1. Background
The basic unit of organization of all living organisms is the cell; most cells carry a genome that encodes all its properties in their DNA. DNA (Deoxyribonucleic acid) is a polymer of two intertwined polynucleotide chains that form a double helix [
1]. Information about an organism’s DNA allows detailed conclusions about its phenotypic characteristics.
In the last 20 years, it has become feasible to sequence entire genomes rather than short fragments. Current third-generation sequencing methods, such as nanopore sequencing, have led to surging numbers of available genome sequences [
2]. It is essential to incorporate this information into the design of future studies. Especially in the world of ever-changing pathogens, all available sequences should be integrated into the design of detection oligos or primers to ensure broad applicability.
Comparison and analysis of complete genomes is now becoming the gold standard for genotyping, and knowledge of full-length genome sequences is becoming the basis for developing various molecular assays for detection and typing. Compared to other biologically relevant macromolecules, DNA is a long-term stable molecule and can be amplified exponentially (PCR, isothermal amplifications), enabling highly sensitive assay formats [
3]. The specific Watson–Crick base pair binding [
4] in DNA—DNA and DNA—RNA hybridizations allows precise detection of nucleic acid sequences even in very low concentration ranges and under stringent conditions. Therefore, molecular assays can quickly be developed, manufactured, stored, delivered, multiplexed, and compared between different labs with a reasonable and constantly dropping price per test. However, there are also challenges in assay development. The design of oligonucleotide primers and probes is a crucial task for many biomedical studies and molecular applications aiming, e.g., to identify pathogenic organisms and their resistance and virulence genes. Other applications benefitting from high-quality consensus oligonucleotides include FISH analysis, Northern blots, microarray-based analyses, isothermal amplification procedures, and all types of molecular assays utilizing linear or exponential amplifications [
5]. To ensure the widespread usability of oligonucleotide sequences, detecting high-quality consensus regions among relevant target sequences is a necessary pre-requisite. Given the inherent variability of, e.g., bacterial or viral target genes and the massive increase in the number of published target sequences, it becomes necessary to automatize the process of aligning these sequences and detecting suitable consensus regions.
For the performance and applicability of predicted primers, selecting the target sequences that serve as the basis for the alignment plays a decisive role. This is because functional oligonucleotide sequences can only be predicted if the selected sequences are also representative of the sequences to be examined in a given sample type. The basis for a representative consensus sequence is a high-quality multiple-sequence alignment. The alignment quality depends on several factors, such as the type and length of the input sequences, parameters such as gap opening/extension costs, and the algorithm itself. Therefore, the parameters for calculating the alignments vary based on the particular alignment requirements.
Consensus primer design is not a new problem. There are already publications on programs for the design of consensus primers. Unfortunately, these tools and their online services are often no longer available, such as CODEHOP [
6]. Another tool for consensus primer design is PrimerDesign-M [
7]. Unfortunately, this tool does not allow extensive parameter settings for experimental design, such as setting specific Tm values.
A significant advantage of the ConsensusPrime pipeline is the various possibilities to filter the input alignment and thus improve the quality of the predicted primers.
2. Materials and Methods
With MAFFT [
8] for generating multiple sequence alignments and Primer3 [
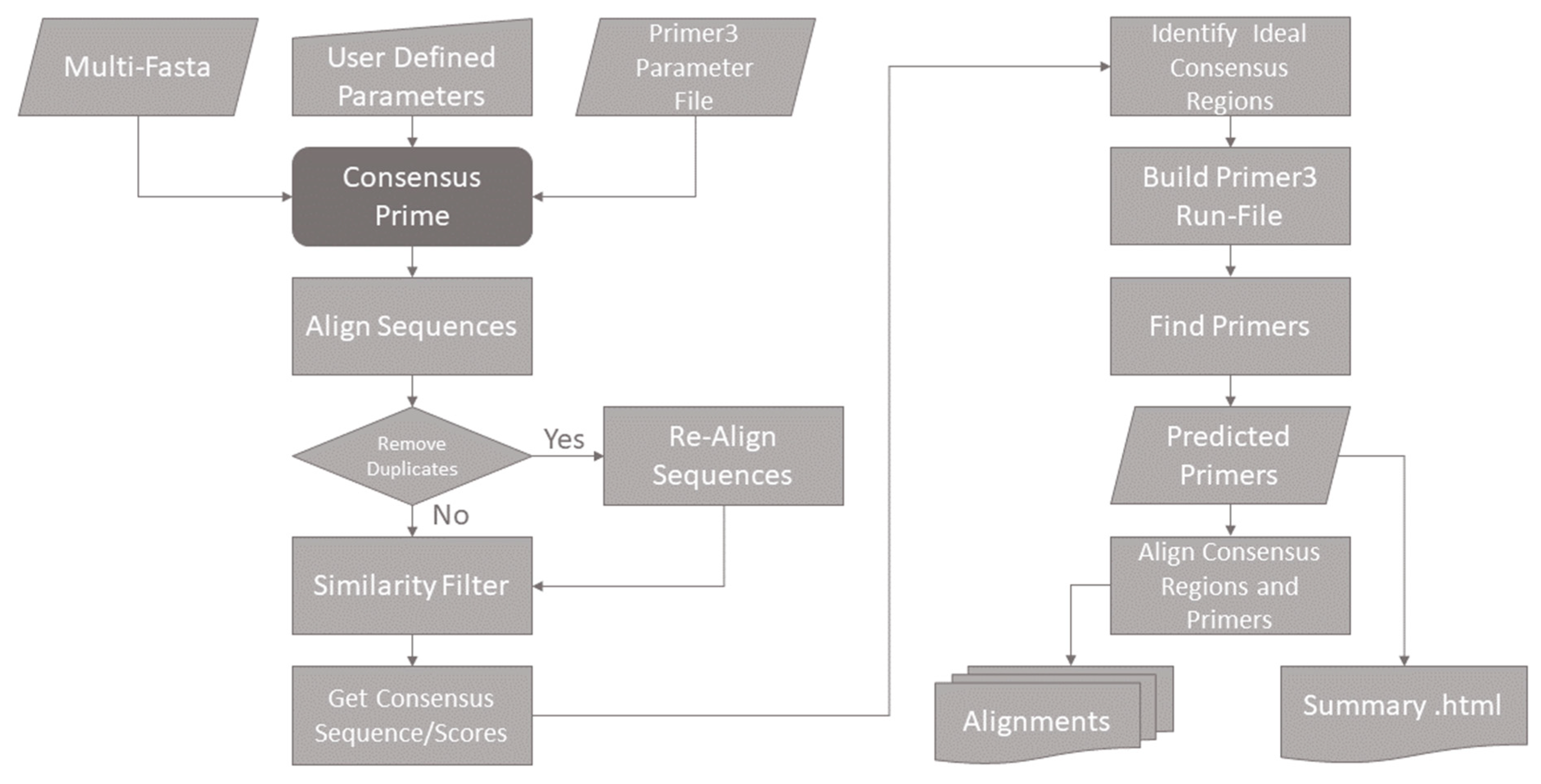
9] for predicting primers and probes, the ConsensusPrime pipeline uses well-established tools. The pipeline needs three different pieces of input information. First, the sequences have to be provided in multi-fasta format. Second is the Primer3 parameter file containing the physicochemical primer/probe parameters for Primer3, and thirdly the ConsensusPrime parameters that influence the pipeline itself are given via the command line. The ConsensusPrime pipeline processes the input data in several successive filter and alignment steps to identify suitable consensus regions. The regions found in this way are automatically written into the Primer3 parameter file, and the primer prediction is started using Primer3. Afterward, the output of Primer3, in addition to the details of the individual filter steps, is written into a concise HTML file. Predicted primers are also added to a final alignment to be displayed with any alignment visualization tool.
The entire pipeline is based on multiple sequence alignments, so their quality is vital for functional assays. Therefore, the parameters of MAFFT were adjusted to fulfill the alignment requirements in every alignment step. This is of particular importance when aligning the short primer sequences for visualization. In this case, the ‘--addfragments’ parameter of MAFFT is used to align the short primers to their origin properly. MAFFT also allows the automated adjustment of the strand direction of a sequence. Another important parameter is ‘--adjustdirectioin’, which allows the automated detection and adjustment of the strand direction in which sequences are provided, as well as the mapping of the reverse primers.
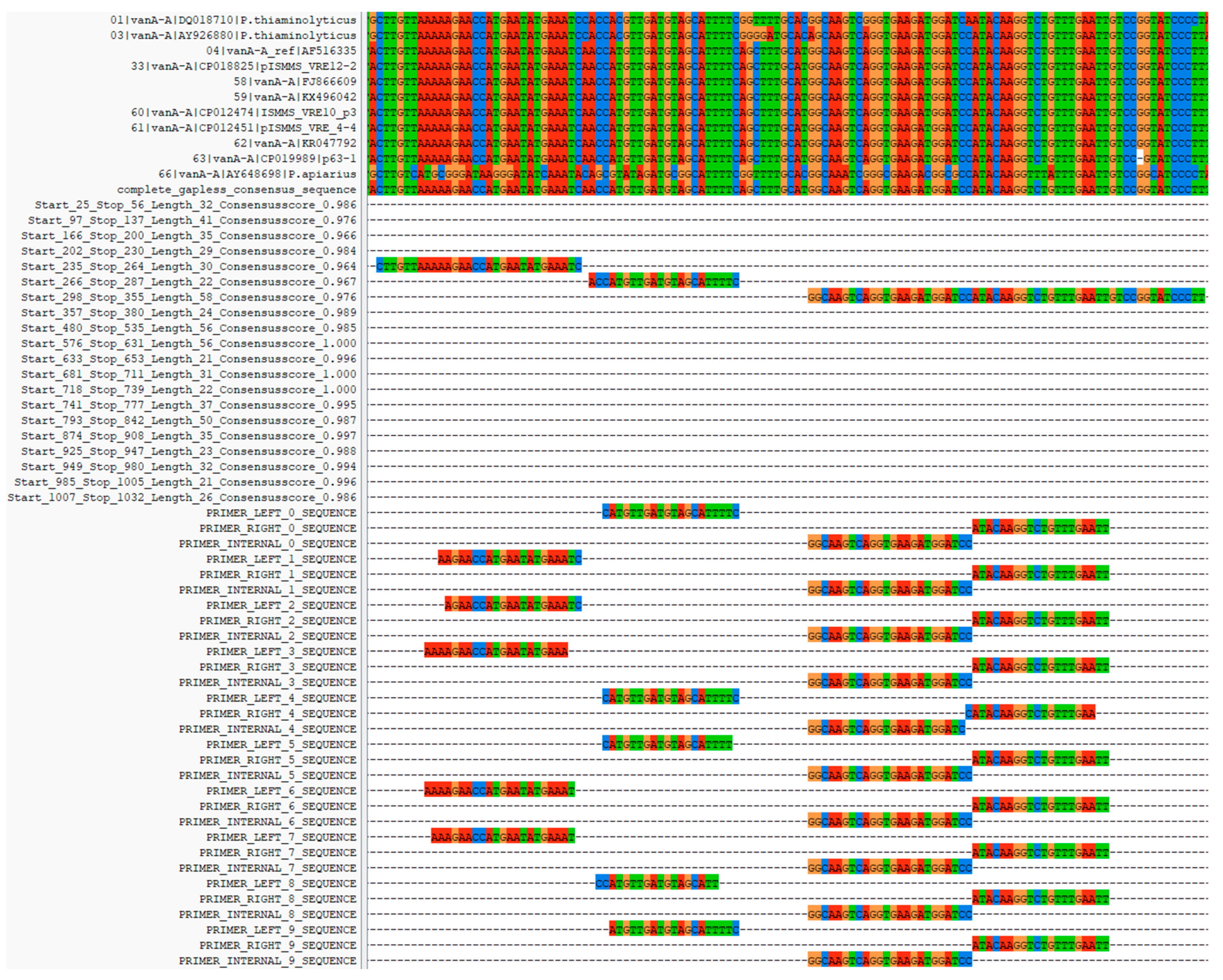
To avoid unwanted distortions of the consensus scores caused by overrepresented sequences, identical sequences are removed from the alignment unless specified differently using the ‘--keepduplicates’ parameter. The ‘--consensussimilarity’ parameter defines the similarity cutoff for each sequence in comparison to the consensus sequence of the alignment. The default value of, e.g., 0.8 means that a sequence has to have at least 80% of its aligned nucleotides in common with the consensus sequence. Otherwise, the sequence is removed from the alignment for primer prediction. The filtered sequences are then re-aligned. Therefore, the pipeline uses MAFFT to align all input sequences in a global multiple-sequence alignment, and it identifies the most common nucleotide for every position in the alignment. In addition, a consensus score is calculated for every alignment position, which is the ratio of the respective count/number of most common nucleotide or gap symbol (−) at that position to the total number of sequences. All letters that are not ATGC are treated as a gap. A perfectly conserved region in which all sequences at a given position are identical is thus assigned a consensus score of 1. The pipeline allows the user to control the quality values of the consensus sequence used for primer prediction via the ‘--consensusthreshold’ parameter. The default value of 0.95 ensures that the most abundant nucleotide occurs in at least 95% of the sequences at the given position. In addition, the regions above the threshold must have a contiguous minimum length of at least 20 nucleotides. All regions below these values are excluded from the subsequent primer prediction.
Before the consensus regions are identified for primer design, gaps are removed from the consensus sequence and the corresponding value from the consensus scores. This removal is necessary because gaps are not encoded by nucleotides and are irrelevant to primer design. Gaps in the consensus sequence are caused by insertions in one or more related sequences of the alignment for better visualization.
From this “gapless consensus sequence”, the regions relevant to the primer design are identified.
As Primer3 searches for the primer pair in a contiguous sequence section, instead of using the area in which primers are to be searched (SEQUENCE_INCLUDED_REGION and SEQUENCE_INTERNAL_INCLUDED_REGION), all areas in which primers are not to be searched are excluded by the pipeline (SEQUENCE_EXCLUDED_REGION and SEQUENCE_INTERNAL_EXCLUDED_REGION). This allows the prediction of primers in non-consecutive sequence segments. Furthermore, the gapless consensus sequence is automatically written into the Primer3 parameter file (SEQUENCE_TEMPLATE). All other parameters, such as melting temperatures or primer lengths, are taken from the user-defined Primer3 parameter file. (For a detailed parameter description, check out the Primer3 manual (
https://primer3.org/manual.html, accessed on 7 October 2022). From this, Primer3 predicts the optimal consensus primers and displays the results in a plain text file. The ConsensusPrime pipeline reads the Primer3 output and creates a comprehensive output including all details of previous filter steps in HTML format. The predicted primers are added to a final alignment to be visualized using ClustalX [
10] and can be manually inspected for logical errors. The structure of the final alignment is always in the order of filtered sequences, consensus sequence, sequence parts used for the primer prediction, and predicted primer/probe pairs. This alignment enables the user to check the alignment of the filtered input sequences and the resulting consensus regions considered for the actual primer prediction. It also gives an excellent overview of the predicted primers to choose the best pair if multiple predictions have been made. Note that the reverse primer is added to the final alignment as the reverse complementary sequence.