652–660. [Google Scholar] [CrossRef]
- Zhao, Y.; Meng, Q.; Gao, X.; Zhang, L.; An, L. Down-Regulation of Mediator Complex Subunit 19 (Med19) Induces Apoptosis in Human Laryngocarcinoma HEp2 Cells in an Apaf-1-Dependent Pathway. Am. J. Transl. Res. 2017, 9, 755–761. [Google Scholar]
- Cinege, G.; Zsámboki, J.; Vidal-Quadras, M.; Uv, A.; Csordás, G.; Honti, V.; Gábor, E.; Hegedűs, Z.; Varga, G.I.B.; Kovács, A.L.; et al. Genes Encoding Cuticular Proteins Are Components of the Nimrod Gene Cluster in Drosophila. Insect Biochem. Mol. Biol. 2017, 87, 45–54. [Google Scholar] [CrossRef]
- Seeds, A.M.; Sandquist, J.C.; Spana, E.P.; York, J.D. A molecular basis for inositol polyphosphate synthesis in Drosophila melanogaster. J. Biol. Chem. 2004, 279, 47222–47232. [Google Scholar] [CrossRef]
- Eichers, E.R.; Green, J.S.; Stockton, D.W.; Jackman, C.S.; Whelan, J.; McNamara, J.A.; Johnson, G.J.; Lupski, J.R.; Katsanis, N. Newfoundland rod-cone dystrophy, an early-onset retinal dystrophy, is caused by splice-junction mutations in RLBP1. Am. J. Hum. Genet. 2002, 70, 955–964. [Google Scholar] [CrossRef] [PubMed]
- Burstedt, M.S.; Sandgren, O.; Holmgren, G.; Forsman-Semb, K. Bothnia dystrophy caused by mutations in the cellular retinaldehyde-binding protein gene (RLBP1) on chromosome 15q26. Investig. Ophthal. Vis. Sci. 1999, 40, 995–1000. [Google Scholar]
- Storey, J.D.; Xiao, W.; Leek, J.T.; Tompkins, R.G.; Davis, R.W. Significance Analysis of Time Course Microarray Experiments. Proc. Natl. Acad. Sci. USA 2005, 102, 12837–12842. [Google Scholar] [CrossRef]
- Talsness, D.M.; Owings, K.G.; Coelho, E.; Mercenne, G.; Pleinis, J.M.; Partha, R.; Hope, K.A.; Zuberi, A.R.; Clark, N.L.; Lutz, C.M.; et al. A Drosophila Screen Identifies NKCC1 as a Modifier of NGLY1 Deficiency. eLife 2020, 9, e57831. [Google Scholar] [CrossRef] [PubMed]
- Palu, R.A.S.; Ong, E.; Stevens, K.; Chung, S.; Owings, K.G.; Goodman, A.G.; Chow, C.Y. Natural Genetic Variation Screen in Drosophila Identifies Wnt Signaling, Mitochondrial Metabolism, and Redox Homeostasis Genes as Modifiers of Apoptosis. G3 Genes Genomes Genet. 2019, 9, 3995–4005. [Google Scholar] [CrossRef]
- Lavoy, S.; Chittoor-Vinod, V.G.; Chow, C.Y.; Martin, I. Genetic Modifiers of Neurodegeneration in a Drosophila Model of Parkinson’s Disease. Genetics 2018, 209, 1345–1356. [Google Scholar] [CrossRef]
- He, B.Z.; Ludwig, M.Z.; Dickerson, D.A.; Barse, L.; Arun, B.; Vilhjálmsson, B.J.; Jiang, P.; Park, S.-Y.; Tamarina, N.A.; Selleck, S.B.; et al. Effect of Genetic Variation in a Drosophila Model of Diabetes-Associated Misfolded Human Proinsulin. Genetics 2014, 196, 557–567. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).