Abstract
The adoption of “artificial intelligence (AI) in drug discovery”, where AI is used in the process of pharmaceutical research and development, is progressing. By using the ability to process large amounts of data, which is a characteristic of AI, and achieving advanced data analysis and inference, there are benefits such as shortening development time, reducing costs, and reducing the workload of researchers. There are various problems in drug development, but the following two issues are particularly problematic: (1) the yearly increases in development time and cost of drugs and (2) the difficulty in finding highly accurate target genes. Therefore, screening and simulation using AI are expected. Researchers have high demands for data collection and the utilization of infrastructure for AI analysis. In the field of drug discovery, for example, interest in data use increases with the amount of chemical or biological data available. The application of AI in drug discovery is becoming more active due to improvement in computer processing power and the development and spread of machine-learning frameworks, including deep learning. To evaluate performance, various statistical indices have been introduced. However, the factors affected in performance have not been revealed completely. In this study, we summarized and reviewed the applications of deep learning for drug discovery with BigData.
1. Introduction
Originally, artificial intelligence (AI) was used in the field of drug discovery for processes such as the pattern matching of molecular structure correlations and the automatic registration of case reports [1,2,3,4]. However, due to advances made in recent years, AI can also be used in target search, target selection, lead compound search, and phase optimization. To make medicine, researchers need to know what the keyhole that opens the door that causes the disease looks like, which is the process of target search and selection. To develop a key that fits the keyhole, a compound is developed and optimized. In addition, animal studies and clinical trials are repeated to check whether the door can be unlocked and if there are any problems [5,6,7]. If the active ingredient is then approved through an application for marketing authorization and testing, it can finally be marketed as a drug. Therefore, the development of a drug usually takes a very long time—more than 10 years on average—and involves enormous costs. However, the probability of success in discovering a drug can be as low as 0.004%, so a tremendous amount of trial and error is required, resulting in increased labor, time, and cost. It is also a daunting task to select drug discovery target proteins from more than one hundred thousand types of in vivo proteins and to try combinations with active compounds. Even when animal studies are successful, significant side effects often occur in human clinical trials [8]. Due to advances in medical science, research topics have shifted to complex diseases with unknown causes, and the hurdles of drug discovery are becoming higher by the year [9,10,11]. In addition, tightening pharmaceutical regulations, pressure to raise drug prices, and generic competition lead to an ever-widening innovation gap. Therefore, it is understandable that transforming conventional methods into more efficient drug discovery systems is a great challenge, and AI drug discovery is also attracting increasing attention. However, it is not easy to discover target genes that lead to successful drug development. The failure rate is particularly high in the proof-of-concept (PoC) stage, the proof of superiority assumed at the beginning of drug discovery, and in phase II of small-scale clinical trials [12,13]. The main reasons for this difficulty are the selection of low-accuracy target genes based mainly on animal studies and the start of the drug discovery process with insufficient stratification of patients to be treated. Therefore, efforts are being made in target gene searches to promote new drug discovery targets based on human or other data [14,15,16]. It is expected that the use of human data such, as omics data and clinical information, can reduce failures due to reliance on animal testing [17,18]. Another benefit is the ability to leverage search results from other disease predictors that share drug discovery targets. In the search for lead compounds, efforts have been made toward an AI system performing the screening of a large number of active compounds. In particular, AI is expected to improve development efficiency in the search for lead compounds, such as biopharmaceuticals that have larger molecular sizes and are more complex than low-molecular-weight drugs [19]. Although AI drug discovery is starting to deliver results, it is fair to say that it is still a developing field. For example, aggregated data cannot be used in original form for AI machine learning, and format standardization is insufficient [20,21,22]. However, as a platform for AI drug discovery that combines genomes and AI, a data-driven drug discovery target search approach is being promoted.
Laboratory automation, including research automation, is gaining attention due to advances in AI technology and data-driven research and development. Alternatively, the development of measurement technology, such as robotic automated experimentation technology, has produced large-scale biological image data that cannot be processed by humans [23,24,25]. As a result, analyzing and quantifying large amounts of high-resolution biological image data have become important issues in biology. Therefore, “bioinformatics”, which solves these problems using information science methods, has attracted much attention in recent years. Particularly in bioimaging, bioinformatics has been pushing forward and various research has been conducted, such as database development for biological images and the extraction of information from images, including the segmentation of cells and tissues, as well as the visualization of image information [26,27,28]. Among these, research on the classification and clustering of biological images using machine learning has been the most-studied area. Applied research, such as lesion detection from medical images, has also actively been conducted [29,30,31]. Since image classification is an important research step in the analysis of biological image data, a method for classifying biological image data with high accuracy and high throughput is desired. In this case, if there are few data, it is possible to classify images visually. However, when there are a lot of data, it is difficult to classify all the images manually. Therefore, it is common to automatically classify image data using a machine-learning method called supervised learning. As methods for learning, multilayer neural networks, such as deep learning, which are currently widely used, are attracting attention due to their amazing prediction performances [32,33,34,35]. Deep learning is an AI technology that automatically extracts and learns features based on large amounts of data [36,37]. It essentially consists of a multilayer neural network and an algorithm that mimics human neurons, and it automatically processes and learns input data and passes them to the next layer, which consists of three or more layers; in these layers, it is possible to deepen the characteristics of the data to be learned using multiple layers of this neural network, which result in deep-learning models with extremely high accuracy, sometimes surpassing human recognition accuracy [38,39,40,41,42,43]. As a deep-learning method, the structure of a neural network is often used. A neural network has what is called a “hidden layer”, which is about two to three layers in conventional neural networks. However, deep-learning models can have up to 150 hidden layers. For this reason, deep-learning models are also referred to as deep neural networks. Traditionally, feature extraction has been performed manually. However, deep-learning models are trained using large-scale, labeled data and neural network structures, where it is possible to learn features directly from the data, eliminating the need for feature extraction. Convolutional neural networks (CNNs or ConvNets) are used in deep neural networks (Figure 1 and Figure 2) [44,45,46,47,48,49,50]. CNNs do not require manual feature extraction and have the advantage of eliminating the need to search for features during image classification. In recent years, deep learning has become an indispensable method for AI development that has been trained with deep learning and has attracted great attention because it has a higher performance than networks trained with other methods. What should be focused on now is thinking about “What kind of data do we collect and for what purpose do we use it”. As for “What can be learned from the data”, we are entering an era where the power of AI can be harnessed. Even in drug discovery research, the amount of data that can be collected and processed is exploding. This is driven by various advantages, such as the proliferation of comprehensive biomolecular measurement devices, including next-generation sequences, or the digitization of medical information [51,52,53,54,55]. In contrast, the pharmaceutical industry faces challenges that complicate the development of new drugs, such as rising research and development costs and the depletion of drug targets that can be developed using conventional approaches. It can be said that the development and introduction of AI systems that search for drug discovery targets are urgently needed as a way out of this situation. Given the rapid decline in data collection and processing costs, the demand for AI, which refers to all attempts to make computers realize the information processing of the human brain, is likely to continue to increase in drug discovery research. Alternatively, machine learning refers to a branch of technology that enables such efforts in AI. One type of machine learning is a method called a neural network, which imitates how information is transmitted from neurons in the brain. In recent years, an extended approach called deep learning has been developed, and it is expanding its range of activities in many fields. For example, one of the reasons why deep learning has become so popular is that a computer figures out “what kind of data should be prepared for computer learning, feature design”, instead of a data analyst. Conversely, deep learning is designed to automatically obtain useful feature values for prediction because it repeats the process of combining input feature values to create new feature values [56,57,58]. Such features are particularly useful when one is trying to complete a task in a data-driven manner without relying on known information or when large numbers of feature values in the input data make it difficult to determine which feature values are important for prediction.
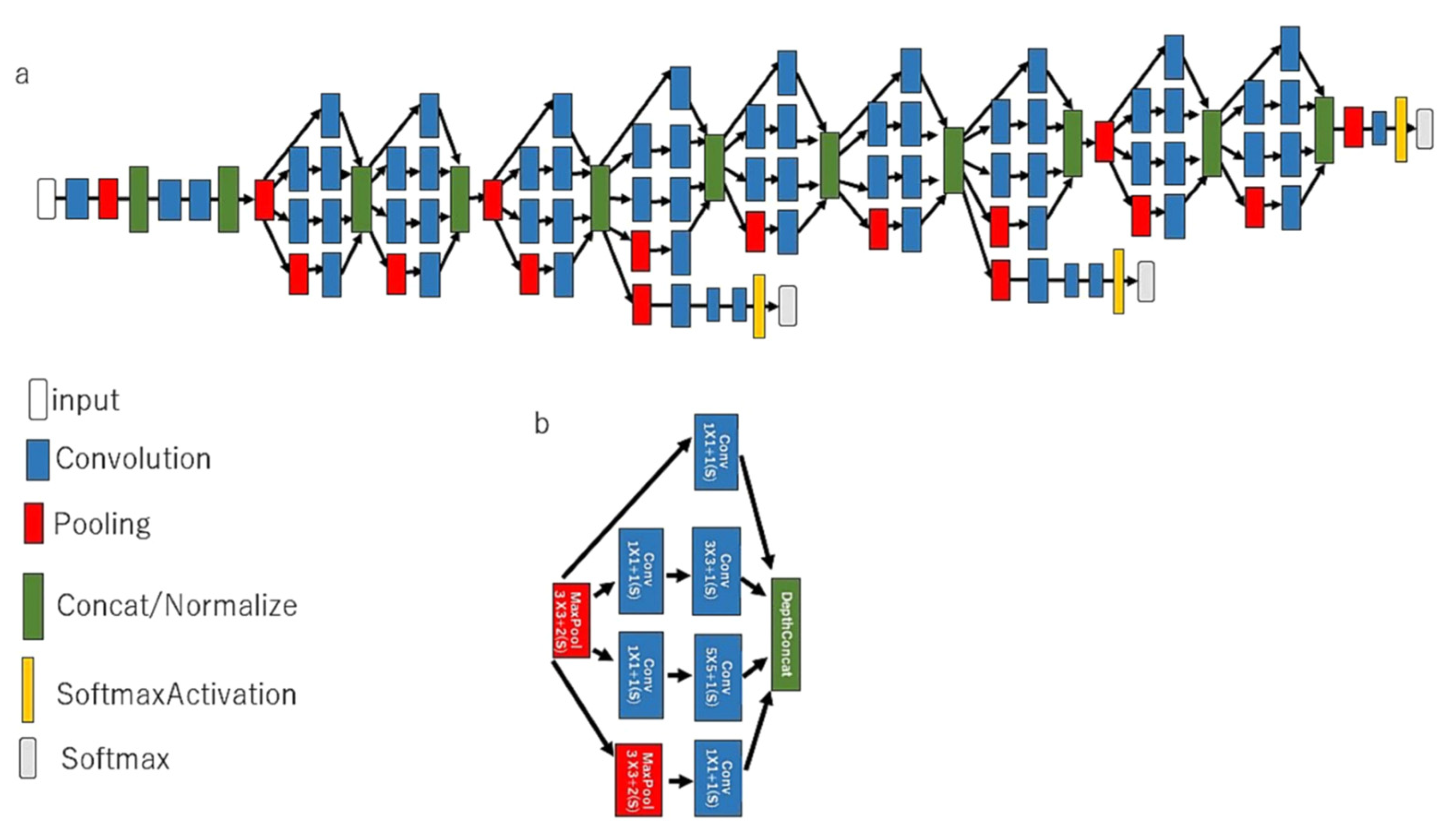
Figure 1.
The architecture of a CNN model in GoogLeNet. The pretrained CNN comprises a 22-layer DNN, both (a) implemented with a novel element dubbed an inception module and (b) implemented with batch normalization, image distortion, and RMSprop, including a total of 4 million parameters.
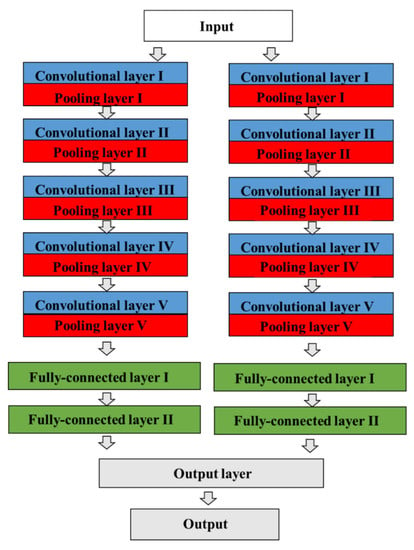
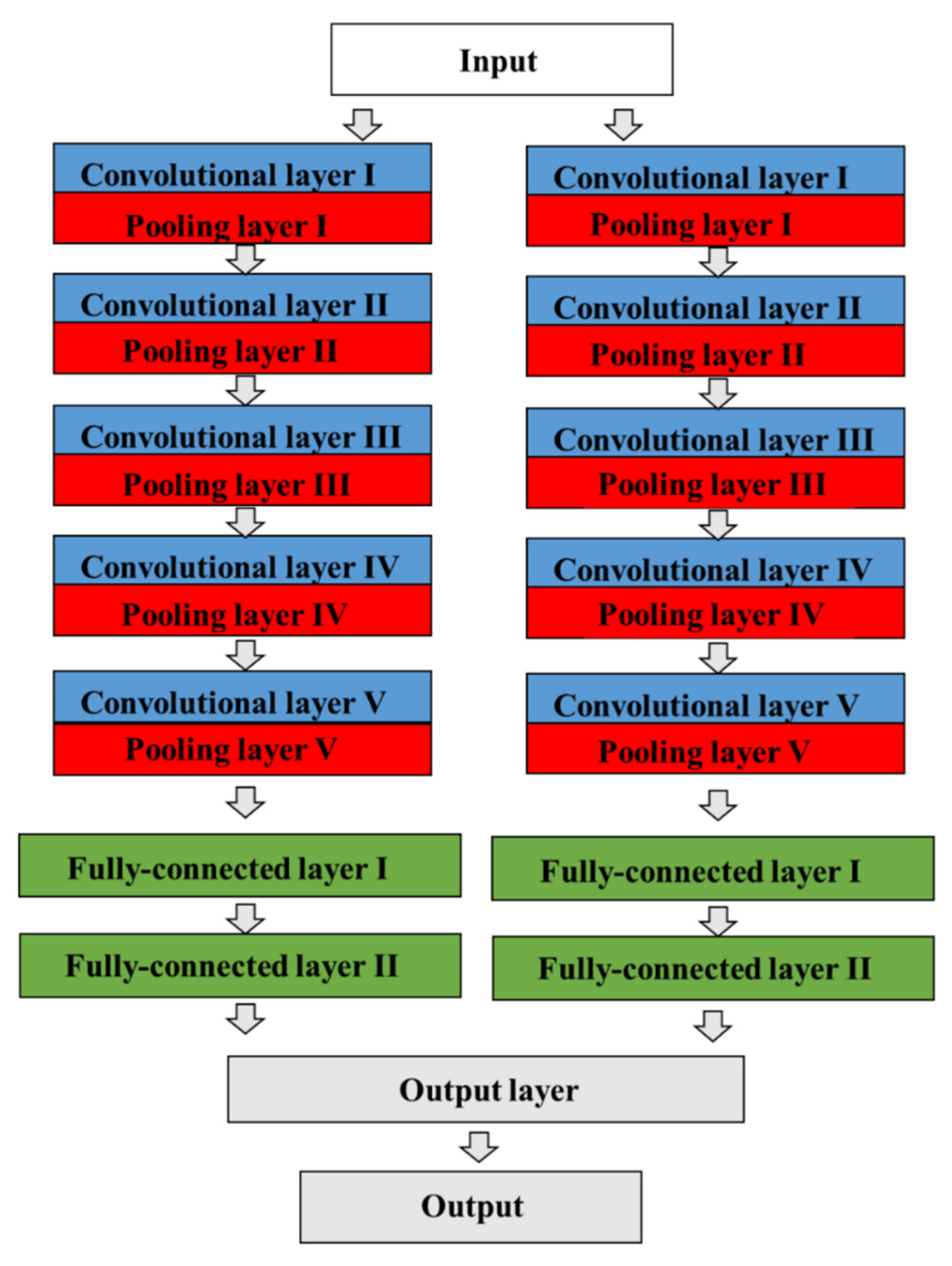
Figure 2.
The architecture of a CNN model in AlexNet. The CNN contains a total of eight prelearned layers, which consist of five convolutional and max-pooling layers, three fully connected layers, dropout, data augmentation, rectified linear unit activations, and a stochastic gradient descent with momentum, including a total of 60 million parameters. The two adjacent convolutional and pooling layers are finally combined into a third fully connected layer.
Deep learning has a long history, and the theory itself was proposed in the 1980s [59,60]. One of the reasons why the theory, which has existed for a long time, is attracting attention in modern times is that research on neural networks has made progress, but that is not the only reason. In other words, the reason deep learning is attracting attention today is that it has become easier to prepare the data used for learning. This is because the development of the Internet has made it easier to obtain the data used in deep learning. Another reason is the development of technologies to process unstructured data, such as that of images, voice, and text. In the 1980s, when the theory of deep learning was proposed, it was not yet possible to collect and store large amounts of data. However, in modern times, improved technology has made it possible to collect large amounts of labeled data [61,62,63,64,65]. With this background, it has become possible to realize deep learning, which previously had problems in terms of data. Another reason why deep learning is attracting attention is that the processing power of computers has improved [66,67,68]. To perform deep learning, a high-performance graphics processing unit (GPU) is essential so that an AI can learn while comparing a large amount of data [69,70]. At the time deep learning was proposed, this advanced technology did not exist. In modern times, the technology has evolved, and it has become possible to improve the accuracy and efficiency of deep learning. In recent years, GPUs have a “parallel configuration” that enables efficient deep learning. In this way, improving the computing power of computers has become an important factor for the development of deep learning in recent years.
On the other hand, GPUs cause high calculation costs compared to those of some conventional processes of machine learning. However, a GPU is the main cause of high performances in CNNs due to the high processing ability of graphical data. CNNs represent an upgrade by improving structural disadvantages. In this review, we summarize the applications of deep learning for drug discovery with BigData.
2. Types of Deep-Learning Network Models
2.1. Networks in Deep Learning
Deep learning can be classified into different types depending on the learning method and structure. First, CNNs are mainly used in image recognition [71,72,73]. Images that contain meaningful information for humans are nothing but numerical data for machines. Therefore, it is not easy for machines to extract the information that humans want to know from images. However, CNNs have high recognition accuracy and learn features directly without requiring humans to extract them. In addition, a generative adversarial network (GAN) consists of two networks, a generator (generative network) and a discriminator (discriminator network), which are characterized by competing with each other to increase accuracy [74,75,76,77,78]. This can be used, for example, to correct blurred photos or to restore old, low-resolution images to high resolution. It can also create images of non-existent people by capturing images of multiple faces [79,80]. It is mainly used in the world of image processing, as it is possible to generate or transform nonexistent data by learning features from the data. In addition, a recurrent neural network’s (RNN’s) algorithm is particularly suitable for processing data collected in a timeseries or data that has an ordered meaning [81,82]. By applying various kinds of filters, CNN-based machine-learning models are able to capture high-level representations of the input data, making them popular in computer vision tasks. Examples of CNN applications include image classification (e.g., AlexNet, VGG network, ResNet, and MobileNet) and object detection (e.g., Fast R-CNN, Mask R-CNN, YOLO, and SSD). As shown in Figure 3, the performances of popular CNNs applied to AI vision tasks have gradually improved over the years, surpassing human vision [83]. These models can extract and classify the structural features of images, such as chemical compounds, leading to the discovery of novel drug seeds.
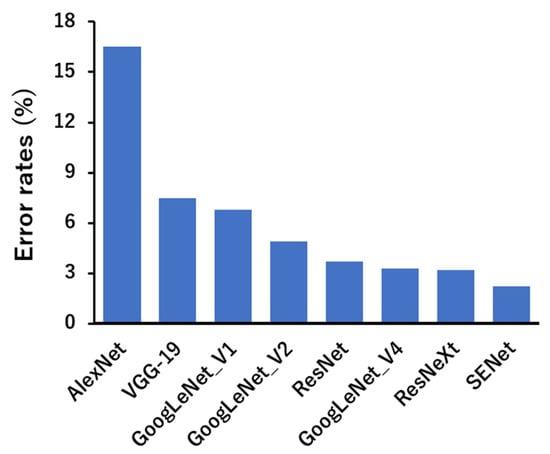
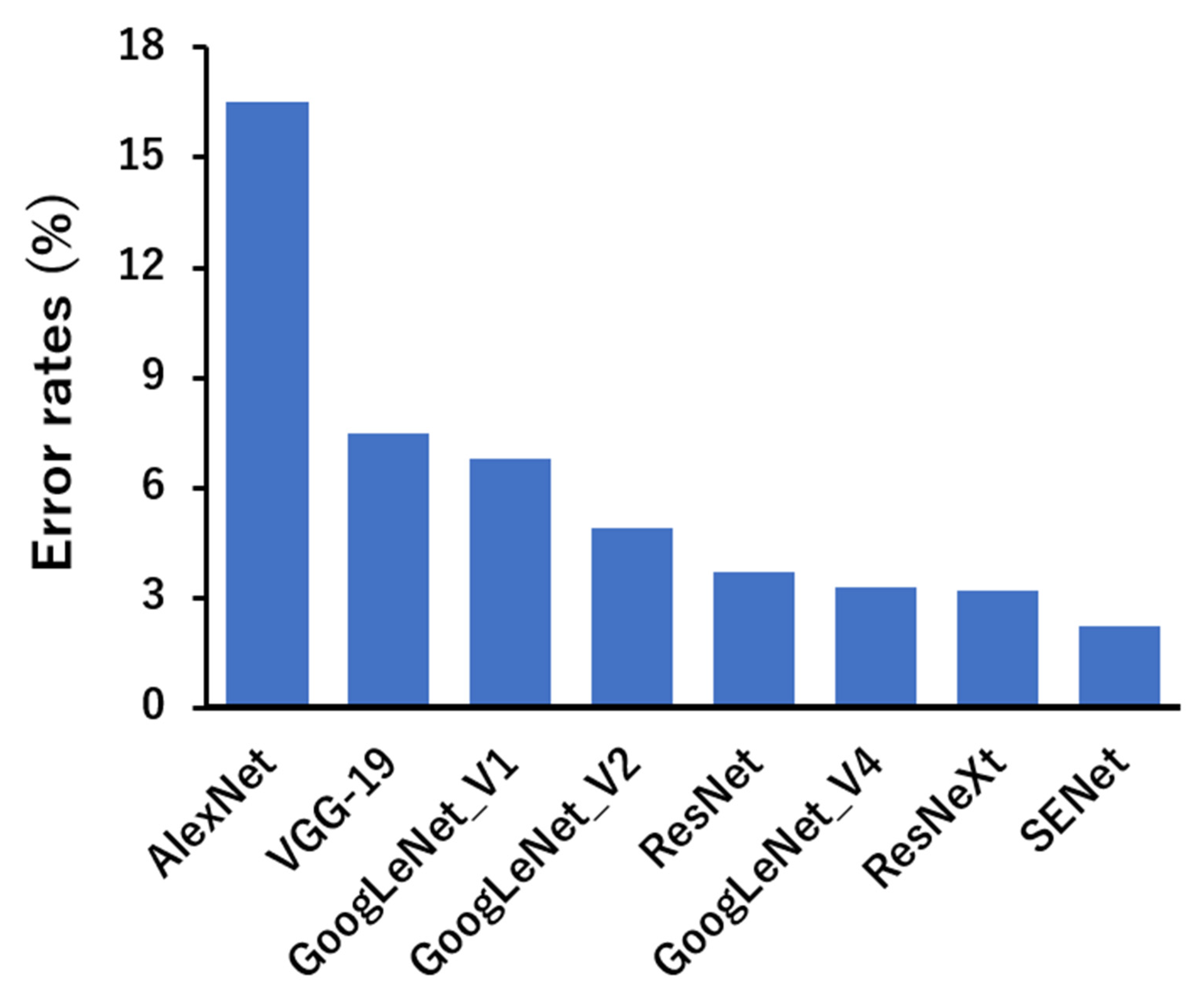
Figure 3.
Performances of current popular deep neural networks on ImageNet [83].
2.2. Technological Application in BigData and Deep Learning
The term AI was first used in the summer of 1956 at the Dartmouth Conference by John McCarthy, an American computer scientist [84]. The concept of AI is now widely known and has triggered the entry of many researchers into research, alternating between boom and bust periods [85]. Initially, the first AI boom began in the 1950s and lasted until the 1960s, when “inference” and “exploration” were the main research areas, and it became a boom because it became possible to present solutions to specific problems. However, the problems that people faced in their lives did not choose patterns within specific rules. It became easier to find an optimal answer within set rules, but it was difficult to solve problems that could arise in the real world, and disappointment with AI spread. As a result, the boom ended, and AI research entered a winter period. After that, there were advances in research that used computer processing to reason and solve problems. The second AI boom that followed is said to have occurred in the 1980s, when attention was primarily focused on expert systems, systems in which a computer acts like an expert by taking the “knowledge” of a specialized field into the computer and drawing conclusions. It became possible to solve complex problems by inputting specialized information into computers and incorporating conditional expressions that provided answers when the conditions were met. The use of expert systems was expected to make it possible to solve practical problems that could not be solved in the first AI boom. In addition, many attempts were made to reproduce intellectual judgments made by humans using manually written rules. However, manual rule writing was limited in terms of expanding scope and maintaining quality, and the scope of its application was not expanded. Although expert systems seemed to be a great approach, they proved to have difficulty with handling all cases accurately. The reason was that the amount of “knowledge” became enormous and contradictory, there was a lack of consistency among the “knowledge”, it was difficult to assess ambiguous cases, and the huge amount of “knowledge” needed to be written and edited by humans. Because of the limitations of such expert systems, expectations for AI again declined. Subsequently, AI research was actively pursued in terms of the memory that stores knowledge in computers. The recent AI boom is referred to as the third AI boom, which was triggered by the spread of the Internet and the advancement of technology, and has continued since the 2000s until today. In 2012, a team using a neural network won an image recognition software competition with a large margin of accuracy [86]. “Machine learning”, in which AI itself acquires knowledge by using a large amount of data known as “BigData”, has been put to practical use. Through a machine-learning approach, it became possible to inductively extract rules from observed data based on case studies, overcoming the limitations of manual rule descriptions.
In addition to technological innovations related to data collection (such as next-generation sequencers) and dramatic improvements in data-processing technology (such as computer specifications), omics analysis, which comprehensively measures tens of thousands of biomolecules (such as transcripts and proteins) in a single experiment, is now essential in the field of life science research [87,88,89,90,91]. Against this backdrop, the economic cost of omics analysis is decreasing, and multi-omics analysis, in which multiple types of omics analyses are performed under certain experimental conditions, is becoming increasingly popular as an attempt to systematically understand life phenomena. The problem at this point is that it is difficult for humans to interpret the data because a large number of elements are measured under specific experimental conditions, and the computational cost is high. To deal with this problem, possible approaches include (1) reducing the feature quantity to only important features or (2) replacing the feature quantity with a smaller amount of feature quantity by replacing the data with a new feature set. For example, a principal components analysis (PCA) plot reduces the high-dimensional (D) data to 2D or 3D, and the data are visualized by plotting the newly obtained feature values on the x-axis and the y-axis, as well as the z-axis (dimensionality reduction) [92,93,94,95,96,97]. In addition, multilayered autoencoders are also used for dimensionality reduction and are a type of deep learning that compress data that are “recoverably” used for learning [98]. For example, when high-dimensional multi-omics data with tens of thousands of feature values are input, these feature values are added together after they are weighted differently, and then a new feature value is generated through nonlinear transformation using an activation function in a hidden layer (encoder). After this process, the output layer restores the original input data (decoder). In recent years, a model for predicting biological or pathological states of RNA-seq data has been developed by quantifying RNA, miRNAseq, and methylated DNA data using next-generation sequencers. In this analysis pipeline, an autoencoder with three hidden layers is introduced for the dimensionality reduction of omics data. Then, clustering is performed with the new features generated in the intermediate layer, and it can be said that the “omics data are integrated” because new feature values are generated by combining feature values derived from the heterogeneous omics data. However, in the autoencoder, the feature that contributes to the predictions is a black box [99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121,122,123,124,125,126,127,128]. Even in multi-omics analysis, knowing “which feature value contributed to the prediction” is expected to be useful in drug discovery target search. The development of deep learning, in which the prediction criteria are not black boxes but are interpretable, is also progressing. With the widespread use of high-throughput measurement instruments, such as next-generation sequencers, the barriers to comprehensive data collection at multiple levels, such as genes, transcripts, proteins, etc., in biological and pathological conditions have already fallen. Conversely, research is still underway on how to integrate heterogeneous but highly interrelated multi-omics data to gain biologically useful insights. Moreover, machine-learning methods, including deep learning, can only perform as expected if the datasets containing multi-omics data are sufficiently large.
The availability of BigData is what makes this approach so effective. In other words, large amounts of data are now collected from various services on the Internet and various sensors in the real world. In addition, improvements in computing power have made it possible to process large amounts of data at high speeds, leading to advances in machine-learning technology [129]. In addition, deep learning has emerged where an AI system learns the elements that define knowledge on its own, which has improved performances in various fields, such as image recognition, voice recognition, and translation. Where will the third AI boom lead? One problem currently being considered is that it is not possible to understand the process by which an AI system makes decisions. The application of machine-learning technology is currently spreading in many fields, along with the use of BigData. Even if the so-called boom comes to an end, there is no doubt that machine-learning technology will continue to spread and permeate society as an effective means of solving various problems.
3. Deep Learning and Technical Problems
3.1. Black Box Problem
Deep learning has become the mainstream of machine learning, and with the expansion of its applications, technical problems from the perspective of real-world problem solving have also emerged, primarily the black box problem: neural network machine-learning in deep learning is a black box [130,131,132,133,134,135,136,137,138,139,140,141,142,143,144]. The learning results are reflected in the node weights, and the obtained regularities and models are not represented in a form that humans can directly understand. In other words, it is not possible to explain the reasons for the results of the discrimination, classification, prediction, and detection of anomalies. In industrial applications, the data analysts using machine learning and the decision makers using the results of analyses are usually not the same, and data analysts must explain the prediction results and reasons to decision makers. Additionally, in situations such as medical care where people’s lives and health are at stake, it is necessary to be able to check the consistency between the results derived from machine learning and medical evidence, as well as to be able to use them according to the correct judgment. Because of these societal demands, transparency and accountability have been discussed as important requirements in AI development guidelines. However, it is difficult to meet these demands and requirements with black box machine learning.
3.2. Gap between Machine Learning and Decisions
There is a gap between machine learning and decision making. Discrimination, classification, prediction, and anomaly detection by machine learning often do not necessarily solve real-world problems. There is a need for solutions that go as far as making decisions based on the results obtained by machine learning. This is true not only for deep learning, but also for general data analysis through machine learning. There are four stages of data analysis: (i) descriptive analysis, (ii) diagnostic analysis, (iii) predictive analysis, and (iv) prescriptive analysis. For these, a solution method is required to be used in decision making.
3.3. Amounts of Data and Computational Power
The amounts of data and computational power required for learning have increased. Deep learning uses DNNs with hundreds of layers and a large number of parameters related to structure [145,146,147,148,149,150,151,152,153,154,155,156,157,158,159,160,161]. Therefore, it is prone to overfitting, which is a condition where the learning data are overfitted, generalization is not possible, and high accuracy cannot be achieved with unknown data. To suppress overfitting and achieve high accuracy, a large amount of training data is required, but in practice, it is not always easy to collect a large amount of training data [162,163,164,165,166,167,168,169,170,171,172,173,174,175,176,177,178]. Moreover, it takes a long time to input a large amount of training data into a large-scale DNN and execute the learning process. In addition, network structures tend to become multilayered and complex, so the time required for learning keeps increasing.
3.4. Theoretical Explanation
There is difficulty in developing analytical processes. There is no theoretical explanation for why deep learning achieves such high accuracy. The structural design of a DNN and the method of learning to achieve high accuracy depend on the accumulation of expertise and empirical rules, and it is still difficult to fully exploit deep learning. In addition, when applying machine-learning technology, which is not limited to deep learning, data analysts usually hypothesize and test by repeated trial and error to select methods and models and determine feature values and parameters, as well as the learning method.
4. Approaches to Technical Problems in Deep Learning
4.1. Interpretability of Learning Results
The previous section addressed four technical issues facing deep learning today. Some approaches to technical challenges that have presented to solve them are introduced, with one approach being ensuring the interpretability of learning results [179,180,181,182,183,184,185,186,187,188,189,190,191,192,193,194,195,196,197,198,199]. In response to the black box problem, the US Defense Advanced Research Projects Agency (DARPA) announced an investment program for explainable AI in 2016. The plan states that machine-learning technology represents a trade-off between accuracy and interpretability and that research and development are required to achieve both. This study presents two approaches to achieving both accuracy and interpretability. The first approach is to give interpretability to highly accurate black box methods, such as deep learning. For example, this approach attempts to investigate what input patterns a trained neural network responds to and to infer from the results what it focuses on. Another approach is to develop white box thinking, with interpretability such as decision trees and linear regression to achieve high accuracy. Heterogeneous mixture learning and factorized asymptotic Bayesian inference [200,201], have shown that this approach can achieve both high accuracy and good interpretability, which is high when the target data can be expressed in a simple format, such as a linear sum of feature quantities or explanatory variables. However, when target data with complex behavior are expressed in a simple format, approximation becomes rough and accuracy deteriorates. Therefore, by dividing the target data into several parts and applying a simple format to each part, one can achieve both high accuracy and good interpretability. Heterogeneous mixture learning uses this strategy to automatically calculate an optimal data division method and apply it to each division of data. In addition, with deep learning, there is no theoretical explanation as to why high accuracy is achieved in the first place. Deep learning is known to be a non-convex optimization learning method, and the theory of convex optimization learning, which has been theoretically elucidated, cannot be applied. Efforts to fundamentally and theoretically clarify the black box problem are also important.
4.2. Interpretability of Learning Results
Realization of solutions from machine learning to decision making is needed. For the problem of the gap between machine learning and decision making, solution methods have been researched and developed that take the path from machine learning to decision-making. There are mainly three types of approaches. (II-a) Deep reinforcement learning is a machine-learning algorithm that deals with a type of problem where a learner performs a certain state and receives a certain reward [202,203,204,205,206,207,208,209,210,211,212,213]. By repeatedly selecting actions and receiving rewards, it learns decision-making strategies for selecting actions that yield more rewards in the future. In reinforcement learning, it is necessary to find a value function that expresses how good it is to choose a certain action in a certain state, and learning this value function and policy through deep learning is deep reinforcement learning. This approach is suitable for problems where a large number of examples can be easily collected or where simulation can generate a large number of examples. (II-b) The second approach is predictive decision-making optimization based on machine learning. Deep reinforcement learning is suitable for problems where a large number of examples can be collected or created, i.e., problems where it is easy to repeatedly try decision making. In contrast, decision-making problems such as those addressed in classical operation research (OR) suffer great damage when decision making fails, and it is difficult to repeatedly try decision making. In a classical OR problem, it is assumed that the data are static, but machine learning has made it possible to generate a large number of predictions dynamically, with uncertain information that can be wrong. A new development is born when considering this as an OR problem based on a large amount of output from machine learning. A concrete approach to this is a machine learning–OR pipeline framework called predictive decision-making optimization, which is based on generating a large number of predictors through heterogeneous mixture learning.
4.3. Acceleration and Efficiency of Deep Learning
For deep learning, which requires large amounts of data and computational power, efforts are being made to accelerate processing and improve computational efficiency, both in terms of hardware and algorithms. In terms of hardware, the development of a GPU that can perform high-speed deep learning is underway. A processor suitable for deep learning called a tensor processing unit (TPU) was developed, which could provide 10 times more computational power, especially in inference processing, per power consumption compared to GPU [214]. This new hardware achieves high-speed learning by performing floating-point calculations in single precision or half precision instead of double precision, as in the past. In contrast, as an algorithmic improvement, deep residual networks (ResNets) should be mentioned, whose features allow efficient learning, even with deep hierarchies, due to a network structure with detours [215,216]. It has attracted attention due to its overwhelming victory at ILSVRC2015, which used a ResNet with 152 layers, which was about eight times deeper than any network before. Moreover, it is not necessarily easy to collect a large amount of training data sufficient to improve accuracy. To solve this problem, various strategies have been considered: (1) active learning, in which only a part of the data is efficiently labeled to improve the accuracy; (2) semi-supervised learning, in which some labeled data are used as clues for learning unlabeled data; and (3) transfer learning and domain adaptation, in which large amounts of data and learning results from similar data are used for learning, considering the imbalance in the number of samples per class, data generation by physics simulation, etc.
4.4. Establishment of Machine-Learning System Development Methodology
Attempts to directly improve the difficulty developing analytical procedures include automating the development of analytical procedures through the development and extension of machine-learning technology. Finally, given data and a specific purpose, there is a need to automatically determine an optimal analysis method or procedure to obtain results. Although there are still limitations in terms of purpose, method, scale, etc., several trials have just begun. In terms of neural network structure design, a technique to automatically generate an intermediate layer using the as-if-silent (Ais) method has been reported. In addition, predictive analysis automation technology that automatically selects feature values from relational databases and analysis methods, i.e., prediction models, and combines them according to a purpose has been reported. Alternatively, more and more machine-learning technologies have been integrated into various systems, which may undoubtedly lead to a paradigm shift in system development methods. In the past, the paradigm of system development shifted from wired logic, i.e., hardware logic circuits assembled from physical connections, to software-defined systems that define the behavior of a system by writing program code. However, systems based on machine-learning technology define the behavior of a system inductively by learning a large amount of data, which is also referred to as BigData-defined system development. It is necessary to establish and maintain new methodologies, such as system requirement definition, operation, and quality assurance concepts, and boundary optimization with software-defined, operational optimization as a “learning factory”, including data collection and input efficiency. By the application of these approaches, efforts toward drug discovery have proposed some novel systems using deep learning (Table 1) [217,218,219,220,221,222,223,224,225,226,227,228,229,230,231,232,233,234,235,236,237,238,239,240,241,242,243,244,245,246,247,248,249,250,251].

Table 1.
Deep learning based drug discovery programs.
5. Conclusions
In order for society to enjoy the benefits of extracting knowledge from data, it is necessary not only to develop AI technologies, but also to organize many processes, such as data collection, cleaning, analysis, interpretation, and evaluation. There are still some issues regarding AI application in practical and medical fields. For example, the generality of performances in various datasets has not been applied completely. The need for the development of AI technologies is high in many fields, and it is already being vigorously pursued, but these efforts are not enough to bring AI into drug discovery research. The introduction of AI cannot achieve a breakthrough in the area of drug discovery target search unless legal and cultural issues are resolved. AI technology may be suitable for the screening of seeds for new drugs. Therefore, to enable better use of data, which is expected to increase in the future, it is desirable to invest in infrastructure development as early as possible, not only from scientific and technological perspectives, but also from a humanities perspective.
Author Contributions
Writing—review and editing, Y.M.; supervision, R.Y.; funding acquisition, Y.M. All authors have read and agreed to the published version of the manuscript.
Funding
This review was funded by the Fukuda Foundation for Medical Technology, and the APC was funded by the Fukuda Foundation for Medical Technology.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Jiménez-Luna, J.; Grisoni, F.; Schneider, G. Drug discovery with explainable artificial intelligence. Nat. Mach. Intell. 2020, 2, 573–584. [Google Scholar] [CrossRef]
- Tripathi, A.; Misra, K.; Dhanuka, R.; Singh, J.P. Artificial Intelligence in Accelerating Drug Discovery and Development. Recent Pat. Biotechnol. 2022; in press. [Google Scholar] [CrossRef]
- Vijayan, R.S.K.; Kihlberg, J.; Cross, J.B.; Poongavanam, V. Enhancing preclinical drug discovery with artificial intelligence. Drug Discov. Today 2022, 27, 967–984. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.R. Application of Artificial Intelligence and Machine Learning in Drug Discovery. Methods Mol. Biol. 2022, 2390, 113–124. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Yang, Z.; Rodrigues, A.D. Cynomolgus Monkey as an Emerging Animal Model to Study Drug Transporters: In Vitro, In Vivo, In Vitro-to-In Vivo Translation. Drug Metab. Dispos. 2022, 50, 299–319. [Google Scholar] [CrossRef]
- Spreafico, A.; Hansen, A.R.; Abdul Razak, A.R.; Bedard, P.L.; Siu, L.L. The Future of Clinical Trial Design in Oncology. Cancer Discov. 2021, 11, 822–837. [Google Scholar] [CrossRef]
- Van Norman, G.A. Limitations of Animal Studies for Predicting Toxicity in Clinical Trials: Is it Time to Rethink Our Current Approach? JACC Basic Transl. Sci. 2019, 4, 845–854. [Google Scholar] [CrossRef]
- Bracken, M.B. Why animal studies are often poor predictors of human reactions to exposure. J. R. Soc. Med. 2009, 102, 120–122. [Google Scholar] [CrossRef]
- Singh, A.K.; Kumar, A.; Singh, H.; Sonawane, P.; Paliwal, H.; Thareja, S.; Pathak, P.; Grishina, M.; Jaremko, M.; Emwas, A.H.; et al. Concept of Hybrid Drugs and Recent Advancements in Anticancer Hybrids. Concept of Hybrid Drugs and Recent Advancements in Anticancer Hybrids. Pharmaceuticals 2022, 15, 1071. [Google Scholar] [CrossRef]
- Feldmann, C.; Bajorath, J. Advances in Computational Polypharmacology. Mol. Inform. 2022, 24, e2200190. [Google Scholar] [CrossRef]
- Ding, P.; Pan, Y.; Wang, Q.; Xu, R. Prediction and evaluation of combination pharmacotherapy using natural language processing, machine learning and patient electronic health records. J. Biomed. Inform. 2022, 133, 104164. [Google Scholar] [CrossRef]
- Van Norman, G.A. Phase II Trials in Drug Development and Adaptive Trial Design. JACC Basic Transl. Sci. 2019, 4, 428–437. [Google Scholar] [CrossRef] [PubMed]
- Takebe, T.; Imai, R.; Ono, S. The Current Status of Drug Discovery and Development as Originated in United States Academia: The Influence of Industrial and Academic Collaboration on Drug Discovery and Development. Clin. Transl. Sci. 2018, 11, 597–606. [Google Scholar] [CrossRef] [PubMed]
- Cong, Y.; Shintani, M.; Imanari, F.; Osada, N.; Endo, T. A New Approach to Drug Repurposing with Two-Stage Prediction, Machine Learning, and Unsupervised Clustering of Gene Expression. OMICS 2022, 26, 339–347. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.; Manuel, A.M.; Dai, Y.; Fernandes, B.S.; Enduru, N.; Jia, P.; Zhao, Z. Identifying candidate genes and drug targets for Alzheimer’s disease by an integrative network approach using genetic and brain region-specific proteomic data. Hum. Mol. Genet. 2022, 28, ddac124. [Google Scholar] [CrossRef]
- Tolios, A.; De Las Rivas, J.; Hovig, E.; Trouillas, P.; Scorilas, A.; Mohr, T. Computational approaches in cancer multidrug resistance research: Identification of potential biomarkers, drug targets and drug-target interactions. Drug Resist. Updates 2020, 48, 100662. [Google Scholar] [CrossRef]
- Zhao, K.; Shi, Y.; So, H.C. Prediction of Drug Targets for Specific Diseases Leveraging Gene Perturbation Data: A Machine Learning Approach. Pharmaceutics 2022, 14, 234. [Google Scholar] [CrossRef]
- Pestana, R.C.; Roszik, J.; Groisberg, R.; Sen, S.; Van Tine, B.A.; Conley, A.P.; Subbiah, V. Discovery of targeted expression data for novel antibody-based and chimeric antigen receptor-based therapeutics in soft tissue sarcomas using RNA-sequencing: Clinical implications. Curr. Probl. Cancer 2021, 45, 100794. [Google Scholar] [CrossRef]
- Puranik, A.; Dandekar, P.; Jain, R. Exploring the potential of machine learning for more efficient development and production of biopharmaceuticals. Biotechnol. Prog. 2022, 2, e3291. [Google Scholar] [CrossRef]
- Yu, L.; Qiu, W.; Lin, W.; Cheng, X.; Xiao, X.; Dai, J. HGDTI: Predicting drug-target interaction by using information aggregation based on heterogeneous graph neural network. BMC Bioinform. 2022, 23, 126. [Google Scholar] [CrossRef]
- Bemani, A.; Björsell, N. Aggregation Strategy on Federated Machine Learning Algorithm for Collaborative Predictive Maintenance. Sensors 2022, 22, 6252. [Google Scholar] [CrossRef]
- Garcia, B.J.; Urrutia, J.; Zheng, G.; Becker, D.; Corbet, C.; Maschhoff, P.; Cristofaro, A.; Gaffney, N.; Vaughn, M.; Saxena, U.; et al. A toolkit for enhanced reproducibility of RNASeq analysis for synthetic biologists. Synth. Biol. 2022, 7, ysac012. [Google Scholar] [CrossRef]
- Nambiar, A.M.K.; Breen, C.P.; Hart, T.; Kulesza, T.; Jamison, T.F.; Jensen, K.F. Bayesian Optimization of Computer-Proposed Multistep Synthetic Routes on an Automated Robotic Flow Platform. ACS Cent. Sci. 2022, 8, 825–836. [Google Scholar] [CrossRef] [PubMed]
- Qadeer, N.; Shah, J.H.; Sharif, M.; Khan, M.A.; Muhammad, G.; Zhang, Y.D. Intelligent Tracking of Mechanically Thrown Objects by Industrial Catching Robot for Automated In-Plant Logistics 4.0. Sensors 2022, 22, 2113. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Prieto, P.L.; Zepel, T.; Grunert, S.; Hein, J.E. Automated Experimentation Powers Data Science in Chemistry. Acc. Chem. Res. 2021, 54, 546–555. [Google Scholar] [CrossRef]
- Liu, G.H.; Zhang, B.W.; Qian, G.; Wang, B.; Mao, B.; Bichindaritz, I. Bioimage-Based Prediction of Protein Subcellular Location in Human Tissue with Ensemble Features and Deep Networks. IEEE/ACM Trans. Comput. Biol. Bioinform. 2020, 17, 1966–1980. [Google Scholar] [CrossRef]
- Haase, R.; Fazeli, E.; Legland, D.; Doube, M.; Culley, S.; Belevich, I.; Jokitalo, E.; Schorb, M.; Klemm, A.; Tischer, C. A Hitchhiker’s guide through the bio-image analysis software universe. FEBS Lett. 2022; in press. [Google Scholar] [CrossRef]
- Chessel, A. An Overview of data science uses in bioimage informatics. Methods 2017, 115, 110–118. [Google Scholar] [CrossRef]
- Mendes, J.; Domingues, J.; Aidos, H.; Garcia, N.; Matela, N. AI in Breast Cancer Imaging: A Survey of Different Applications. J. Imaging 2022, 8, 228. [Google Scholar] [CrossRef]
- Harris, R.J.; Baginski, S.G.; Bronstein, Y.; Schultze, D.; Segel, K.; Kim, S.; Lohr, J.; Towey, S.; Shahi, N.; Driscoll, I.; et al. Detection Of Critical Spinal Epidural Lesions on CT Using Machine Learning. Spine, 2022; in press. [Google Scholar] [CrossRef]
- Survarachakan, S.; Prasad, P.J.R.; Naseem, R.; Pérez de Frutos, J.; Kumar, R.P.; Langø, T.; Alaya Cheikh, F.; Elle, O.J.; Lindseth, F. Deep learning for image-based liver analysis—A comprehensive review focusing on malignant lesions. Artif. Intell. Med. 2022, 130, 102331. [Google Scholar] [CrossRef]
- Karakaya, M.; Aygun, R.S.; Sallam, A.B. Collaborative Deep Learning for Privacy Preserving Diabetic Retinopathy Detection. In Proceedings of the 44th Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Glasgow, UK, 11–15 July 2022; pp. 2181–2184. [Google Scholar] [CrossRef]
- Zeng, L.; Huang, M.; Li, Y.; Chen, Q.; Dai, H.N. Progressive Feature Fusion Attention Dense Network for Speckle Noise Removal in OCT Images. IEEE/ACM Trans. Comput. Biol. Bioinform. 2022; in press. [Google Scholar] [CrossRef]
- Fritz, B.; Yi, P.H.; Kijowski, R.; Fritz, J. Radiomics and Deep Learning for Disease Detection in Musculoskeletal Radiology: An Overview of Novel MRI- and CT-Based Approaches. Investig. Radiol. 2022; in press. [Google Scholar] [CrossRef]
- Gao, W.; Li, X.; Wang, Y.; Cai, Y. Medical Image Segmentation Algorithm for Three-Dimensional Multimodal Using Deep Reinforcement Learning and Big Data Analytics. Front. Public Health 2022, 10, 879639. [Google Scholar] [CrossRef]
- Zhou, Y. Antistroke Network Pharmacological Prediction of Xiaoshuan Tongluo Recipe Based on Drug-Target Interaction Based on Deep Learning. Comput. Math. Methods Med. 2022, 2022, 6095964. [Google Scholar] [CrossRef]
- Zheng, Z. The Classification of Music and Art Genres under the Visual Threshold of Deep Learning. Comput. Intell. Neurosci. 2022, 2022, 4439738. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Chen, D.; Wang, H.; Pan, Y.; Peng, X.; Liu, X.; Liu, Y. Automatic BASED scoring on scalp EEG in children with infantile spasms using convolutional neural network. Front. Mol. Biosci. 2022, 9, 931688. [Google Scholar] [CrossRef] [PubMed]
- Feng, T.; Fang, Y.; Pei, Z.; Li, Z.; Chen, H.; Hou, P.; Wei, L.; Wang, R.; Wang, S. A Convolutional Neural Network Model for Detecting Sellar Floor Destruction of Pituitary. Front. Neurosci. 2022, 16, 900519. [Google Scholar] [CrossRef]
- Stofa, M.M.; Zulkifley, M.A.; Zainuri, M.A.A.M. Micro-Expression-Based Emotion Recognition Using Waterfall Atrous Spatial Pyramid Pooling Networks. Sensors 2022, 22, 4634. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Wang, H.; Feng, M.; Chen, H.; Zhang, W.; Wei, L.; Pei, Z.; Wang, R.; Wang, S. Application of Convolutional Neural Network in the Diagnosis of Cavernous Sinus Invasion in Pituitary Adenoma. Front. Oncol. 2022, 12, 835047. [Google Scholar] [CrossRef] [PubMed]
- Gan, S.; Zhuang, Q.; Gong, B. Human-computer interaction based interface design of intelligent health detection using PCANet and multi-sensor information fusion. Comput. Methods Programs Biomed. 2022, 216, 106637. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Sun, Z.; Xie, J.; Yu, J.; Li, J.; Wang, J. Identification of autism spectrum disorder based on short-term spontaneous hemodynamic fluctuations using deep learning in a multi-layer neural network. Clin. Neurophysiol. 2021, 132, 457–468. [Google Scholar] [CrossRef]
- Cho, Y.; Cho, H.; Shim, J.; Choi, J.I.; Kim, Y.H.; Kim, N.; Oh, Y.W.; Hwang, S.H. Efficient Segmentation for Left Atrium With Convolution Neural Network Based on Active Learning in Late Gadolinium Enhancement Magnetic Resonance Imaging. J. Korean Med. Sci. 2022, 37, e271. [Google Scholar] [CrossRef]
- Islam, M.M.; Nooruddin, S.; Karray, F.; Muhammad, G. Human activity recognition using tools of convolutional neural networks: A state of the art review, data sets, challenges, and future prospects. Comput. Biol. Med. 2022, 149, 106060. [Google Scholar] [CrossRef]
- Khan, S.W.; Hafeez, Q.; Khalid, M.I.; Alroobaea, R.; Hussain, S.; Iqbal, J.; Almotiri, J.; Ullah, S.S. Anomaly Detection in Traffic Surveillance Videos Using Deep Learning. Sensors 2022, 22, 6563. [Google Scholar] [CrossRef]
- Farahani, H.; Boschman, J.; Farnell, D.; Darbandsari, A.; Zhang, A.; Ahmadvand, P.; Jones, S.J.M.; Huntsman, D.; Köbel, M.; Gilks, C.B.; et al. Deep learning-based histotype diagnosis of ovarian carcinoma whole-slide pathology images. Mod. Pathol. 2022; in press. [Google Scholar] [CrossRef]
- Agrawal, P.; Nikhade, P. Artificial Intelligence in Dentistry: Past, Present, and Future. Cureus 2022, 14, e27405. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Zaccagna, F.; Rundo, L.; Testa, C.; Agati, R.; Lodi, R.; Manners, D.N.; Tonon, C. Convolutional Neural Network Techniques for Brain Tumor Classification (from 2015 to 2022): Review, Challenges, and Future Perspectives. Diagnostics 2022, 12, 1850. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, L.; Luo, R.; Wang, S.; Wang, H.; Gao, F.; Wang, D. A deep learning model based on dynamic contrast-enhanced magnetic resonance imaging enables accurate prediction of benign and malignant breast lessons. Front. Oncol. 2022, 12, 943415. [Google Scholar] [CrossRef] [PubMed]
- Dotolo, S.; Esposito Abate, R.; Roma, C.; Guido, D.; Preziosi, A.; Tropea, B.; Palluzzi, F.; Giacò, L.; Normanno, N. Bioinformatics: From NGS Data to Biological Complexity in Variant Detection and Oncological Clinical Practice. Biomedicines 2022, 10, 2074. [Google Scholar] [CrossRef]
- Srivastava, R. Applications of artificial intelligence multiomics in precision oncology. J. Cancer Res. Clin. Oncol. 2022, in press. [Google Scholar] [CrossRef]
- Gim, J.A. A Genomic Information Management System for Maintaining Healthy Genomic States and Application of Genomic Big Data in Clinical Research. Int. J. Mol. Sci. 2022, 23, 5963. [Google Scholar] [CrossRef]
- Qiao, S.; Li, X.; Olatosi, B.; Young, S.D. Utilizing Big Data analytics and electronic health record data in HIV prevention, treatment, and care research: A literature review. AIDS Care 2021, 14, 1–21. [Google Scholar] [CrossRef]
- Orthuber, W. Information Is Selection-A Review of Basics Shows Substantial Potential for Improvement of Digital Information Representation. Int. J. Environ. Res. Public Health 2020, 17, 2975. [Google Scholar] [CrossRef]
- Aslan, M.F.; Sabanci, K.; Durdu, A.; Unlersen, M.F. COVID-19 diagnosis using state-of-the-art CNN architecture features and Bayesian Optimization. Comput. Biol. Med. 2022, 142, 105244. [Google Scholar] [CrossRef]
- Li, J.; Wang, P.; Zhou, Y.; Liang, H.; Lu, Y.; Luan, K. A novel classification method of lymph node metastasis in colorectal cancer. Bioengineered 2021, 12, 2007–2021. [Google Scholar] [CrossRef]
- Khosravi, P.; Lysandrou, M.; Eljalby, M.; Li, Q.; Kazemi, E.; Zisimopoulos, P.; Sigaras, A.; Brendel, M.; Barnes, J.; Ricketts, C.; et al. A Deep Learning Approach to Diagnostic Classification of Prostate Cancer Using Pathology-Radiology Fusion. J. Magn. Reson. Imaging 2021, 54, 462–471. [Google Scholar] [CrossRef] [PubMed]
- Emmert-Streib, F.; Yang, Z.; Feng, H.; Tripathi, S.; Dehmer, M. An Introductory Review of Deep Learning for Prediction Models With Big Data. Front. Artif. Intell. 2020, 3, 4. [Google Scholar] [CrossRef] [PubMed]
- Adami, C. A Brief History of Artificial Intelligence Research. Artif. Life 2021, 27, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Ren, P.; Sun, H.; Hao, J.; Qi, Q.; Wang, J.; Liao, J. A Dual-Branch Self-Boosting Framework for Self-Supervised 3D Hand Pose Estimation. IEEE Trans. Image Process. 2022, 31, 5052–5066. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Yang, X.; Huang, Y.; Li, H.; He, S.; Hu, X.; Chen, Z.; Xue, W.; Cheng, J.; Ni, D. Sketch guided and progressive growing GAN for realistic and editable ultrasound image synthesis. Med. Image Anal. 2022, 79, 102461. [Google Scholar] [CrossRef]
- Wang, H.; Li, Q.; Yuan, Y.; Zhang, Z.; Wang, K.; Zhang, H. Inter-subject registration-based one-shot segmentation with alternating union network for cardiac MRI images. Med. Image Anal. 2022, 79, 102455. [Google Scholar] [CrossRef]
- Li, Y.; Zhu, M.; Sun, G.; Chen, J.; Zhu, X.; Yang, J. Weakly supervised training for eye fundus lesion segmentation in patients with diabetic retinopathy. Math. Biosci. Eng. 2022, 19, 5293–5311. [Google Scholar] [CrossRef]
- Shi, Z.; Ma, Y.; Fu, M. Fuzzy Support Tensor Product Adaptive Image Classification for the Internet of Things. Comput. Intell. Neurosci. 2022, 2022, 3532605. [Google Scholar] [CrossRef]
- Rashmi, R.; Prasad, K.; Udupa, C.B.K. Breast histopathological image analysis using image processing techniques for diagnostic puposes: A methodological review. J. Med. Syst. 2021, 46, 7. [Google Scholar] [CrossRef]
- Li, W.; Shi, P.; Yu, H. Gesture Recognition Using Surface Electromyography and Deep Learning for Prostheses Hand: State-of-the-Art, Challenges, and Future. Front. Neurosci. 2021, 15, 621885. [Google Scholar] [CrossRef]
- Gültekin, İ.B.; Karabük, E.; Köse, M.F. “Hey Siri! Perform a type 3 hysterectomy. Please watch out for the ureter!” What is autonomous surgery and what are the latest developments? J. Turk. Ger. Gynecol. Assoc. 2021, 22, 58–70. [Google Scholar] [CrossRef] [PubMed]
- Pandey, M.; Fernandez, M.; Gentile, F.; Isayev, O.; Tropsha, A.; Stern, A.C.; Cherkasov, A. The transformational role of GPU computing and deep learning in drug discovery. Nat. Mach. Intell. 2022, 4, 211–221. [Google Scholar] [CrossRef]
- Shi, S.; Wang, Q.; Chu, X. Performance Modeling and Evaluation of Distributed Deep Learning Frameworks on GPUs. arXiv. 2017. arXiv:1711.05979. Available online: https://arxiv.org/abs/1711.05979 (accessed on 20 August 2018).
- Baur, D.; Kroboth, K.; Heyde, C.E.; Voelker, A. Convolutional Neural Networks in Spinal Magnetic Resonance Imaging: A Systematic Review. World Neurosurg. 2022, 166, 60–70. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.Y.; Mao, X.L.; Chen, Y.H.; You, N.N.; Song, Y.Q.; Zhang, L.H.; Cai, Y.; Ye, X.N.; Ye, L.P.; Li, S.W. The Feasibility of Applying Artificial Intelligence to Gastrointestinal Endoscopy to Improve the Detection Rate of Early Gastric Cancer Screening. Front. Med. 2022, 9, 886853. [Google Scholar] [CrossRef]
- Dhaka, V.S.; Meena, S.V.; Rani, G.; Sinwar, D.; Kavita Ijaz, M.F.; Woźniak, M. A Survey of Deep Convolutional Neural Networks Applied for Prediction of Plant Leaf Diseases. Sensors 2021, 21, 4749. [Google Scholar] [CrossRef]
- Zhao, J.; Hou, X.; Pan, M.; Zhang, H. Attention-based generative adversarial network in medical imaging: A narrative review. Comput. Biol. Med. 2022, 149, 105948. [Google Scholar] [CrossRef]
- Rani, G.; Misra, A.; Dhaka, V.S.; Zumpano, E.; Vocaturo, E. Spatial feature and resolution maximization GAN for bone suppression in chest radiographs. Comput. Methods Programs Biomed. 2022, 224, 107024. [Google Scholar] [CrossRef]
- Shokraei Fard, A.; Reutens, D.C.; Vegh, V. From CNNs to GANs for cross-modality medical image estimation. Comput. Biol. Med. 2022, 146, 105556. [Google Scholar] [CrossRef]
- You, A.; Kim, J.K.; Ryu, I.H.; Yoo, T.K. Application of generative adversarial networks (GAN) for ophthalmology image domains: A survey. Eye Vis. 2022, 9, 6. [Google Scholar] [CrossRef]
- Lan, L.; You, L.; Zhang, Z.; Fan, Z.; Zhao, W.; Zeng, N.; Chen, Y.; Zhou, X. Generative Adversarial Networks and Its Applications in Biomedical Informatics. Front. Public Health 2020, 8, 164. [Google Scholar] [CrossRef]
- Karras, T.; Laine, S.; Aila, T. A Style-Based Generator Architecture for Generative Adversarial Networks. arXiv. 2018. arXiv:1812.04948. Available online: https://arxiv.org/abs/1812.04948 (accessed on 29 March 2019).
- Nickabadi, A.; Fard, M.S.; Farid, N.M.; Mohammadbagheri, N. A comprehensive survey on semantic facial attribute editing using generative adversarial networks. arXiv. 2022. arXiv:2205.10587. Available online: https://arxiv.org/abs/2205.10587v1 (accessed on 21 May 2022).
- Feghali, J.; Jimenez, A.E.; Schilling, A.T.; Azad, T.D. Overview of Algorithms for Natural Language Processing and Time Series Analyses. Acta Neurochir. Suppl. 2022, 134, 221–242. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; An, S.Y.; Guan, P.; Huang, D.S.; Zhou, B.S. Time series analysis of human brucellosis in mainland China by using Elman and Jordan recurrent neural networks. BMC Infect. Dis. 2019, 19, 414. [Google Scholar] [CrossRef] [PubMed]
- Boesch, G. Deep Neural Network: The 3 Popular Types (MLP, CNN, and RNN). Available online: https://viso.ai/deep-learning/deep-neural-network-three-popular-types/ (accessed on 29 September 2022).
- Moor, J. The Dartmouth College Artificial Intelligence Conference: The Next Fifty Years. AI Mag. 2006, 27, 2006. [Google Scholar]
- Chauhan, V.; Negi, S.; Jain, D.; Singh, P.; Sagar, A.K.; Sharma, A.H. Quantum Computers: A Review on How Quantum Computing Can Boom AI. In Proceedings of the 2nd International Conference on Advance Computing and Innovative Technologies in Engineering (ICACITE), Greater Noida, India, 28–29 April 2022. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. In Proceedings of the 25th International Conference on Neural Information Processing Systems, Carson, NV, USA, 3–6 December 2012; pp. 1097–1105. [Google Scholar] [CrossRef]
- Aborode, A.T.; Awuah, W.A.; Mikhailova, T.; Abdul-Rahman, T.; Pavlock, S.; Nansubuga, E.P.; Kundu, M.; Yarlagadda, R.; Pustake, M.; Correia, I.F.S.; et al. OMICs Technologies for Natural Compounds-based Drug Development. Curr. Top Med. Chem. 2022; in press. [Google Scholar] [CrossRef]
- Park, Y.; Heider, D.; Hauschild, A.C. Integrative Analysis of Next-Generation Sequencing for Next-Generation Cancer Research toward Artificial Intelligence. Cancers 2021, 13, 3148. [Google Scholar] [CrossRef]
- Ristori, M.V.; Mortera, S.L.; Marzano, V.; Guerrera, S.; Vernocchi, P.; Ianiro, G.; Gardini, S.; Torre, G.; Valeri, G.; Vicari, S.; et al. Proteomics and Metabolomics Approaches towards a Functional Insight onto AUTISM Spectrum Disorders: Phenotype Stratification and Biomarker Discovery. Int. J. Mol. Sci. 2020, 21, 6274. [Google Scholar] [CrossRef]
- Liu, X.N.; Cui, D.N.; Li, Y.F.; Liu, Y.H.; Liu, G.; Liu, L. Multiple “Omics” data-based biomarker screening for hepatocellular carcinoma diagnosis. World J. Gastroenterol. 2019, 25, 4199–4212. [Google Scholar] [CrossRef]
- Harakalova, M.; Asselbergs, F.W. Systems analysis of dilated cardiomyopathy in the next generation sequencing era. Wiley Interdiscip. Rev. Syst. Biol. Med. 2018, 10, e1419. [Google Scholar] [CrossRef]
- Lee, J.; Choe, Y. Robust PCA Based on Incoherence with Geometrical Interpretation. IEEE Trans. Image Process. 2018, 27, 1939–1950. [Google Scholar] [CrossRef]
- Spencer, A.P.C.; Goodfellow, M. Using deep clustering to improve fMRI dynamic functional connectivity analysis. Neuroimage 2022, 257, 119288. [Google Scholar] [CrossRef]
- Li, Z.; Peleato, N.M. Comparison of dimensionality reduction techniques for cross-source transfer of fluorescence contaminant detection models. Chemosphere 2021, 276, 130064. [Google Scholar] [CrossRef] [PubMed]
- Fromentèze, T.; Yurduseven, O.; Del Hougne, P.; Smith, D.R. Lowering latency and processing burden in computational imaging through dimensionality reduction of the sensing matrix. Sci. Rep. 2021, 11, 3545. [Google Scholar] [CrossRef] [PubMed]
- Donnarumma, F.; Prevete, R.; Maisto, D.; Fuscone, S.; Irvine, E.M.; van der Meer, M.A.A.; Kemere, C.; Pezzulo, G. A framework to identify structured behavioral patterns within rodent spatial trajectories. Sci. Rep. 2021, 11, 468. [Google Scholar] [CrossRef] [PubMed]
- Sakai, T.; Niu, G.; Sugiyama, M. Information-Theoretic Representation Learning for Positive-Unlabeled Classification. Neural Comput. 2021, 33, 244–268. [Google Scholar] [CrossRef]
- Singh, A.; Ogunfunmi, T. An Overview of Variational Autoencoders for Source Separation, Finance, and Bio-Signal Applications. Entropy 2021, 24, 55. [Google Scholar] [CrossRef]
- Ran, X.; Chen, W.; Yvert, B.; Zhang, S. A hybrid autoencoder framework of dimensionality reduction for brain-computer interface decoding. Comput. Biol. Med. 2022, 148, 105871. [Google Scholar] [CrossRef]
- Zhu, X.; Li, J.; Lin, Y.; Zhao, L.; Wang, J.; Peng, X. Dimensionality Reduction of Single-Cell RNA Sequencing Data by Combining Entropy and Denoising AutoEncoder. J. Comput. Biol. 2022; in press. [Google Scholar] [CrossRef]
- Gomari, D.P.; Schweickart, A.; Cerchietti, L.; Paietta, E.; Fernandez, H.; Al-Amin, H.; Suhre, K.; Krumsiek, J. Variational autoencoders learn transferrable representations of metabolomics data. Commun. Biol. 2022, 5, 645. [Google Scholar] [CrossRef]
- Kamikokuryo, K.; Haga, T.; Venture, G.; Hernandez, V. Adversarial Autoencoder and Multi-Armed Bandit for Dynamic Difficulty Adjustment in Immersive Virtual Reality for Rehabilitation: Application to Hand Movement. Sensors 2022, 22, 4499. [Google Scholar] [CrossRef]
- Seyboldt, R.; Lavoie, J.; Henry, A.; Vanaret, J.; Petkova, M.D.; Gregor, T.; François, P. Latent space of a small genetic network: Geometry of dynamics and information. Proc. Natl. Acad. Sci. USA 2022, 119, e2113651119. [Google Scholar] [CrossRef]
- Zhao, S.; Zhang, L.; Liu, X. DAE-TPGM: A deep autoencoder network based on a two-part-gamma model for analyzing single-cell RNA-seq data. Comput. Biol. Med. 2022, 146, 105578. [Google Scholar] [CrossRef]
- Lin, Y.K.; Lee, C.Y.; Chen, C.Y. Robustness of autoencoders for establishing psychometric properties based on small sample sizes: Results from a Monte Carlo simulation study and a sports fan curiosity study. PeerJ. Comput. Sci. 2022, 8, e782. [Google Scholar] [CrossRef] [PubMed]
- Ausmees, K.; Nettelblad, C. A deep learning framework for characterization of genotype data. G3 2022, 12, jkac020. [Google Scholar] [CrossRef] [PubMed]
- Walbech, J.S.; Kinalis, S.; Winther, O.; Nielsen, F.C.; Bagger, F.O. Interpretable Autoencoders Trained on Single Cell Sequencing Data Can Transfer Directly to Data from Unseen Tissues. Cells 2021, 11, 85. [Google Scholar] [CrossRef] [PubMed]
- Gayathri, J.L.; Abraham, B.; Sujarani, M.S.; Nair, M.S. A computer-aided diagnosis system for the classification of COVID-19 and non-COVID-19 pneumonia on chest X-ray images by integrating CNN with sparse autoencoder and feed forward neural network. Comput. Biol. Med. 2022, 141, 105134. [Google Scholar] [CrossRef]
- Belkacemi, Z.; Gkeka, P.; Lelièvre, T.; Stoltz, G. Chasing Collective Variables Using Autoencoders and Biased Trajectories. J. Chem. Theory Comput. 2022, 18, 59–78. [Google Scholar] [CrossRef]
- Fong, Y.; Xu, J. Forward Stepwise Deep Autoencoder-based Monotone Nonlinear Dimensionality Reduction Methods. J. Comput. Graph Stat. 2021, 30, 519–529. [Google Scholar] [CrossRef]
- Geenjaar, E.; Lewis, N.; Fu, Z.; Venkatdas, R.; Plis, S.; Calhoun, V. Fusing multimodal neuroimaging data with a variational autoencoder. In Proceedings of the 43rd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Guadalajara, Mexico, 1–5 November 2021; pp. 3630–3633. [Google Scholar] [CrossRef]
- Pintelas, E.; Livieris, I.E.; Pintelas, P.E. A Convolutional Autoencoder Topology for Classification in High-Dimensional Noisy Image Datasets. Sensors 2021, 21, 7731. [Google Scholar] [CrossRef]
- Ghorbani, M.; Prasad, S.; Klauda, J.B.; Brooks, B.R. Variational embedding of protein folding simulations using Gaussian mixture variational autoencoders. J. Chem. Phys. 2021, 155, 194108. [Google Scholar] [CrossRef]
- Luo, Z.; Xu, C.; Zhang, Z.; Jin, W. A topology-preserving dimensionality reduction method for single-cell RNA-seq data using graph autoencoder. Sci. Rep. 2021, 11, 20028. [Google Scholar] [CrossRef]
- Harefa, E.; Zhou, W. Performing sequential forward selection and variational autoencoder techniques in soil classification based on laser-induced breakdown spectroscopy. Anal. Methods 2021, 13, 4926–4933. [Google Scholar] [CrossRef]
- Whiteway, M.R.; Biderman, D.; Friedman, Y.; Dipoppa, M.; Buchanan, E.K.; Wu, A.; Zhou, J.; Bonacchi, N.; Miska, N.J.; Noel, J.P.; et al. Partitioning variability in animal behavioral videos using semi-supervised variational autoencoders. PLoS Comput. Biol. 2021, 17, e1009439. [Google Scholar] [CrossRef]
- Zhao, J.; Wang, N.; Wang, H.; Zheng, C.; Su, Y. SCDRHA: A scRNA-Seq Data Dimensionality Reduction Algorithm Based on Hierarchical Autoencoder. Front. Genet. 2021, 12, 733906. [Google Scholar] [CrossRef]
- Hu, H.; Li, Z.; Li, X.; Yu, M.; Pan, X. cCAEs: Deep clustering of single-cell RNA-seq via convolutional autoencoder embedding and soft K-means. Brief Bioinform. 2022, 23, bbab321. [Google Scholar] [CrossRef] [PubMed]
- Naqvi, S.A.A.; Tennankore, K.; Vinson, A.; Roy, P.C.; Abidi, S.S.R. Predicting Kidney Graft Survival Using Machine Learning Methods: Prediction Model Development and Feature Significance Analysis Study. J. Med. Internet. Res. 2021, 23, e26843. [Google Scholar] [CrossRef] [PubMed]
- Frassek, M.; Arjun, A.; Bolhuis, P.G. An extended autoencoder model for reaction coordinate discovery in rare event molecular dynamics datasets. J. Chem. Phys. 2021, 155, 064103. [Google Scholar] [CrossRef]
- Nauwelaers, N.; Matthews, H.; Fan, Y.; Croquet, B.; Hoskens, H.; Mahdi, S.; El Sergani, A.; Gong, S.; Xu, T.; Bronstein, M.; et al. Exploring palatal and dental shape variation with 3D shape analysis and geometric deep learning. Orthod. Craniofac. Res. 2021, 24, 134–143. [Google Scholar] [CrossRef]
- Rosafalco, L.; Manzoni, A.; Mariani, S.; Corigliano, A. An Autoencoder-Based Deep Learning Approach for Load Identification in Structural Dynamics. Sensors 2021, 21, 4207. [Google Scholar] [CrossRef]
- Shen, J.; Li, W.; Deng, S.; Zhang, T. Supervised and unsupervised learning of directed percolation. Phys. Rev. 2021, 103, 052140. [Google Scholar] [CrossRef]
- Hira, M.T.; Razzaque, M.A.; Angione, C.; Scrivens, J.; Sawan, S.; Sarker, M. Integrated multi-omics analysis of ovarian cancer using variational autoencoders. Sci. Rep. 2021, 11, 6265. [Google Scholar] [CrossRef]
- Ahmed, I.; Galoppo, T.; Hu, X.; Ding, Y. Graph Regularized Autoencoder and its Application in Unsupervised Anomaly Detection. IEEE Trans. Pattern Anal. Mach. Intell. 2022, 44, 4110–4124. [Google Scholar] [CrossRef]
- Patel, N.; Patel, S.; Mankad, S.H. Impact of autoencoder based compact representation on emotion detection from audio. J. Ambient Intell. Humaniz. Comput. 2022, 13, 867–885. [Google Scholar] [CrossRef] [PubMed]
- Battey, C.J.; Coffing, G.C.; Kern, A.D. Visualizing population structure with variational autoencoders. G3 2021, 11, jkaa036. [Google Scholar] [CrossRef] [PubMed]
- López-Cortés, X.A.; Matamala, F.; Maldonado, C.; Mora-Poblete, F.; Scapim, C.A. A Deep Learning Approach to Population Structure Inference in Inbred Lines of Maize. Front. Genet. 2020, 11, 543459. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Cui, Y.; Peng, S.; Liao, X.; Yu, Y. MinimapR: A parallel alignment tool for the analysis of large-scale third-generation sequencing data. Comput. Biol. Chem. 2022, 99, 107735. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Jeon, J.; Son, J.; Cho, K.; Kim, S. NAND and NOR logic-in-memory comprising silicon nanowire feedback field-effect transistors. Sci. Rep. 2022, 12, 3643. [Google Scholar] [CrossRef] [PubMed]
- Hanussek, M.; Bartusch, F.; Krüger, J. Performance and scaling behavior of bioinformatic applications in virtualization environments to create awareness for the efficient use of compute resources. PLoS Comput. Biol. 2021, 17, e1009244. [Google Scholar] [CrossRef]
- Dong, Z.; Gray, H.; Leggett, C.; Lin, M.; Pascuzzi, V.R.; Yu, K. Porting HEP Parameterized Calorimeter Simulation Code to GPUs. Front. Big Data 2021, 4, 665783. [Google Scholar] [CrossRef]
- Linse, C.; Alshazly, H.; Martinetz, T. A walk in the black-box: 3D visualization of large neural networks in virtual reality. Neural Comput. Appl. 2022, 34, 21237–21252. [Google Scholar] [CrossRef]
- Yu, J.; Liu, G. Knowledge Transfer-Based Sparse Deep Belief Network. IEEE Trans. Cybern. 2022; in press. [Google Scholar] [CrossRef]
- Li, C.; Liu, J.; Chen, J.; Yuan, Y.; Yu, J.; Gou, Q.; Guo, Y.; Pu, X. An Interpretable Convolutional Neural Network Framework for Analyzing Molecular Dynamics Trajectories: A Case Study on Functional States for G-Protein-Coupled Receptors. J. Chem. Inf. Model 2022, 62, 1399–1410. [Google Scholar] [CrossRef]
- Zhao, Z.; Li, T.; An, B.; Wang, S.; Ding, B.; Yan, R.; Chen, X. Model-driven deep unrolling: Towards interpretable deep learning against noise attacks for intelligent fault diagnosis. ISA Trans. 2022, 129, 644–662. [Google Scholar] [CrossRef]
- Zhao, M.; Xin, J.; Wang, Z.; Wang, X.; Wang, Z. Interpretable Model Based on Pyramid Scene Parsing Features for Brain Tumor MRI Image Segmentation. Comput. Math. Methods Med. 2022, 2022, 8000781. [Google Scholar] [CrossRef] [PubMed]
- Jin, D.; Sergeeva, E.; Weng, W.H.; Chauhan, G.; Szolovits, P. Explainable deep learning in healthcare: A methodological survey from an attribution view. WIREs Mech. Dis. 2022, 14, e1548. [Google Scholar] [CrossRef] [PubMed]
- Marcinowski, M. Top interpretable neural network for handwriting identification. J. Forensic Sci. 2022, 67, 1140–1148. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liu, A.; Bai, X.; Liu, X. Universal Adversarial Patch Attack for Automatic Checkout Using Perceptual and Attentional Bias. IEEE Trans. Image Process. 2022, 31, 598–611. [Google Scholar] [CrossRef]
- Withnell, E.; Zhang, X.; Sun, K.; Guo, Y. XOmiVAE: An interpretable deep learning model for cancer classification using high-dimensional omics data. Brief Bioinform. 2021, 22, bbab315. [Google Scholar] [CrossRef]
- Auzina, I.A.; Tomczak, J.M. Approximate Bayesian Computation for Discrete Spaces. Entropy 2021, 23, 312. [Google Scholar] [CrossRef]
- Yu, J.; Liu, G. Extracting and inserting knowledge into stacked denoising auto-encoders. Neural Netw. 2021, 137, 31–42. [Google Scholar] [CrossRef]
- Lee, C.K.; Samad, M.; Hofer, I.; Cannesson, M.; Baldi, P. Development and validation of an interpretable neural network for prediction of postoperative in-hospital mortality. NPJ Digit Med. 2021, 4, 8. [Google Scholar] [CrossRef]
- Chu, W.M.; Kristiani, E.; Wang, Y.C.; Lin, Y.R.; Lin, S.Y.; Chan, W.C.; Yang, C.T.; Tsan, Y.T. A model for predicting fall risks of hospitalized elderly in Taiwan-A machine learning approach based on both electronic health records and comprehensive geriatric assessment. Front. Med. 2022, 9, 937216. [Google Scholar] [CrossRef]
- Aslam, A.R.; Hafeez, N.; Heidari, H.; Altaf, M.A.B. Channels and Features Identification: A Review and a Machine-Learning Based Model With Large Scale Feature Extraction for Emotions and ASD Classification. Front. Neurosci. 2022, 16, 844851. [Google Scholar] [CrossRef]
- Chen, C.; Zheng, S.; Guo, L.; Yang, X.; Song, Y.; Li, Z.; Zhu, Y.; Liu, X.; Li, Q.; Zhang, H.; et al. Identification of misdiagnosis by deep neural networks on a histopathologic review of breast cancer lymph node metastases. Sci. Rep. 2022, 12, 13482. [Google Scholar] [CrossRef] [PubMed]
- Catal, C.; Giray, G.; Tekinerdogan, B.; Kumar, S.; Shukla, S. Applications of deep learning for phishing detection: A systematic literature review. Knowl. Inf. Syst. 2022, 64, 1457–1500. [Google Scholar] [CrossRef]
- Zhao, B.; Kurgan, L. Deep learning in prediction of intrinsic disorder in proteins. Comput. Struct. Biotechnol. J. 2022, 20, 1286–1294. [Google Scholar] [CrossRef] [PubMed]
- Scalco, E.; Rizzo, G.; Mastropietro, A. The stability of oncologic MRI radiomic features and the potential role of deep learning: A review. Phys. Med. Biol. 2022; in press. [Google Scholar] [CrossRef]
- Ma, J.K.; Wrench, A.A. Automated assessment of hyoid movement during normal swallow using ultrasound. Int. J. Lang. Commun. Disord. 2022, 57, 615–629. [Google Scholar] [CrossRef] [PubMed]
- Fkirin, A.; Attiya, G.; El-Sayed, A.; Shouman, M.A. Copyright protection of deep neural network models using digital watermarking: A comparative study. Multimed. Tools Appl. 2022, 81, 15961–15975. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.; Khan, M.A.; Jan, S.U.; Ahmad, J.; Jamal, S.S.; Shah, A.A.; Pitropakis, N.; Buchanan, W.J. A Deep Learning-Based Intrusion Detection System for MQTT Enabled IoT. Sensors 2021, 21, 7016. [Google Scholar] [CrossRef]
- Ghods, A.; Cook, D.J. A Survey of Deep Network Techniques All Classifiers Can Adopt. Data Min. Knowl. Discov. 2021, 35, 46–87. [Google Scholar] [CrossRef]
- Meng, Z.; Ravi, S.N.; Singh, V. Physarum Powered Differentiable Linear Programming Layers and Applications. Proc. Conf. AAAI Artif. Intell. 2021, 35, 8939–8949. [Google Scholar]
- Grant, L.L.; Sit, C.S. De novo molecular drug design benchmarking. RSC Med. Chem. 2021, 12, 1273–1280. [Google Scholar] [CrossRef]
- Gao, Y.; Ascoli, G.A.; Zhao, L. BEAN: Interpretable and Efficient Learning With Biologically-Enhanced Artificial Neuronal Assembly Regularization. Front. Neurorobot. 2021, 15, 567482. [Google Scholar] [CrossRef]
- Talukder, A.; Barham, C.; Li, X.; Hu, H. Interpretation of deep learning in genomics and epigenomics. Brief Bioinform. 2021, 22, bbaa177. [Google Scholar] [CrossRef] [PubMed]
- Ashat, M.; Klair, J.S.; Singh, D.; Murali, A.R.; Krishnamoorthi, R. Impact of real-time use of artificial intelligence in improving adenoma detection during colonoscopy: A systematic review and meta-analysis. Endosc. Int. Open 2021, 9, E513–E521. [Google Scholar] [CrossRef]
- Kumar, A.; Kini, S.G.; Rathi, E. A Recent Appraisal of Artificial Intelligence and In Silico ADMET Prediction in the Early Stages of Drug Discovery. Mini Rev. Med. Chem. 2021, 21, 2788–2800. [Google Scholar] [CrossRef] [PubMed]
- Koktzoglou, I.; Huang, R.; Ankenbrandt, W.J.; Walker, M.T.; Edelman, R.R. Super-resolution head and neck MRA using deep machine learning. Magn. Reson. Med. 2021, 86, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Zheng, X.; Liu, D.; Ai, L.; Tang, P.; Wang, B.; Wang, Y. WBC Image Segmentation Based on Residual Networks and Attentional Mechanisms. Comput. Intell. Neurosci. 2022, 2022, 1610658. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Zhang, J.; Pham, H.A.; Li, Z.; Lyu, J. Virtual Conjugate Coil for Improving KerNL Reconstruction. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2022, 2022, 599–602. [Google Scholar] [CrossRef]
- Sukegawa, S.; Tanaka, F.; Nakano, K.; Hara, T.; Yoshii, K.; Yamashita, K.; Ono, S.; Takabatake, K.; Kawai, H.; Nagatsuka, H.; et al. Effective deep learning for oral exfoliative cytology classification. Sci. Rep. 2022, 12, 13281. [Google Scholar] [CrossRef]
- Xi, B.; Li, J.; Li, Y.; Song, R.; Hong, D.; Chanussot, J. Few-Shot Learning With Class-Covariance Metric for Hyperspectral Image Classification. IEEE Trans. Image Process. 2022, 31, 5079–5092. [Google Scholar] [CrossRef]
- Yan, H.; Liu, Z.; Chen, J.; Feng, Y.; Wang, J. Memory-augmented skip-connected autoencoder for unsupervised anomaly detection of rocket engines with multi-source fusion. ISA Trans. 2022; in press. [Google Scholar] [CrossRef]
- Ni, P.; Sun, L.; Yang, J.; Li, Y. Multi-End Physics-Informed Deep Learning for Seismic Response Estimation. Sensors 2022, 22, 3697. [Google Scholar] [CrossRef]
- Nishiura, H.; Miyamoto, A.; Ito, A.; Harada, M.; Suzuki, S.; Fujii, K.; Morifuji, H.; Takatsuka, H. Machine-learning-based quality-level-estimation system for inspecting steel microstructures. Microscopy 2022, 71, 214–221. [Google Scholar] [CrossRef]
- Wang, C.; Li, Y.; Tsuboshita, Y.; Sakurai, T.; Goto, T.; Yamaguchi, H.; Yamashita, Y.; Sekiguchi, A.; Tachimori, H. Alzheimer’s Disease Neuroimaging Initiative. A high-generalizability machine learning framework for predicting the progression of Alzheimer’s disease using limited data. NPJ Digit Med. 2022, 5, 43. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Wu, H.; Zhou, K. Design of Fault Prediction System for Electromechanical Sensor Equipment Based on Deep Learning. Comput. Intell. Neurosci. 2022, 2022, 3057167. [Google Scholar] [CrossRef] [PubMed]
- Elazab, A.; Elfattah, M.A.; Zhang, Y. Novel multi-site graph convolutional network with supervision mechanism for COVID-19 diagnosis from X-ray radiographs. Appl. Soft Comput. 2022, 114, 108041. [Google Scholar] [CrossRef] [PubMed]
- Hou, R.; Peng, Y.; Grimm, L.J.; Ren, Y.; Mazurowski, M.A.; Marks, J.R.; King, L.M.; Maley, C.C.; Hwang, E.S.; Lo, J.Y. Anomaly Detection of Calcifications in Mammography Based on 11,000 Negative Cases. IEEE Trans. Biomed. Eng. 2022, 69, 1639–1650. [Google Scholar] [CrossRef]
- Zhao, F.; Zeng, Y.; Bai, J. Toward a Brain-Inspired Developmental Neural Network Based on Dendritic Spine Dynamics. Neural Comput. 2021, 34, 172–189. [Google Scholar] [CrossRef]
- Mori, T.; Terashi, G.; Matsuoka, D.; Kihara, D.; Sugita, Y. Efficient Flexible Fitting Refinement with Automatic Error Fixing for De Novo Structure Modeling from Cryo-EM Density Maps. J. Chem. Inf. Model 2021, 61, 3516–3528. [Google Scholar] [CrossRef]
- Li, H.; Weng, J.; Mao, Y.; Wang, Y.; Zhan, Y.; Cai, Q.; Gu, W. Adaptive Dropout Method Based on Biological Principles. IEEE Trans. Neural Netw. Learn Syst. 2021, 32, 4267–4276. [Google Scholar] [CrossRef]
- Umezawa, E.; Ishihara, D.; Kato, R. A Bayesian approach to diffusional kurtosis imaging. Magn. Reson. Med. 2021, 86, 1110–1124. [Google Scholar] [CrossRef]
- da Costa-Luis, C.O.; Reader, A.J. Micro-Networks for Robust MR-Guided Low Count PET Imaging. IEEE Trans. Radiat. Plasma Med. Sci. 2020, 5, 202–212. [Google Scholar] [CrossRef]
- Xu, Y.; Xie, L.; Dai, W.; Zhang, X.; Chen, X.; Qi, G.J.; Xiong, H.; Tian, Q. Partially-Connected Neural Architecture Search for Reduced Computational Redundancy. IEEE Trans. Pattern Anal. Mach. Intell. 2021, 43, 2953–2970. [Google Scholar] [CrossRef]
- Rasheed, K.; Qayyum, A.; Ghaly, M.; Al-Fuqaha, A.; Razi, A.; Qadir, J. Explainable, trustworthy, and ethical machine learning for healthcare: A survey. Comput. Biol. Med. 2022, 149, 106043. [Google Scholar] [CrossRef] [PubMed]
- Bhowmik, A.; Eskreis-Winkler, S. Deep learning in breast imaging. BJR Open 2022, 4, 20210060. [Google Scholar] [CrossRef] [PubMed]
- Andreini, C.; Rosato, A. Structural Bioinformatics and Deep Learning of Metalloproteins: Recent Advances and Applications. Int. J. Mol. Sci. 2022, 23, 7684. [Google Scholar] [CrossRef]
- Teng, Q.; Liu, Z.; Song, Y.; Han, K.; Lu, Y. A survey on the interpretability of deep learning in medical diagnosis. Multimed. Syst. 2022, 25, 1–21. [Google Scholar] [CrossRef]
- Livesey, B.J.; Marsh, J.A. Interpreting protein variant effects with computational predictors and deep mutational scanning. Dis. Model Mech. 2022, 15, dmm049510. [Google Scholar] [CrossRef] [PubMed]
- Stafford, I.S.; Gosink, M.M.; Mossotto, E.; Ennis, S.; Hauben, M. A Systematic Review of Artificial Intelligence and Machine Learning Applications to Inflammatory Bowel Disease, with Practical Guidelines for Interpretation. Inflamm. Bowel Dis. 2022, 14, izac115. [Google Scholar] [CrossRef]
- Foroughi Pour, A.; White, B.S.; Park, J.; Sheridan, T.B.; Chuang, J.H. Deep learning features encode interpretable morphologies within histological images. Sci. Rep. 2022, 12, 9428. [Google Scholar] [CrossRef]
- Bowler, A.L.; Pound, M.P.; Watson, N.J. A review of ultrasonic sensing and machine learning methods to monitor industrial processes. Ultrasonics 2022, 124, 106776. [Google Scholar] [CrossRef]
- Gamble, P.; Jaroensri, R.; Wang, H.; Tan, F.; Moran, M.; Brown, T.; Flament-Auvigne, I.; Rakha, E.A.; Toss, M.; Dabbs, D.J.; et al. Determining breast cancer biomarker status and associated morphological features using deep learning. Commun. Med. 2021, 1, 14. [Google Scholar] [CrossRef]
- van der Velden, B.H.M.; Kuijf, H.J.; Gilhuijs, K.G.A.; Viergever, M.A. Explainable artificial intelligence (XAI) in deep learning-based medical image analysis. Med. Image Anal. 2022, 79, 102470. [Google Scholar] [CrossRef]
- Fan, F.L.; Xiong, J.; Li, M.; Wang, G. On Interpretability of Artificial Neural Networks: A Survey. IEEE Trans. Radiat. Plasma Med. Sci. 2021, 5, 741–760. [Google Scholar] [CrossRef] [PubMed]
- Padash, S.; Mohebbian, M.R.; Adams, S.J.; Henderson, R.D.E.; Babyn, P. Pediatric chest radiograph interpretation: How far has artificial intelligence come? A systematic literature review. Pediatr. Radiol. 2022, 52, 1568–1580. [Google Scholar] [CrossRef] [PubMed]
- Barragán-Montero, A.; Bibal, A.; Dastarac, M.H.; Draguet, C.; Valdés, G.; Nguyen, D.; Willems, S.; Vandewinckele, L.; Holmström, M.; Löfman, F.; et al. Towards a safe and efficient clinical implementation of machine learning in radiation oncology by exploring model interpretability, explainability and data-model dependency. Phys. Med. Biol. 2022; in press. [Google Scholar] [CrossRef]
- Aljabri, M.; AlAmir, M.; AlGhamdi, M.; Abdel-Mottaleb, M.; Collado-Mesa, F. Towards a better understanding of annotation tools for medical imaging: A survey. Multimed. Tools Appl. 2022, 81, 25877–25911. [Google Scholar] [CrossRef]
- Kagiyama, N.; Tokodi, M.; Sengupta, P.P. Machine Learning in Cardiovascular Imaging. Heart Fail. Clin. 2022, 18, 245–258. [Google Scholar] [CrossRef] [PubMed]
- Adnan, N.; Umer, F. Understanding deep learning—Challenges and prospects. J. Pak. Med. Assoc. 2022, 72, S59–S63. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, J.; Morse, R.; Taylor, A.; Phillips, D.; Higham, H.; Burckett-St Laurent, D.; Bowness, J. Artificial Intelligence: Innovation to Assist in the Identification of Sono-anatomy for Ultrasound-Guided Regional Anaesthesia. Adv. Exp. Med. Biol. 2022, 1356, 117–140. [Google Scholar] [CrossRef] [PubMed]
- Alouani, D.J.; Ransom, E.M.; Jani, M.; Burnham, C.A.; Rhoads, D.D.; Sadri, N. Deep Convolutional Neural Networks Implementation for the Analysis of Urine Culture. Clin. Chem. 2022, 68, 574–583. [Google Scholar] [CrossRef]
- Lim, W.X.; Chen, Z.; Ahmed, A. The adoption of deep learning interpretability techniques on diabetic retinopathy analysis: A review. Med. Biol. Eng. Comput. 2022, 60, 633–642. [Google Scholar] [CrossRef]
- Treppner, M.; Binder, H.; Hess, M. Interpretable generative deep learning: An illustration with single cell gene expression data. Hum. Genet. 2022, 141, 1481–1498. [Google Scholar] [CrossRef]
- Huang, J.; Shlobin, N.A.; DeCuypere, M.; Lam, S.K. Deep Learning for Outcome Prediction in Neurosurgery: A Systematic Review of Design, Reporting, and Reproducibility. Neurosurgery 2022, 90, 16–38. [Google Scholar] [CrossRef]
- Hayashi, N. The exact asymptotic form of Bayesian generalization error in latent Dirichlet allocation. Neural Netw. 2021, 137, 127–137. [Google Scholar] [CrossRef] [PubMed]
- van Zanten, H. Nonparametric Bayesian methods for one-dimensional diffusion models. Math. Biosci. 2013, 243, 215–222. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yu, Z.; Xu, X.; Zhou, W. Task Offloading and Resource Allocation Strategy Based on Deep Learning for Mobile Edge Computing. Comput. Intell. Neurosci. 2022, 2022, 1427219. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Yue, L.; Wang, C.; Sun, J.; Zhang, S.; Wei, A.; Xie, G. Leveraging Imitation Learning on Pose Regulation Problem of a Robotic Fish. IEEE Trans. Neural Netw. Learn Syst. 2022; in press. [Google Scholar] [CrossRef]
- Yao, S.; Liu, X.; Zhang, Y.; Cui, Z. An approach to solving optimal control problems of nonlinear systems by introducing detail-reward mechanism in deep reinforcement learning. Math. Biosci. Eng. 2022, 19, 9258–9290. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Li, X.; Liu, Q.; Li, Z.; Yang, F.; Luan, T. Multi-Agent Decision-Making Modes in Uncertain Interactive Traffic Scenarios via Graph Convolution-Based Deep Reinforcement Learning. Sensors 2022, 22, 4586. [Google Scholar] [CrossRef]
- Rupprecht, T.; Wang, Y. A survey for deep reinforcement learning in markovian cyber-physical systems: Common problems and solutions. Neural Netw. 2022, 153, 13–36. [Google Scholar] [CrossRef]
- Suhaimi, A.; Lim, A.W.H.; Chia, X.W.; Li, C.; Makino, H. Representation learning in the artificial and biological neural networks underlying sensorimotor integration. Sci. Adv. 2022, 8, eabn0984. [Google Scholar] [CrossRef]
- Ecoffet, P.; Fontbonne, N.; André, J.B.; Bredeche, N. Policy search with rare significant events: Choosing the right partner to cooperate with. PLoS ONE 2022, 17, e0266841. [Google Scholar] [CrossRef]
- Rajendran, S.K.; Zhang, F. Design, Modeling, and Visual Learning-Based Control of Soft Robotic Fish Driven by Super-Coiled Polymers. Front. Robot AI 2022, 8, 809427. [Google Scholar] [CrossRef]
- Fintz, M.; Osadchy, M.; Hertz, U. Using deep learning to predict human decisions and using cognitive models to explain deep learning models. Sci. Rep. 2022, 12, 4736. [Google Scholar] [CrossRef]
- Shen, Y.; Jia, Q.; Huang, Z.; Wang, R.; Fei, J.; Chen, G. Reinforcement Learning-Based Reactive Obstacle Avoidance Method for Redundant Manipulators. Entropy 2022, 24, 279. [Google Scholar] [CrossRef] [PubMed]
- Ivoghlian, A.; Salcic, Z.; Wang, K.I. Adaptive Wireless Network Management with Multi-Agent Reinforcement Learning. Sensors 2022, 22, 1019. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.G.; Tan, C.; Libedinsky, C.; Yen, S.C.; Tan, A.Y.Y. A nonlinear hidden layer enables actor-critic agents to learn multiple paired association navigation. Cereb. Cortex 2022, 32, 3917–3936. [Google Scholar] [CrossRef] [PubMed]
- Perešíni, P.; Boža, V.; Brejová, B.; Vinař, T. Nanopore Base Calling on the Edge. Bioinformatics 2021, 37, 4661–4667. [Google Scholar] [CrossRef] [PubMed]
- Gholamiankhah, F.; Mostafapour, S.; Abdi Goushbolagh, N.; Shojaerazavi, S.; Layegh, P.; Tabatabaei, S.M.; Arabi, H. Automated Lung Segmentation from Computed Tomography Images of Normal and COVID-19 Pneumonia Patients. Iran. J. Med. Sci. 2022, 47, 440–449. [Google Scholar] [CrossRef]
- Qian, X.; Qiu, Y.; He, Q.; Lu, Y.; Lin, H.; Xu, F.; Zhu, F.; Liu, Z.; Li, X.; Cao, Y.; et al. A Review of Methods for Sleep Arousal Detection Using Polysomnographic Signals. Brain Sci. 2021, 11, 1274. [Google Scholar] [CrossRef]
- Atance, S.R.; Diez, J.V.; Engkvist, O.; Olsson, S.; Mercado, R. De Novo Drug Design Using Reinforcement Learning with Graph-Based Deep Generative Models. J. Chem. Inf. Model 2022, 62, 4863–4872. [Google Scholar] [CrossRef]
- Wang, L.; Yu, Z.; Wang, S.; Guo, Z.; Sun, Q.; Lai, L. Discovery of novel SARS-CoV-2 3CL protease covalent inhibitors using deep learning-based screen. Eur. J. Med. Chem. 2022, 244, 114803. [Google Scholar] [CrossRef]
- Qian, Y.; Wu, J.; Zhang, Q. CAT-CPI: Combining CNN and transformer to learn compound image features for predicting compound-protein interactions. Front. Mol. Biosci. 2022, 9, 963912. [Google Scholar] [CrossRef]
- Gu, Y.; Zheng, S.; Yin, Q.; Jiang, R.; Li, J. REDDA: Integrating multiple biological relations to heterogeneous graph neural network for drug-disease association prediction. REDDA: Integrating multiple biological relations to heterogeneous graph neural network for drug-disease association prediction. Comput. Biol. Med. 2022, 150, 106127. [Google Scholar] [CrossRef]
- Pandiyan, S.; Wang, L. A comprehensive review on recent approaches for cancer drug discovery associated with artificial intelligence. Comput. Biol. Med. 2022, 150, 106140. [Google Scholar] [CrossRef] [PubMed]
- Metzger, J.J.; Pereda, C.; Adhikari, A.; Haremaki, T.; Galgoczi, S.; Siggia, E.D.; Brivanlou, A.H.; Etoc, F. Deep-learning analysis of micropattern-based organoids enables high-throughput drug screening of Huntington’s disease models. Cell Rep. Methods 2022, 2, 100297. [Google Scholar] [CrossRef] [PubMed]
- Ye, Q.; Zhang, X.; Lin, X. Drug-target Interaction Prediction Via Graph Auto-encoder and Multi-subspace Deep Neural Networks. IEEE/ACM Trans. Comput. Biol. Bioinform. 2022; in press. [Google Scholar] [CrossRef]
- Dutta, A. Predicting Drug Mechanics by Deep Learning on Gene and Cell Activities. In Proceedings of the 44th Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Glasgow, UK, 11–15 July 2022; pp. 2916–2919. [Google Scholar] [CrossRef]
- Li, Y.; Liang, W.; Peng, L.; Zhang, D.; Yang, C.; Li, K.C. Predicting Drug-Target Interactions via Dual-Stream Graph Neural Network. IEEE/ACM Trans. Comput. Biol. Bioinform. 2022; in press. [Google Scholar] [CrossRef]
- Lin, S.; Shi, C.; Chen, J. GeneralizedDTA: Combining pre-training and multi-task learning to predict drug-target binding affinity for unknown drug discovery. BMC Bioinform. 2022, 23, 367. [Google Scholar] [CrossRef]
- Liu, G.; Stokes, J.M. A brief guide to machine learning for antibiotic discovery. Curr. Opin. Microbiol. 2022, 69, 102190. [Google Scholar] [CrossRef]
- Wu, Y.; Liu, Q.; Qiu, Y.; Xie, L. Deep learning prediction of chemical-induced dose-dependent and context-specific multiplex phenotype responses and its application to personalized alzheimer’s disease drug repurposing. PLoS Comput. Biol. 2022, 18, e1010367. [Google Scholar] [CrossRef]
- Kwapien, K.; Nittinger, E.; He, J.; Margreitter, C.; Voronov, A.; Tyrchan, C. Implications of Additivity and Nonadditivity for Machine Learning and Deep Learning Models in Drug Design. ACS Omega 2022, 7, 26573–26581. [Google Scholar] [CrossRef] [PubMed]
- Mukaidaisi, M.; Vu, A.; Grantham, K.; Tchagang, A.; Li, Y. Multi-Objective Drug Design Based on Graph-Fragment Molecular Representation and Deep Evolutionary Learning. Front. Pharmacol. 2022, 13, 920747. [Google Scholar] [CrossRef] [PubMed]
- Wilman, W.; Wróbel, S.; Bielska, W.; Deszynski, P.; Dudzic, P.; Jaszczyszyn, I.; Kaniewski, J.; Młokosiewicz, J.; Rouyan, A.; Satława, T.; et al. Machine-designed biotherapeutics: Opportunities, feasibility and advantages of deep learning in computational antibody discovery. Brief Bioinform. 2022, 23, bbac267. [Google Scholar] [CrossRef]
- Aziz, M.; Ejaz, S.A.; Zargar, S.; Akhtar, N.; Aborode, A.T.; Wani, T.A.; Batiha, G.E.; Siddique, F.; Alqarni, M.; Akintola, A.A. Deep Learning and Structure-Based Virtual Screening for Drug Discovery against NEK7: A Novel Target for the Treatment of Cancer. Molecules 2022, 27, 4098. [Google Scholar] [CrossRef]
- Zheng, J.; Xiao, X.; Qiu, W.R. DTI-BERT: Identifying Drug-Target Interactions in Cellular Networking Based on BERT and Deep Learning Method. Front. Genet. 2022, 13, 859188. [Google Scholar] [CrossRef]
- Yeh, S.J.; Yeh, T.Y.; Chen, B.S. Systems Drug Discovery for Diffuse Large B Cell Lymphoma Based on Pathogenic Molecular Mechanism via Big Data Mining and Deep Learning Method. Int. J. Mol. Sci. 2022, 23, 6732. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Hsieh, C.Y.; Wang, J.; Wang, D.; Weng, G.; Shen, C.; Yao, X.; Bing, Z.; Li, H.; Cao, D.; et al. RELATION: A Deep Generative Model for Structure-Based De Novo Drug Design. J. Med. Chem. 2022, 65, 9478–9492. [Google Scholar] [CrossRef] [PubMed]
- Tayebi, A.; Yousefi, N.; Yazdani-Jahromi, M.; Kolanthai, E.; Neal, C.J.; Seal, S.; Garibay, O.O. UnbiasedDTI: Mitigating Real-World Bias of Drug-Target Interaction Prediction by Using Deep Ensemble-Balanced Learning. Molecules 2022, 27, 2980. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhou, D.; Zhang, X.; Shi, Y.; Han, J.; Zhou, L.; Wu, L.; Ma, M.; Li, J.; Peng, S.; et al. D3AI-CoV: A deep learning platform for predicting drug targets and for virtual screening against COVID-19. Brief Bioinform. 2022, 23, bbac147. [Google Scholar] [CrossRef]
- Kurata, H.; Tsukiyama, S. ICAN: Interpretable cross-attention network for identifying drug and target protein interactions. PLoS ONE 2022, 17, e0276609. [Google Scholar] [CrossRef]
- Kawama, K.; Fukushima, Y.; Ikeguchi, M.; Ohta, M.; Yoshidome, T. gr Predictor: A Deep Learning Model for Predicting the Hydration Structures around Proteins. gr Predictor: A Deep Learning Model for Predicting the Hydration Structures around Proteins. J. Chem. Inf. Model. 2022, 62, 4460–4473. [Google Scholar] [CrossRef]
- Li, C.; Sutherland, D.; Hammond, S.A.; Yang, C.; Taho, F.; Bergman, L.; Houston, S.; Warren, R.L.; Wong, T.; Hoang, L.M.N.; et al. AMPlify: Attentive deep learning model for discovery of novel antimicrobial peptides effective against WHO priority pathogens. BMC Genomics 2022, 23, 77. [Google Scholar] [CrossRef]
- Geoffrey, A.S.B.; Madaj, R.; Valluri, P.P. QPoweredCompound2DeNovoDrugPropMax—A novel programmatic tool incorporating deep learning and in silico methods for automated in silico bio-activity discovery for any compound of interest. J. Biomol. Struct. Dyn. 2022, 10, 1–8. [Google Scholar] [CrossRef]
- Li, T.; Tong, W.; Roberts, R.; Liu, Z.; Thakkar, S. DeepCarc: Deep Learning-Powered Carcinogenicity Prediction Using Model-Level Representation. Front. Artif. Intell. 2021, 4, 757780. [Google Scholar] [CrossRef]
- Zan, A.; Xie, Z.R.; Hsu, Y.C.; Chen, Y.H.; Lin, T.H.; Chang, Y.S.; Chang, K.Y. DeepFlu: A deep learning approach for forecasting symptomatic influenza A infection based on pre-exposure gene expression. Comput. Methods Programs Biomed. 2022, 213, 106495. [Google Scholar] [CrossRef]
- Yang, Y.; Zheng, S.; Su, S.; Zhao, C.; Xu, J.; Chen, H. SyntaLinker: Automatic fragment linking with deep conditional transformer neural networks. Chem. Sci. 2020, 11, 8312–8322. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xin, B.; Tan, W.; Xu, Z.; Li, K.; Li, F.; Zhong, W.; Peng, S. DeepR2cov: Deep representation learning on heterogeneous drug networks to discover anti-inflammatory agents for COVID-19. Brief Bioinform. 2021, 22, bbab226. [Google Scholar] [CrossRef] [PubMed]
- Yoshimori, A.; Hu, H.; Bajorath, J. Adapting the DeepSARM approach for dual-target ligand design. J. Comput. Aided Mol. Des. 2021, 35, 587–600. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhang, L.; Feng, H.; Meng, J.; Xie, D.; Yi, L.; Arkin, I.T.; Liu, H. MutagenPred-GCNNs: A Graph Convolutional Neural Network-Based Classification Model for Mutagenicity Prediction with Data-Driven Molecular Fingerprints. Interdiscip. Sci. 2021, 13, 25–33. [Google Scholar] [CrossRef]
- Zeng, B.; Glicksberg, B.S.; Newbury, P.; Chekalin, E.; Xing, J.; Liu, K.; Wen, A.; Chow, C.; Chen, B. OCTAD: An open workspace for virtually screening therapeutics targeting precise cancer patient groups using gene expression features. Nat. Protoc. 2021, 16, 728–753. [Google Scholar] [CrossRef]
- Oh, M.; Park, S.; Lee, S.; Lee, D.; Lim, S.; Jeong, D.; Jo, K.; Jung, I.; Kim, S. DRIM: A Web-Based System for Investigating Drug Response at the Molecular Level by Condition-Specific Multi-Omics Data Integration. Front. Genet. 2020, 11, 564792. [Google Scholar] [CrossRef]
- Rifaioglu, A.S.; Cetin Atalay, R.; Cansen Kahraman, D.; Doğan, T.; Martin, M.; Atalay, V. MDeePred: Novel multi-channel protein featurization for deep learning-based binding affinity prediction in drug discovery. Bioinformatics 2021, 37, 693–704. [Google Scholar] [CrossRef]
- Gentile, F.; Agrawal, V.; Hsing, M.; Ton, A.T.; Ban, F.; Norinder, U.; Gleave, M.E.; Cherkasov, A. Deep Docking: A Deep Learning Platform for Augmentation of Structure Based Drug Discovery. ACS Cent. Sci. 2020, 6, 939–949. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).