Ergosterol Biosynthesis and Regulation Impact the Antifungal Resistance and Virulence of Candida spp.
Abstract
1. Introduction
2. Sterol Biosynthesis
3. Transcription Factors Involved in Ergosterol Biosynthesis Regulation in Candida spp.
4. Sterol Profiles of C. albicans and C. glabrata Ergosterol Biosynthesis Mutants
5. Ergosterol Influence on Membranes and Organelles of Candida spp.
6. Ergosterol Biosynthesis and Antifungal Resistance of Candida spp. Clinical Isolates
7. Ergosterol Biosynthesis Modulations Affect the Virulence of Candida spp.
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Abe, F.; Hiraki, T. Mechanistic role of ergosterol in membrane rigidity and cycloheximide resistance in Saccharomyces cerevisiae. Biochim. Biophys. Acta 2009, 1788, 743–752. [Google Scholar] [CrossRef]
- Guan, X.L.; Souza, C.M.; Pichler, H.; Dewhurst, G.; Schaad, O.; Kajiwara, K.; Wakabayashi, H.; Ivanova, T.; Castillon, G.A.; Piccolis, M.; et al. Functional interactions between sphingolipids and sterols in biological membranes regulating cell physiology. Mol. Biol. Cell 2009, 20, 2083–2085. [Google Scholar] [CrossRef]
- Dupont, S.; Lemetais, G.; Ferreira, T.; Cayot, P.; Gervais, P.; Beney, L. Ergosterol biosynthesis: A fungal pathway for life on land? Evolution 2012, 66, 2961–2968. [Google Scholar] [CrossRef]
- Khmelinskaia, A.; Marquês, J.M.T.; Bastos, A.E.P.; Antunes, C.A.C.; Bento-Oliveira, A.; Scolari, S.; Lobo, G.M.D.S.; Malhó, R.; Herrmann, A.; Marinho, H.S.; et al. Liquid-ordered phase formation by mammalian and yeast sterols: A common feature with organizational differences. Front. Cell Dev. Biol. 2020, 8, 337. [Google Scholar] [CrossRef]
- Suchodolski, J.; Muraszko, J.; Bernat, P.; Krasowska, A. A crucial role for ergosterol in plasma membrane composition, localisation, and activity of Cdr1p and H+-ATPase in Candida albicans. Microorganisms 2019, 7, 378. [Google Scholar] [CrossRef]
- Urbanek, A.K.; Muraszko, J.; Derkacz, D.; Łukaszewicz, M.; Bernat, P.; Krasowska, A. The role of ergosterol and sphingolipids in the localization and activity of Candida albicans’ multidrug transporter Cdr1p and plasma membrane ATPase Pma1p. Int. J. Mol. Sci. 2022, 23, 9975. [Google Scholar] [CrossRef]
- Spira, F.; Mueller, N.S.; Beck, G.; von Olshausen, P.; Beig, J.; Wedlich-Söldner, R. Patchwork organization of the yeast plasma membrane into numerous coexisting domains. Nat. Cell Biol. 2012, 14, 640–648. [Google Scholar] [CrossRef] [PubMed]
- Weete, J.D.; Abril, M.; Blackwell, M. Phylogenetic distribution of fungal sterols. PLoS ONE 2010, 5, e10899. [Google Scholar] [CrossRef]
- Kodedová, M.; Sychrová, H. Changes in the sterol composition of the plasma membrane affect membrane potential, salt tolerance and the activity of multidrug resistance pumps in Saccharomyces cerevisiae. PLoS ONE 2015, 10, e0139306. [Google Scholar] [CrossRef]
- Liu, G.; Chen, Y.; Færgeman, N.J.; Nielsen, J. Elimination of the last reactions in ergosterol biosynthesis alters the resistance of Saccharomyces cerevisiae to multiple stresses. FEMS Yeast Res. 2017, 17, fox063. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, S.; Esquivel, B.D.; White, T.C. Overexpression or deletion of ergosterol biosynthesis genes alters doubling time, response to stress agents, and drug susceptibility in Saccharomyces cerevisiae. mBio 2018, 9, e01291-18. [Google Scholar] [CrossRef] [PubMed]
- Parks, L.W.; Smith, S.J.; Crowley, J.H. Biochemical and physiological effects of sterol alterations in yeast—A review. Lipids 1995, 30, 227–230. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, S.; Sae-Tia, S.; Fries, B.C. Candidiasis and mechanisms of antifungal resistance. Antibiotics 2020, 9, 312. [Google Scholar] [CrossRef] [PubMed]
- Houst, J.; Spizek, J.; Havlicek, V. Antifungal Drugs. Metabolites 2020, 10, 106. [Google Scholar] [CrossRef]
- Singh, A.; Yadav, V.; Prasad, R. Comparative lipidomics in clinical isolates of Candida albicans reveal crosstalk between mitochondria, cell wall integrity and azole resistance. PLoS ONE 2012, 7, e39812. [Google Scholar] [CrossRef]
- Rella, A.; Farnoud, A.M.; Del Poeta, M. Plasma membrane lipids and their role in fungal virulence. Prog. Lipid Res. 2016, 61, 63–72. [Google Scholar] [CrossRef]
- Arendrup, M.C.; Patterson, T.F. Multidrug-resistant Candida: Epidemiology, molecular mechanisms, and treatment. J. Infect. Dis. 2017, 216, S445–S451. [Google Scholar] [CrossRef]
- Rayens, E.; Norris, K.A. Prevalence and healthcare burden of fungal infections in the United States, 2018. Open Forum Infect. Dis. 2022, 9, ofab593. [Google Scholar] [CrossRef] [PubMed]
- Pappas, P.G.; Lionakis, M.S.; Arendrup, M.C.; Ostrosky-Zeichner, L.; Kullberg, B.J. Invasive candidiasis. Nat. Rev. Dis. Primers 2018, 4, 18026. [Google Scholar] [CrossRef]
- Galocha, M.; Pais, P.; Cavalheiro, M.; Pereira, D.; Viana, R.; Teixeira, M.C. Divergent approaches to virulence in C. albicans and C. glabrata: Two sides of the same coin. Int. J. Mol. Sci. 2019, 20, 2345. [Google Scholar] [CrossRef]
- Quindós, G.; Marcos-Arias, C.; San-Millán, R.; Mateo, E.; Eraso, E. The continuous changes in the aetiology and epidemiology of invasive candidiasis: From familiar Candida albicans to multiresistant Candida auris. Int. Microbiol. 2018, 21, 107–119. [Google Scholar] [CrossRef] [PubMed]
- Kristan, K.; Rizner, T.L. Steroid-transforming enzymes in fungi. J. Steroid Biochem. Mol. Biol. 2012, 129, 79–91. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; He, B.; Ma, L.; Sun, Y.; Niu, Y.; Zeng, B. Recent advances in ergosterol biosynthesis and regulation mechanisms in Saccharomyces cerevisiae. Indian J. Microbiol. 2017, 57, 270–277. [Google Scholar] [CrossRef]
- Johnston, E.J.; Moses, T.; Rosser, S.J. The wide-ranging phenotypes of ergosterol biosynthesis mutants, and implications for microbial cell factories. Yeast 2020, 37, 27–44. [Google Scholar] [CrossRef]
- Jordá, T.; Puig, S. Regulation of Ergosterol Biosynthesis in Saccharomyces cerevisiae. Genes 2020, 11, 795. [Google Scholar] [CrossRef]
- Lv, Q.Z.; Yan, L.; Jiang, Y.Y. The synthesis, regulation, and functions of sterols in Candida albicans: Well-known but still lots to learn. Virulence 2016, 7, 649–659. [Google Scholar] [CrossRef] [PubMed]
- Chau, A.S.; Gurnani, M.; Hawkinson, R.; Laverdiere, M.; Cacciapuoti, A.; McNicholas, P.M. Inactivation of sterol Delta5,6-desaturase attenuates virulence in Candida albicans. Antimicrob. Agents Chemother. 2005, 49, 3646–3651. [Google Scholar] [CrossRef]
- Pierson, C.A.; Eckstein, J.; Barbuch, R.; Bard, M. Ergosterol gene expression in wild-type and ergosterol-deficient mutants of Candida albicans. Med. Mycol. 2004, 42, 385–389. [Google Scholar] [CrossRef][Green Version]
- Geber, A.; Hitchcock, C.A.; Swartz, J.E.; Pullen, F.S.; Marsden, K.E.; Kwon-Chung, K.J.; Bennett, J.E. Deletion of the Candida glabrata ERG3 and ERG11 genes: Effect on cell viability, cell growth, sterol composition, and antifungal susceptibility. Antimicrob. Agents Chemother. 1995, 39, 2708–2717. [Google Scholar] [CrossRef] [PubMed]
- Jensen-Pergakes, K.L.; Kennedy, M.A.; Lees, N.D.; Barbuch, R.; Koegel, C.; Bard, M. Sequencing, disruption, and characterization of the Candida albicans sterol methyltransferase (ERG6) gene: Drug susceptibility studies in erg6 mutants. Antimicrob. Agents Chemother. 1998, 42, 1160–1167. [Google Scholar] [CrossRef]
- Sanglard, D.; Ischer, F.; Parkinson, T.; Falconer, D.; Bille, J. Candida albicans mutations in the ergosterol biosynthetic pathway and resistance to several antifungal agents. Antimicrob. Agents Chemother. 2003, 47, 2404–2412. [Google Scholar] [CrossRef] [PubMed]
- Bard, M.; Lees, N.D.; Turi, T.; Craft, D.; Cofrin, L.; Barbuch, R.; Koegel, C.; Loper, J.C. Sterol synthesis and viability of erg11 (cytochrome P450 lanosterol demethylase) mutations in Saccharomyces cerevisiae and Candida albicans. Lipids 1993, 28, 963–967. [Google Scholar] [CrossRef] [PubMed]
- Elias, D.; Toth Hervay, N.; Jacko, J.; Morvova, M.; Valachovic, M.; Gbelska, Y. Erg6p is essential for antifungal drug resistance, plasma membrane properties and cell wall integrity in Candida glabrata. FEMS Yeast Res. 2022, 21, foac045. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, T.; Miyazaki, Y.; Izumikawa, K.; Kakeya, H.; Miyakoshi, S.; Bennett, J.E.; Kohno, S. Fluconazole treatment is effective against a Candida albicans erg3/erg3 mutant in vivo despite in vitro resistance. Antimicrob. Agents Chemother. 2006, 50, 580–586. [Google Scholar] [CrossRef]
- Regan, J.; DeJarnette, C.; Luna-Tapia, A.; Parker, J.E.; Reitler, P.; Barnett, S.; Tucker, K.M.; Kelly, S.L.; Palmer, G.E. Titration of C-5 sterol desaturase activity reveals its relationship to Candida albicans virulence and antifungal susceptibility is dependent upon host immune status. mBio 2022, 13, e0011522. [Google Scholar] [CrossRef]
- Martel, C.M.; Parker, J.E.; Bader, O.; Weig, M.; Gross, U.; Warrilow, A.G.; Kelly, D.E.; Kelly, S.L. A clinical isolate of Candida albicans with mutations in ERG11 (encoding sterol 14alpha-demethylase) and ERG5 (encoding C22 desaturase) is cross resistant to azoles and amphotericin B. Antimicrob. Agents Chemother. 2010, 54, 3578–3583. [Google Scholar] [CrossRef]
- Becker, J.M.; Kauffman, S.J.; Hauser, M.; Huang, L.; Lin, M.; Sillaots, S.; Jiang, B.; Xu, D.; Roemer, T. Pathway analysis of Candida albicans survival and virulence determinants in a murine infection model. Proc. Natl. Acad. Sci. USA 2010, 107, 22044–22049. [Google Scholar] [CrossRef]
- Nakayama, H.; Izuta, M.; Nakayama, N.; Arisawa, M.; Aoki, Y. Depletion of the squalene synthase (ERG9) gene does not impair growth of Candida glabrata in mice. Antimicrob. Agents Chemother. 2000, 44, 2411–2418. [Google Scholar] [CrossRef]
- Pasrija, R.; Krishnamurthy, S.; Prasad, T.; Ernst, J.F.; Prasad, R. Squalene epoxidase encoded by ERG1 affects morphogenesis and drug susceptibilities of Candida albicans. J. Antimicrob. Chemother. 2005, 55, 905–913. [Google Scholar] [CrossRef]
- Tsai, H.F.; Bard, M.; Izumikawa, K.; Krol, A.A.; Sturm, A.M.; Culbertson, N.T.; Pierson, C.A.; Bennett, J.E. Candida glabrata erg1 mutant with increased sensitivity to azoles and to low oxygen tension. Antimicrob. Agents Chemother. 2004, 48, 2483–2489. [Google Scholar] [CrossRef]
- Okamoto, M.; Takahashi-Nakaguchi, A.; Tejima, K.; Sasamoto, K.; Yamaguchi, M.; Aoyama, T.; Nagi, M.; Tanabe, K.; Miyazaki, Y.; Nakayama, H.; et al. Erg25 controls host-cholesterol uptake mediated by Aus1p-associated sterol-rich membrane domains in Candida glabrata. Front. Cell Dev. Biol. 2022, 10, 820675. [Google Scholar] [CrossRef] [PubMed]
- Stefanek, M.; Garaiova, M.; Valcek, A.; Jordao, L.; Bujdakova, H. Comparative analysis of two Candida parapsilosis isolates originating from the same patient harbouring the Y132F and R398I mutations in the ERG11 gene. Cells 2023, 12, 1579. [Google Scholar] [CrossRef] [PubMed]
- Jia, N.; Arthington-Skaggs, B.; Lee, W.; Pierson, C.A.; Lees, N.D.; Eckstein, J.; Barbuch, R.; Bard, M. Candida albicans sterol C-14 reductase, encoded by the ERG24 gene, as a potential antifungal target site. Antimicrob. Agents Chemother. 2002, 46, 947–957. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, M.A.; Johnson, T.A.; Lees, N.D.; Barbuch, R.; Eckstein, J.A.; Bard, M. Cloning and sequencing of the Candida albicans C-4 sterol methyl oxidase gene (ERG25) and expression of an ERG25 conditional lethal mutation in Saccharomyces cerevisiae. Lipids 2000, 35, 257–262. [Google Scholar] [CrossRef] [PubMed]
- Aaron, K.E.; Pierson, C.A.; Lees, N.D.; Bard, M. The Candida albicans ERG26 gene encoding the C-3 sterol dehydrogenase (C-4 decarboxylase) is essential for growth. FEMS Yeast Res. 2001, 1, 93–101. [Google Scholar] [CrossRef]
- Pierson, C.A.; Jia, N.; Mo, C.; Lees, N.D.; Sturm, A.M.; Eckstein, J.; Barbuct, R.; Bard, M. Isolation, characterization, and regulation of the Candida albicans ERG27 gene encoding the sterol 3-keto reductase. Med. Mycol. 2004, 42, 461–473. [Google Scholar] [CrossRef]
- Luna-Tapia, A.; Peters, B.M.; Eberle, K.E.; Kerns, M.E.; Foster, T.P.; Marrero, L.; Noverr, M.C.; Fidel, P.L., Jr.; Palmer, G.E. ERG2 and ERG24 are required for normal vacuolar physiology as well as Candida albicans pathogenicity in a murine model of disseminated but not vaginal candidiasis. Eukaryot. Cell. 2015, 14, 1006–1016. [Google Scholar] [CrossRef]
- Ahmad, S.; Joseph, L.; Parker, J.E.; Asadzadeh, M.; Kelly, S.L.; Meis, J.F.; Khan, Z. ERG6 and ERG2 are major targets conferring reduced susceptibility to amphotericin B in clinical Candida glabrata isolates in Kuwait. Antimicrob. Agents Chemother. 2019, 63, e01900-18. [Google Scholar] [CrossRef]
- Branco, J.; Ola, M.; Silva, R.M.; Fonseca, E.; Gomes, N.C.; Martins-Cruz, C.; Silva, A.P.; Silva-Dias, A.; Pina-Vaz, C.; Erraught, C.; et al. Impact of ERG3 mutations and expression of ergosterol genes controlled by UPC2 and NDT80 in Candida parapsilosis azole resistance. Clin. Microbiol. Infect. 2017, 23, 575.e1–575.e8. [Google Scholar] [CrossRef]
- Feng, W.; Yang, J.; Xi, Z.; Qiao, Z.; Lv, Y.; Wang, Y.; Ma, Y.; Wang, Y.; Cen, W. Mutations and/or overexpressions of ERG4 and ERG11 genes in clinical azoles-resistant isolates of Candida albicans. Microb. Drug Resist. 2017, 23, 563–570. [Google Scholar] [CrossRef]
- Skrzypek, M.S.; Binkley, J.; Binkley, G.; Miyasato, S.R.; Simison, M.; Sherlock, G. The Candida Genome Database (CGD): Incorporation of Assembly 22, systematic identifiers and visualization of high throughput sequencing data. Nucleic Acids Res. 2017, 45, D592–D596. [Google Scholar] [CrossRef]
- Alcazar-Fuoli, L.; Mellado, E.; Garcia-Effron, G.; Lopez, J.F.; Grimalt, J.O.; Cuenca-Estrella, J.M.; Rodriguez-Tudela, J.L. Ergosterol biosynthesis pathway in Aspergillus fumigatus. Steroids 2008, 73, 339–347. [Google Scholar] [CrossRef]
- Alcazar-Fuoli, L.; Mellado, E. Ergosterol biosynthesis in Aspergillus fumigatus: Its relevance as an antifungal target and role in antifungal drug resistance. Front. Microbiol. 2013, 3, 439. [Google Scholar] [CrossRef] [PubMed]
- Bauer, K.; Rafael, B.; Vago, B.; Kiss-Vetrab, S.; Molnar, A.; Szebenyi, C.; Varga, M.; Szekeres, A.; Vagvolgyi, C.; Papp, T.; et al. Characterization of the sterol 24-C-methyltransferase genes reveals a network of alternative sterol biosynthetic pathways in Mucor lusitanicus. Microbiol. Spectr. 2023, 11, e0031523. [Google Scholar] [CrossRef] [PubMed]
- Luna-Tapia, A.; Parker, J.E.; Kelly, S.L.; Palmer, G.E. Species-specific differences in C-5 sterol desaturase function influence the outcome of azole antifungal exposure. Antimicrob. Agents Chemother. 2021, 65, e0104421. [Google Scholar] [CrossRef]
- Young, L.Y.; Hull, C.M.; Heitman, J. Disruption of ergosterol biosynthesis confers resistance to amphotericin B in Candida lusitaniae. Antimicrob. Agents Chemother. 2003, 47, 2717–2724. [Google Scholar] [CrossRef]
- Henry, K.W.; Nickels, J.T.; Edlind, T.D. Upregulation of ERG genes in Candida species by azoles and other sterol biosynthesis inhibitors. Antimicrob. Agents Chemother. 2000, 44, 2693–2700. [Google Scholar] [CrossRef]
- Vandeputte, P.; Tronchin, G.; Berges, T.; Hennequin, C.; Chabasse, D.; Bouchara, J.P. Reduced susceptibility to polyenes associated with a missense mutation in the ERG6 gene in a clinical isolate of Candida glabrata with pseudohyphal growth. Antimicrob. Agents Chemother. 2007, 51, 982–990. [Google Scholar] [CrossRef]
- Vandeputte, P.; Tronchin, G.; Larcher, G.; Ernoult, E.; Berges, T.; Chabasse, D.; Bouchara, J.P. A nonsense mutation in the ERG6 gene leads to reduced susceptibility to polyenes in a clinical isolate of Candida glabrata. Antimicrob. Agents Chemother. 2008, 52, 3701–3709. [Google Scholar] [CrossRef]
- Li, Q.Q.; Tsai, H.F.; Mandal, A.; Walker, B.A.; Noble, J.A.; Fukuda, Y.; Bennett, J.E. Sterol uptake and sterol biosynthesis act coordinately to mediate antifungal resistance in Candida glabrata under azole and hypoxic stress. Mol. Med. Rep. 2018, 17, 6585–6597. [Google Scholar] [CrossRef]
- Berkow, E.L.; Manigaba, K.; Parker, J.E.; Barker, K.S.; Kelly, S.L.; Rogers, P.D. Multidrug transporters and alterations in sterol biosynthesis contribute to azole antifungal resistance in Candida parapsilosis. Antimicrob. Agents Chemother. 2015, 59, 5942–5950. [Google Scholar] [CrossRef]
- Vik, A.; Rine, J. Upc2p and Ecm22p, dual regulators of sterol biosynthesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 2001, 21, 6395–6405. [Google Scholar] [CrossRef]
- Woods, K.; Höfken, T. The zinc cluster proteins Upc2 and Ecm22 promote filamentation in Saccharomyces cerevisiae by sterol biosynthesis-dependent and -independent pathways. Mol. Microbiol. 2016, 99, 512–527. [Google Scholar] [CrossRef]
- Montañés, F.M.; Pascual-Ahuir, A.; Proft, M. Repression of ergosterol biosynthesis is essential for stress resistance and is mediated by the Hog1 MAP kinase and the Mot3 and Rox1 transcription factors. Mol. Microbiol. 2011, 79, 1008–1023. [Google Scholar] [CrossRef]
- Davies, B.S.; Wang, H.S.; Rine, J. Dual activators of the sterol biosynthetic pathway of Saccharomyces cerevisiae: Similar activation/regulatory domains but different response mechanisms. Mol. Cell. Biol. 2005, 25, 7375–7385. [Google Scholar] [CrossRef]
- Yang, H.; Tong, J.; Lee, C.W.; Ha, S.; Eom, S.H.; Im, Y.J. Structural mechanism of ergosterol regulation by fungal sterol transcription factor Upc2. Nat. Commun. 2015, 6, 6129. [Google Scholar] [CrossRef]
- Tan, L.; Chen, L.; Yang, H.; Jin, B.; Kim, G.; Im, Y.J. Structural basis for activation of fungal sterol receptor Upc2 and azole resistance. Nat. Chem. Biol. 2022, 18, 1253–1262. [Google Scholar] [CrossRef]
- Silver, P.M.; Oliver, B.G.; White, T.C. Role of Candida albicans transcription factor Upc2p in drug resistance and sterol metabolism. Eukaryot. Cell 2004, 3, 1391–1397. [Google Scholar] [CrossRef]
- MacPherson, S.; Akache, B.; Weber, S.; De Deken, X.; Raymond, M.; Turcotte, B. Candida albicans zinc cluster protein Upc2p confers resistance to antifungal drugs and is an activator of ergosterol biosynthetic genes. Antimicrob. Agents Chemother. 2005, 49, 1745–1752. [Google Scholar] [CrossRef]
- Dunkel, N.; Liu, T.T.; Barker, K.S.; Homayouni, R.; Morschhäuser, J.; Rogers, P.D. A gain-of-function mutation in the transcription factor Upc2p causes upregulation of ergosterol biosynthesis genes and increased fluconazole resistance in a clinical Candida albicans isolate. Eukaryot. Cell 2008, 7, 1180–1190. [Google Scholar] [CrossRef] [PubMed]
- Vasicek, E.M.; Berkow, E.L.; Flowers, S.A.; Barker, K.S.; Rogers, P.D. UPC2 is universally essential for azole antifungal resistance in Candida albicans. Eukaryot. Cell 2014, 13, 933–946. [Google Scholar] [CrossRef] [PubMed]
- Flowers, S.A.; Barker, K.S.; Berkow, E.L.; Toner, G.; Chadwick, S.G.; Gygax, S.E.; Morschhäuser, J.; Rogers, P.D. Gain-of-function mutations in UPC2 are a frequent cause of ERG11 upregulation in azole-resistant clinical isolates of Candida albicans. Eukaryot. Cell 2012, 11, 1289–1299. [Google Scholar] [CrossRef] [PubMed]
- Nagi, M.; Nakayama, H.; Tanabe, K.; Bard, M.; Aoyama, T.; Okano, M.; Higashi, S.; Ueno, K.; Chibana, H.; Niimi, M.; et al. Transcription factors CgUPC2A and CgUPC2B regulate ergosterol biosynthetic genes in Candida glabrata. Genes Cells 2011, 16, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Whaley, S.G.; Caudle, K.E.; Vermitsky, J.P.; Chadwick, S.G.; Toner, G.; Barker, K.S.; Gygax, S.E.; Rogers, P.D. UPC2A is required for high-level azole antifungal resistance in Candida glabrata. Antimicrob. Agents Chemother. 2014, 58, 4543–4554. [Google Scholar] [CrossRef]
- Sellam, A.; Tebbji, F.; Nantel, A. Role of Ndt80p in sterol metabolism regulation and azole resistance in Candida albicans. Eukaryot. Cell 2009, 8, 1174–1183. [Google Scholar] [CrossRef]
- Prasad, T.; Hameed, S.; Manoharlal, R.; Biswas, S.; Mukhopadhyay, C.K.; Goswami, S.K.; Prasad, R. Morphogenic regulator EFG1 affects the drug susceptibilities of pathogenic Candida albicans. FEMS Yeast Res. 2010, 10, 587–596. [Google Scholar] [CrossRef]
- Lo, H.J.; Wang, J.S.; Lin, C.Y.; Chen, C.G.; Hsiao, T.Y.; Hsu, C.T.; Su, C.L.; Fann, M.J.; Ching, Y.T.; Yang, Y.L. Efg1 involved in drug resistance by regulating the expression of ERG3 in Candida albicans. Antimicrob. Agents Chemother. 2005, 49, 1213–1215. [Google Scholar] [CrossRef]
- Zheng, H.; Jiang, Y.Y.; Wang, Y.; Jia, X.M.; Yan, T.H.; Gao, P.H.; Yan, L.; Jiang, L.H.; Ji, H.; Cao, Y.B. TOP2 gene disruption reduces drug susceptibility by increasing intracellular ergosterol biosynthesis in Candida albicans. J. Med. Microbiol. 2010, 59, 797–803. [Google Scholar] [CrossRef][Green Version]
- Yau, K.P.S.; Weerasinghe, H.; Olivier, F.A.B.; Lo, T.L.; Powell, D.R.; Koch, B.; Beilharz, T.H.; Traven, A. The proteasome regulator Rpn4 controls antifungal drug tolerance by coupling protein homeostasis with metabolic responses to drug stress. PLoS Pathog. 2023, 19, e1011338. [Google Scholar] [CrossRef]
- Nobile, C.J.; Nett, J.E.; Hernday, A.D.; Homann, O.R.; Deneault, J.S.; Nantel, A.; Andes, D.R.; Johnson, A.D.; Mitchell, A.P. Biofilm matrix regulation by Candida albicans Zap1. PLoS Biol. 2009, 7, e1000133. [Google Scholar] [CrossRef]
- Ollinger, T.L.; Vu, B.; Murante, D.; Parker, J.E.; Simonicova, L.; Doorley, L.; Stamnes, M.A.; Kelly, S.L.; Rogers, P.D.; Moye-Rowley, W.S.; et al. Loss-of-function ROX1 mutations suppress the fluconazole susceptibility of upc2AΔ mutation in Candida glabrata, implicating additional positive regulators of ergosterol biosynthesis. mSphere 2021, 6, e0083021. [Google Scholar] [CrossRef] [PubMed]
- Pais, P.; Califórnia, R.; Galocha, M.; Viana, R.; Ola, M.; Cavalheiro, M.; Takahashi-Nakaguchi, A.; Chibana, H.; Butler, G.; Teixeira, M.C. Candida glabrata transcription factor rpn4 mediates fluconazole resistance through regulation of ergosterol biosynthesis and plasma membrane permeability. Antimicrob. Agents Chemother. 2020, 64, e00554-20. [Google Scholar] [CrossRef] [PubMed]
- Gaspar-Cordeiro, A.; Afonso, G.; Amaral, C.; da Silva, S.M.; Pimentel, C. Zap1 is required for Candida glabrata response to fluconazole. FEMS Yeast Res. 2022, 22, foab068. [Google Scholar] [CrossRef]
- Dewhurst-Maridor, G.; Abegg, D.; David, F.P.A.; Rougemont, J.; Scott, C.C.; Adibekian, A.; Riezman, H. The SAGA complex, together with transcription factors and the endocytic protein Rvs167p, coordinates the reprofiling of gene expression in response to changes in sterol composition in Saccharomyces cerevisiae. Mol. Biol. Cell 2017, 28, 2637–2649. [Google Scholar] [CrossRef]
- Rashid, S.; Correia-Mesquita, T.O.; Godoy, P.; Omran, R.P.; Whiteway, M. SAGA complex subunits in Candida albicans differentially regulate filamentation, invasiveness, and biofilm formation. Front. Cell. Infect. Microbiol. 2022, 12, 764711. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.C.; Dent, S.Y.R. Conservation and diversity of the eukaryotic SAGA coactivator complex across kingdoms. Epigenetics Chromatin 2021, 14, 26. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.J.; Chang, Y.L.; Chen, Y.L. Deletion of ADA2 increases antifungal drug susceptibility and virulence in Candida glabrata. Antimicrob. Agents Chemother. 2018, 62, e01924-17. [Google Scholar] [CrossRef]
- Bhakt, P.; Raney, M.; Kaur, R. The SET-domain protein CgSet4 negatively regulates antifungal drug resistance via the ergosterol biosynthesis transcriptional regulator CgUpc2a. J. Biol. Chem. 2022, 298, 102485. [Google Scholar] [CrossRef]
- Baker, K.M.; Hoda, S.; Saha, D.; Gregor, J.B.; Georgescu, L.; Serratore, N.D.; Zhang, Y.; Cheng, L.; Lanman, N.A.; Briggs, S.D. The Set1 histone H3K4 methyltransferase contributes to azole susceptibility in a species-specific manner by differentially altering the expression of drug efflux pumps and the ergosterol gene pathway. Antimicrob. Agents Chemother. 2022, 66, e0225021. [Google Scholar] [CrossRef]
- Li, X.; Cai, Q.; Mei, H.; Zhou, X.; Shen, Y.; Li, D.; Liu, W. The Rpd3/Hda1 family of histone deacetylases regulates azole resistance in Candida albicans. J. Antimicrob. Chemother. 2015, 70, 1993–2003. [Google Scholar] [CrossRef]
- Usher, J.; Haynes, K. Attenuating the emergence of anti-fungal drug resistance by harnessing synthetic lethal interactions in a model organism. PLoS Genet. 2019, 15, e1008259. [Google Scholar] [CrossRef]
- Moirangthem, R.; Kumar, K.; Kaur, R. Two functionally redundant FK506-binding proteins regulate multidrug resistance gene expression and govern azole antifungal resistance. Antimicrob. Agents Chemother. 2021, 65, e02415-20. [Google Scholar] [CrossRef]
- Vu, B.G.; Thomas, G.H.; Moye-Rowley, W.S. Evidence that ergosterol biosynthesis modulates activity of the Pdr1 transcription factor in Candida glabrata. mBio 2019, 10, e00934-19. [Google Scholar] [CrossRef] [PubMed]
- Vu, B.G.; Stamnes, M.A.; Li, Y.; Rogers, P.D.; Moye-Rowley, W.S. The Candida glabrata Upc2A transcription factor is a global regulator of antifungal drug resistance pathways. PLoS Genet. 2021, 17, e1009582. [Google Scholar] [CrossRef]
- Prasad, T.; Chandra, A.; Mukhopadhyay, C.K.; Prasad, R. Unexpected link between iron and drug resistance of Candida spp.: Iron depletion enhances membrane fluidity and drug diffusion, leading to drug-susceptible cells. Antimicrob. Agents Chemother. 2006, 50, 3597–3606. [Google Scholar] [CrossRef]
- Hosogaya, N.; Miyazaki, T.; Nagi, M.; Tanabe, K.; Minematsu, A.; Nagayoshi, Y.; Yamauchi, S.; Nakamura, S.; Imamura, Y.; Izumikawa, K.; et al. The heme-binding protein Dap1 links iron homeostasis to azole resistance via the P450 protein Erg11 in Candida glabrata. FEMS Yeast Res. 2013, 13, 411–421. [Google Scholar] [CrossRef]
- Hameed, S.; Dhamgaye, S.; Singh, A.; Goswami, S.K.; Prasad, R. Calcineurin signaling and membrane lipid homeostasis regulates iron mediated multidrug resistance mechanisms in Candida albicans. PLoS ONE 2011, 6, e18684. [Google Scholar] [CrossRef][Green Version]
- Rossignol, T.; Ding, C.; Guida, A.; d’Enfert, C.; Higgins, D.G.; Butler, G. Correlation between biofilm formation and the hypoxic response in Candida parapsilosis. Eukaryot. Cell 2009, 8, 550–559. [Google Scholar] [CrossRef]
- Synnott, J.M.; Guida, A.; Mulhern-Haughey, S.; Higgins, D.G.; Butler, G. Regulation of the hypoxic response in Candida albicans. Eukaryot. Cell 2010, 9, 1734–1746. [Google Scholar] [CrossRef]
- Hoot, S.J.; Oliver, B.G.; White, T.C. Candida albicans UPC2 is transcriptionally induced in response to antifungal drugs and anaerobicity through Upc2p-dependent and -independent mechanisms. Microbiology 2008, 154, 2748–2756. [Google Scholar] [CrossRef] [PubMed]
- Leber, R.; Fuchsbichler, S.; Klobucníková, V.; Schweighofer, N.; Pitters, E.; Wohlfarter, K.; Lederer, M.; Landl, K.; Ruckenstuhl, C.; Hapala, I.; et al. Molecular mechanism of terbinafine resistance in Saccharomyces cerevisiae. Antimicrob. Agents Chemother. 2003, 47, 3890–3900. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, H.; Nakayama, N.; Arisawa, M.; Aoki, Y. In vitro and in vivo effects of 14alpha-demethylase (ERG11) depletion in Candida glabrata. Antimicrob. Agents Chemother. 2001, 45, 3037–3045. [Google Scholar] [CrossRef]
- Taramino, S.; Valachovic, M.; Oliaro-Bosso, S.; Viola, F.; Teske, B.; Bard, M.; Balliano, G. Interactions of oxidosqualene cyclase (Erg7p) with 3-keto reductase (Erg27p) and other enzymes of sterol biosynthesis in yeast. Biochim. Biophys. Acta 2010, 1801, 156–162. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kaneshiro, E.S.; Johnston, L.Q.; Nkinin, S.W.; Romero, B.I.; Giner, J.L. Sterols of Saccharomyces cerevisiae erg6 knockout mutant expressing the pneumocystis carinii S-Adenosylmethionine:Sterol C-24 Methyltransferase. J. Eukaryot. Microbiol. 2015, 62, 298–306. [Google Scholar] [CrossRef]
- Mukherjee, S.; Xu, W.; Hsu, F.F.; Patel, J.; Huang, J.; Zhang, K. Sterol methyltransferase is required for optimal mitochondrial function and virulence in Leishmania major. Mol. Microbiol. 2019, 111, 65–81. [Google Scholar] [CrossRef]
- Pourshafie, M.; Morand, S.; Virion, A.; Rakotomanga, M.; Dupuy, C.; Loiseau, P.M. Cloning of S-adenosyl-L-methionine:C-24-Delta-sterol-methyltransferase (ERG6) from Leishmania donovani and characterization of mRNAs in wild-type and amphotericin B-resistant promastigotes. Antimicrob. Agents Chemother. 2004, 48, 2409–2414. [Google Scholar] [CrossRef]
- Zavrel, M.; Hoot, S.J.; White, T.C. Comparison of sterol import under aerobic and anaerobic conditions in three fungal species, Candida albicans, Candida glabrata, and Saccharomyces cerevisiae. Eukaryot. Cell 2013, 12, 725–738. [Google Scholar] [CrossRef]
- Prasad, R.; Singh, A. Lipids of Candida albicans and their role in multidrug resistance. Curr. Genet. 2013, 59, 243–250. [Google Scholar] [CrossRef]
- Stieger, B.; Steiger, J.; Locher, K.P. Membrane lipids and transporter function. Biochim. Biophys. Acta Mol. Basis Dis. 2021, 1867, 166079. [Google Scholar] [CrossRef]
- Mukhopadhyay, K.; Prasad, T.; Saini, P.; Pucadyil, T.J.; Chattopadhyay, A.; Prasad, R. Membrane sphingolipid-ergosterol interactions are important determinants of multidrug resistance in Candida albicans. Antimicrob. Agents Chemother. 2004, 48, 1778–1787. [Google Scholar] [CrossRef]
- Pasrija, R.; Panwar, S.L.; Prasad, R. Multidrug transporters CaCdr1p and CaMdr1p of Candida albicans display different lipid specificities: Both ergosterol and sphingolipids are essential for targeting of CaCdr1p to membrane rafts. Antimicrob. Agents Chemother. 2008, 52, 694–704. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, H.; Tanabe, K.; Bard, M.; Hodgson, W.; Wu, S.; Takemori, D.; Aoyama, T.; Kumaraswami, N.S.; Metzler, L.; Takano, Y.; et al. The Candida glabrata putative sterol transporter gene CgAUS1 protects cells against azoles in the presence of serum. J. Antimicrob. Chemother. 2007, 60, 1264–1272. [Google Scholar] [CrossRef] [PubMed]
- Nagi, M.; Tanabe, K.; Ueno, K.; Nakayama, H.; Aoyama, T.; Chibana, H.; Yamagoe, S.; Umeyama, T.; Oura, T.; Ohno, H.; et al. The Candida glabrata sterol scavenging mechanism, mediated by the ATP-binding cassette transporter Aus1p, is regulated by iron limitation. Mol. Microbiol. 2013, 88, 371–381. [Google Scholar] [CrossRef]
- Ansari, S.; Prasad, R. Effect of miconazole on the structure and function of plasma membrane of Candida albicans. FEMS Microbiol. Lett. 1993, 114, 93–98. [Google Scholar] [CrossRef][Green Version]
- Panwar, S.L.; Pasrija, R.; Prasad, R. Membrane homoeostasis and multidrug resistance in yeast. Biosci. Rep. 2008, 28, 217–228. [Google Scholar] [CrossRef]
- Wu, Y.; Wu, M.; Wang, Y.; Chen, Y.; Gao, J.; Ying, C. ERG11 couples oxidative stress adaptation, hyphal elongation and virulence in Candida albicans. FEMS Yeast Res. 2018, 18, foy057. [Google Scholar] [CrossRef]
- Mukhopadhyay, K.; Kohli, A.; Prasad, R. Drug susceptibilities of yeast cells are affected by membrane lipid composition. Antimicrob. Agents Chemother. 2002, 46, 3695–3705. [Google Scholar] [CrossRef]
- Luna-Tapia, A.; Butts, A.; Palmer, G.E. Loss of C-5 Sterol Desaturase Activity in Candida albicans: Azole Resistance or Merely Trailing Growth? Antimicrob. Agents Chemother. 2018, 63, e01337-18. [Google Scholar] [CrossRef]
- Luna-Tapia, A.; Willems, H.M.E.; Parker, J.E.; Tournu, H.; Barker, K.S.; Nishimoto, A.T.; Rogers, P.D.; Kelly, S.L.; Peters, B.M.; Palmer, G.E. Loss of Upc2p-inducible ERG3 transcription is sufficient to confer niche-specific azole resistance without compromising Candida albicans pathogenicity. mBio 2018, 9, e00225-18. [Google Scholar] [CrossRef]
- Rybak, J.M.; Dickens, C.M.; Parker, J.E.; Caudle, K.E.; Manigaba, K.; Whaley, S.G.; Nishimoto, A.T.; Luna-Tapia, A.; Roy, S.; Zhang, Q.; et al. Loss of C-5 sterol desaturase activity results in increased resistance to azole and echinocandin antifungals in a clinical isolate of Candida parapsilosis. Antimicrob. Agents Chemother. 2017, 61, e00651-17. [Google Scholar] [CrossRef]
- O’Meara, T.R.; Veri, A.O.; Polvi, E.J.; Li, X.; Valaei, S.F.; Diezmann, S.; Cowen, L.E. Mapping the Hsp90 genetic network reveals ergosterol biosynthesis and phosphatidylinositol-4-kinase signaling as core circuitry governing cellular stress. PLoS Genet. 2016, 12, e1006142. [Google Scholar] [CrossRef]
- Vale-Silva, L.A.; Coste, A.T.; Ischer, F.; Parker, J.E.; Kelly, S.L.; Pinto, E.; Sanglard, D. Azole resistance by loss of function of the sterol Δ⁵,⁶-desaturase gene (ERG3) in Candida albicans does not necessarily decrease virulence. Antimicrob. Agents Chemother. 2012, 56, 1960–1968. [Google Scholar] [CrossRef]
- Martel, C.M.; Parker, J.E.; Bader, O.; Weig, M.; Gross, U.; Warrilow, A.G.; Rolley, N.; Kelly, D.E.; Kelly, S.L. Identification and characterization of four azole-resistant erg3 mutants of Candida albicans. Antimicrob. Agents Chemother. 2010, 54, 4527–4533. [Google Scholar] [CrossRef]
- Flowers, S.A.; Colón, B.; Whaley, S.G.; Schuler, M.A.; Rogers, P.D. Contribution of clinically derived mutations in ERG11 to azole resistance in Candida albicans. Antimicrob. Agents Chemother. 2015, 59, 450–460. [Google Scholar] [CrossRef]
- Paul, S.; Kannan, I.; Mohanram, K. Extensive ERG11 mutations associated with fluconazole-resistant Candida albicans isolated from HIV-infected patients. Curr. Med. Mycol. 2019, 5, 1–6. [Google Scholar] [CrossRef]
- Paul, S.; Dadwal, R.; Singh, S.; Shaw, D.; Chakrabarti, A.; Rudramurthy, S.M.; Ghosh, A.K. Rapid detection of ERG11 polymorphism associated azole resistance in Candida tropicalis. PLoS ONE 2021, 16, e0245160. [Google Scholar] [CrossRef]
- Rosana, Y.; Yasmon, A.; Lestari, D.C. Overexpression and mutation as a genetic mechanism of fluconazole resistance in Candida albicans isolated from human immunodeficiency virus patients in Indonesia. J. Med. Microbiol. 2015, 64, 1046–1052. [Google Scholar] [CrossRef]
- Morais Vasconcelos Oliveira, J.; Conceição Oliver, J.; Latércia Tranches Dias, A.; Barbosa Padovan, A.C.; Siqueira Caixeta, E.; Caixeta Franco Ariosa, M. Detection of ERG11 overexpression in Candida albicans isolates from environmental sources and clinical isolates treated with inhibitory and subinhibitory concentrations of fluconazole. Mycoses 2021, 64, 220–227. [Google Scholar] [CrossRef]
- Whaley, S.G.; Berkow, E.L.; Rybak, J.M.; Nishimoto, A.T.; Barker, K.S.; Rogers, P.D. Azole antifungal resistance in Candida albicans and emerging non-albicans Candida species. Front. Microbiol. 2017, 7, 2173. [Google Scholar] [CrossRef]
- Rybak, J.M.; Sharma, C.; Doorley, L.A.; Barker, K.S.; Palmer, G.E.; Rogers, P.D. Delineation of the direct contribution of Candida auris ERG11 mutations to clinical triazole resistance. Microbiol. Spectr. 2021, 9, e0158521. [Google Scholar] [CrossRef]
- Lockhart, S.R.; Etienne, K.A.; Vallabhaneni, S.; Farooqi, J.; Chowdhary, A.; Govender, N.P.; Colombo, A.L.; Calvo, B.; Cuomo, C.A.; Desjardins, C.A.; et al. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin. Infect. Dis. 2017, 64, 134–140, Erratum in Clin. Infect. Dis. 2018, 67, 987. [Google Scholar] [CrossRef]
- Chow, N.A.; Muñoz, J.F.; Gade, L.; Berkow, E.L.; Li, X.; Welsh, R.M.; Forsberg, K.; Lockhart, S.R.; Adam, R.; Alanio, A.; et al. Tracing the evolutionary history and global expansion of Candida auris using population genomic analyses. mBio 2020, 11, e03364-19. [Google Scholar] [CrossRef]
- Jacobs, S.E.; Jacobs, J.L.; Dennis, E.K.; Taimur, S.; Rana, M.; Patel, D.; Gitman, M.; Patel, G.; Schaefer, S.; Iyer, K.; et al. Candida auris pan-drug-resistant to four classes of antifungal agents. Antimicrob. Agents Chemother. 2022, 66, e0005322. [Google Scholar] [CrossRef]
- Li, J.; Coste, A.T.; Liechti, M.; Bachmann, D.; Sanglard, D.; Lamoth, F. Novel ERG11 and TAC1b mutations associated with azole resistance in Candida auris. Antimicrob. Agents Chemother. 2023, 65, e02663-20. [Google Scholar] [CrossRef]
- Ben Abid, F.; Salah, H.; Sundararaju, S.; Dalil, L.; Abdelwahab, A.H.; Salameh, S.; Ibrahim, E.B.; Almaslmani, M.A.; Tang, P.; Perez-Lopez, A.; et al. Molecular characterization of Candida auris outbreak isolates in Qatar from patients with COVID-19 reveals the emergence of isolates resistant to three classes of antifungal drugs. Clin. Microbiol. Infect. 2023, 29, 1083.e1–1083.e7. [Google Scholar] [CrossRef]
- Carolus, H.; Pierson, S.; Muñoz, J.F.; Subotić, A.; Cruz, R.B.; Cuomo, C.A.; Van Dijck, P. Genome-wide analysis of experimentally evolved Candida auris reveals multiple novel mechanisms of multidrug resistance. mBio 2021, 12, e03333-20. [Google Scholar] [CrossRef]
- Muñoz, J.F.; Gade, L.; Chow, N.A.; Loparev, V.N.; Juieng, P.; Berkow, E.L.; Farrer, R.A.; Litvintseva, A.P.; Cuomo, C.A. Genomic insights into multidrug-resistance, mating and virulence in Candida auris and related emerging species. Nat. Commun. 2018, 9, 5346. [Google Scholar] [CrossRef]
- Bhattacharya, S.; Holowka, T.; Orner, E.P.; Fries, B.C. Gene duplication associated with increased fluconazole tolerance in Candida auris cells of advanced generational age. Sci. Rep. 2019, 9, 5052. [Google Scholar] [CrossRef]
- Kordalewska, M.; Guerrero, K.D.; Mikulski, T.D.; Elias, T.N.; Garcia-Rubio, R.; Berrio, I.; Gardam, D.; Heath, C.H.; Chowdhary, A.; Govender, N.P. Rare modification in the ergosterol biosynthesis pathway leads to amphotericin B resistance in Candida auris clinical isolates. bioRxiv 2021. bioRxiv:2021.10.22.465535. [Google Scholar] [CrossRef]
- Rybak, J.M.; Barker, K.S.; Muñoz, J.F.; Parker, J.E.; Ahmad, S.; Mokaddas, E.; Abdullah, A.; Elhagracy, R.S.; Kelly, S.L.; Cuomo, C.A.; et al. In vivo emergence of high-level resistance during treatment reveals the first identified mechanism of amphotericin B resistance in Candida auris. Clin. Microbiol. Infect. 2022, 28, 838–843. [Google Scholar] [CrossRef]
- Hull, C.M.; Bader, O.; Parker, J.E.; Weig, M.; Gross, U.; Warrilow, A.G.; Kelly, D.E.; Kelly, S.L. Two clinical isolates of Candida glabrata exhibiting reduced sensitivity to amphotericin B both harbor mutations in ERG2. Antimicrob. Agents Chemother. 2012, 56, 6417–6421. [Google Scholar] [CrossRef]
- Asadzadeh, M.; Alfouzan, W.; Parker, J.E.; Meis, J.F.; Kelly, S.L.; Joseph, L.; Ahmad, S. Molecular characterization and sterol profiles identify nonsynonymous mutations in ERG2 as a major mechanism conferring reduced susceptibility to amphotericin B in Candida kefyr. Microbiol. Spectr. 2023, 11, e0147423. [Google Scholar] [CrossRef]
- Castanheira, M.; Deshpande, L.M.; Davis, A.P.; Carvalhaes, C.G.; Pfaller, M.A. Azole resistance in Candida glabrata clinical isolates from global surveillance is associated with efflux overexpression. J. Glob. Antimicrob. Resist. 2022, 29, 371–377. [Google Scholar] [CrossRef]
- Morio, F.; Pagniez, F.; Besse, M.; Gay-andrieu, F.; Miegeville, M.; Le Pape, P. Deciphering azole resistance mechanisms with a focus on transcription factor-encoding genes TAC1, MRR1 and UPC2 in a set of fluconazole-resistant clinical isolates of Candida albicans. Int. J. Antimicrob. Agents 2013, 42, 410–415. [Google Scholar] [CrossRef]
- Jiang, C.; Ni, Q.; Dong, D.; Zhang, L.; Li, Z.; Tian, Y.; Peng, Y. The Role of UPC2 gene in azole-resistant Candida tropicalis. Mycopathologia 2016, 181, 833–838. [Google Scholar] [CrossRef]
- Fan, F.; Liu, Y.; Liu, Y.; Lv, R.; Sun, W.; Ding, W.; Cai, Y.; Li, W.; Liu, X.; Qu, W. Candida albicans biofilms: Antifungal resistance, immune evasion, and emerging therapeutic strategies. Int. J. Antimicrob. Agents 2022, 60, 106673. [Google Scholar] [CrossRef]
- Derkacz, D.; Krasowska, A. Alterations in the level of ergosterol in Candida albicans’ plasma membrane correspond with changes in virulence and result in triggering diversed inflammatory response. Int. J. Mol. Sci. 2023, 24, 3966. [Google Scholar] [CrossRef]
- Jin, X.; Luan, X.; Xie, F.; Chang, W.; Lou, H. Erg6 acts as a downstream effector of the transcription factor Flo8 to regulate biofilm formation in Candida albicans. Microbiol. Spectr. 2023, 11, e0039323. [Google Scholar] [CrossRef]
- Mukherjee, P.K.; Chandra, J.; Kuhn, D.M.; Ghannoum, M.A. Mechanism of fluconazole resistance in Candida albicans biofilms: Phase-specific role of efflux pumps and membrane sterols. Infect. Immun. 2003, 71, 4333–4340. [Google Scholar] [CrossRef]
- Silva, S.; Rodrigues, C.F.; Araújo, D.; Rodrigues, M.E.; Henriques, M. Candida species biofilms’ antifungal resistance. J. Fungi 2017, 3, 8. [Google Scholar] [CrossRef]
- Borecka-Melkusova, S.; Moran, G.P.; Sullivan, D.J.; Kucharikova, S.; Chorvat, D., Jr.; Bujdakova, H. The expression of genes involved in the ergosterol biosynthesis pathway in Candida albicans and Candida dubliniensis biofilms exposed to fluconazole. Mycoses 2009, 52, 118–128. [Google Scholar] [CrossRef]
- Finkel, J.S.; Mitchell, A.P. Genetic control of Candida albicans biofilm development. Nat. Rev. Microbiol. 2011, 9, 109–118. [Google Scholar] [CrossRef]
- Fernandes, T.; Silva, S.; Henriques, M. Effect of voriconazole on Candida tropicalis biofilms: Relation with ERG genes expression. Mycopathologia 2016, 181, 643–651. [Google Scholar] [CrossRef]
- Ajdidi, A.; Sheehan, G.; Abu Elteen, K.; Kavanagh, K. Assessment of the in vitro and in vivo activity of atorvastatin against Candida albicans. J. Med. Microbiol. 2019, 68, 1497–1506. [Google Scholar] [CrossRef]
- Derkacz, D.; Bernat, P.; Krasowska, A. K143R amino acid substitution in 14-α-demethylase (Erg11p) changes plasma membrane and cell wall structure of Candida albicans. Int. J. Mol. Sci. 2022, 23, 1631. [Google Scholar] [CrossRef]
- Suchodolski, J.; Muraszko, J.; Korba, A.; Bernat, P.; Krasowska, A. Lipid composition and cell surface hydrophobicity of Candida albicans influence the efficacy of fluconazole-gentamicin treatment. Yeast 2020, 37, 117–129. [Google Scholar] [CrossRef]
- Talapko, J.; Juzbašić, M.; Matijević, T.; Pustijanac, E.; Bekić, S.; Kotris, I.; Škrlec, I. Candida albicans—The virulence factors and clinical manifestations of infection. J. Fungi 2021, 7, 79. [Google Scholar] [CrossRef]
- McCourt, P.; Liu, H.Y.; Parker, J.E.; Gallo-Ebert, C.; Donigan, M.; Bata, A.; Giordano, C.; Kelly, S.L.; Nickels, J.T., Jr. Proper sterol distribution is required for Candida albicans hyphal formation and virulence. G3 2016, 6, 3455–3465. [Google Scholar] [CrossRef]
- Morio, F.; Pagniez, F.; Lacroix, C.; Miegeville, M.; Le Pape, P. Amino acid substitutions in the Candida albicans sterol Δ5,6-desaturase (Erg3p) confer azole resistance: Characterization of two novel mutants with impaired virulence. J. Antimicrob. Chemother. 2012, 67, 2131–2138. [Google Scholar] [CrossRef]
- Hirayama, T.; Miyazaki, T.; Sumiyoshi, M.; Ashizawa, N.; Takazono, T.; Yamamoto, K.; Imamura, Y.; Izumikawa, K.; Yanagihara, K.; Kohno, S.; et al. ERG3-encoding sterol C5,6-desaturase in Candida albicans is required for virulence in an enterically infected invasive candidiasis mouse model. Pathogens 2020, 10, 23. [Google Scholar] [CrossRef]
- Nagi, M.; Tanabe, K.; Tanaka, K.; Ueno, K.; Nakayama, H.; Ishikawa, J.; Abe, M.; Yamagoe, S.; Umeyama, T.; Nakamura, S.; et al. Exhibition of antifungal resistance by sterol-auxotrophic strains of Candida glabrata with intact virulence. JAC Antimicrob. Resist. 2022, 4, dlac018. [Google Scholar] [CrossRef]
- Nagi, M.; Tanabe, K.; Nakayama, H.; Yamagoe, S.; Umeyama, T.; Oura, T.; Ohno, H.; Kajiwara, S.; Miyazaki, Y. Serum cholesterol promotes the growth of Candida glabrata in the presence of fluconazole. J. Infect. Chemother. 2013, 19, 138–143. [Google Scholar] [CrossRef]
- Bard, M.; Sturm, A.M.; Pierson, C.A.; Brown, S.; Rogers, K.M.; Nabinger, S.; Eckstein, J.; Barbuch, R.; Lees, N.D.; Howell, S.A.; et al. Sterol uptake in Candida glabrata: Rescue of sterol auxotrophic strains. Diagn. Microbiol. Infect. Dis. 2005, 52, 285–293. [Google Scholar] [CrossRef]
- Hazen, K.C.; Stei, J.; Darracott, C.; Breathnach, A.; May, J.; Howell, S.A. Isolation of cholesterol-dependent Candida glabrata from clinical specimens. Diagn. Microbiol. Infect. Dis. 2005, 52, 35–37. [Google Scholar] [CrossRef]
- Dumitru, R.; Hornby, J.M.; Nickerson, K.W. Defined anaerobic growth medium for studying Candida albicans basic biology and resistance to eight antifungal drugs. Antimicrob. Agents Chemother. 2004, 48, 2350–2354. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Liao, M.; Zhu, C.; Hu, Y.; Tong, T.; Peng, X.; Li, M.; Feng, M.; Cheng, L.; Ren, B.; et al. ERG3 and ERG11 genes are critical for the pathogenesis of Candida albicans during the oral mucosal infection. Int. J. Oral Sci. 2018, 10, 9. [Google Scholar] [CrossRef]
- Zangl, I.; Beyer, R.; Gattesco, A.; Labuda, R.; Pap, I.J.; Strauss, J.; Schüller, C. Limosilactobacillus fermentum limits Candida glabrata growth by ergosterol depletion. Microbiol. Spectr. 2023, 11, e0332622. [Google Scholar] [CrossRef]
- Kan, V.L.; Geber, A.; Bennett, J.E. Enhanced oxidative killing of azole-resistant Candida glabrata strains with ERG11 deletion. Antimicrob. Agents Chemother. 1996, 40, 1717–1719. [Google Scholar] [CrossRef]
- Elias, D.; Tóth Hervay, N.; Bujdos, M.; Gbelska, Y. Essential role of CgErg6p in maintaining oxidative stress tolerance and iron homeostasis in Candida glabrata. J. Fungi 2023, 9, 579. [Google Scholar] [CrossRef]
- Netea, M.G.; Joosten, L.A.; van der Meer, J.W.; Kullberg, B.J.; van de Veerdonk, F.L. Immune defence against Candida fungal infections. Nat. Rev. Immunol. 2015, 15, 630–642. [Google Scholar] [CrossRef]
- Arce Miranda, J.E.; Baronetti, J.L.; Sotomayor, C.E.; Paraje, M.G. Oxidative and nitrosative stress responses during macrophage-Candida albicans biofilm interaction. Med. Mycol. 2019, 57, 101–113. [Google Scholar] [CrossRef] [PubMed]
- Mailänder-Sánchez, D.; Braunsdorf, C.; Grumaz, C.; Müller, C.; Lorenz, S.; Stevens, P.; Wagener, J.; Hebecker, B.; Hube, B.; Bracher, F.; et al. Antifungal defense of probiotic Lactobacillus rhamnosus GG is mediated by blocking adhesion and nutrient depletion. PLoS ONE 2017, 12, e0184438. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, V.M.; Santos, S.S.; Silva, C.R.; Jorge, A.O.; Leão, M.V. Lactobacillus is able to alter the virulence and the sensitivity profile of Candida albicans. J. Appl. Microbiol. 2016, 121, 1737–1744. [Google Scholar] [CrossRef]
- Sobel, J.D. Vulvovaginal candidosis. Lancet 2007, 369, 1961–1971. [Google Scholar] [CrossRef] [PubMed]
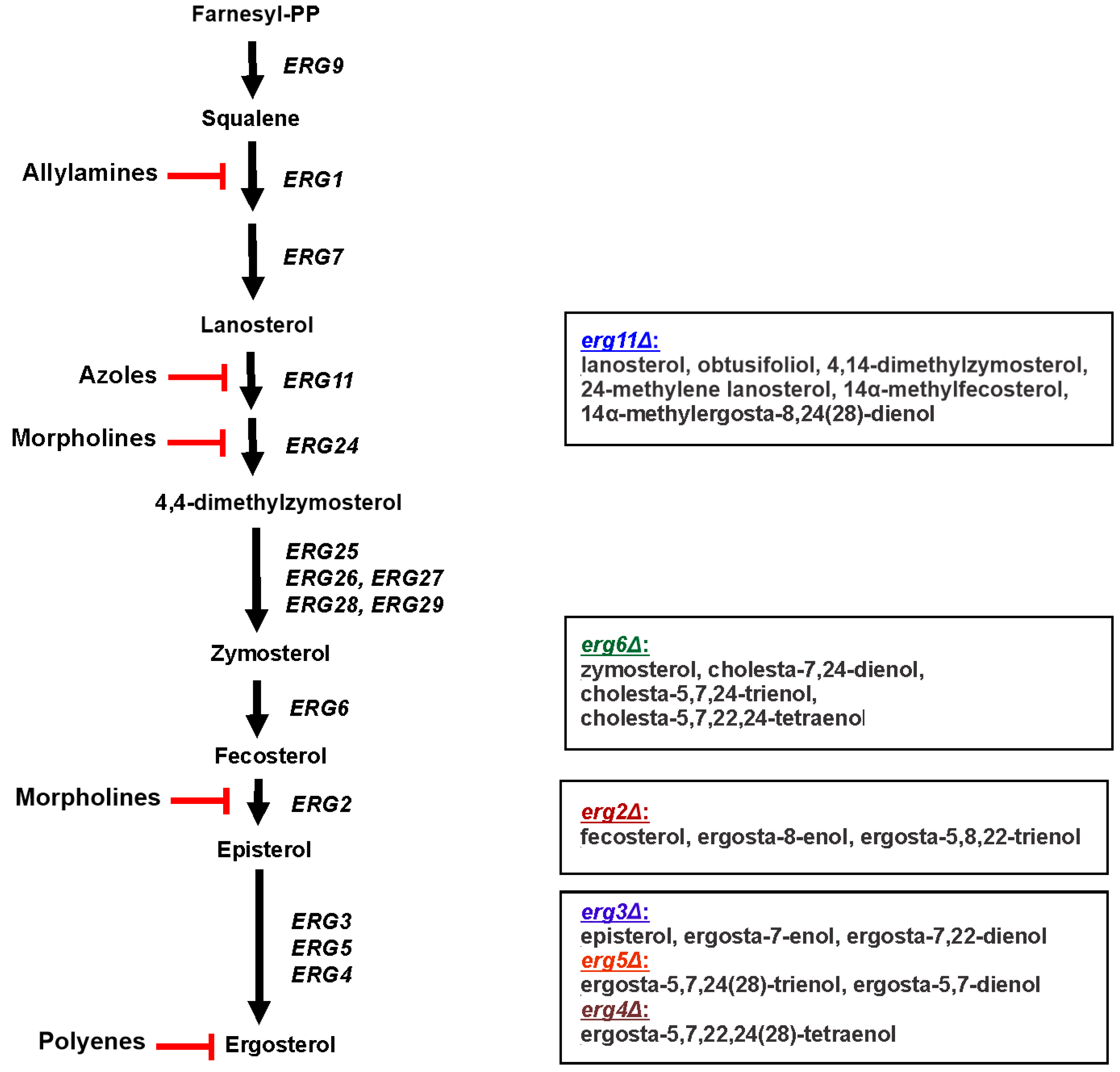

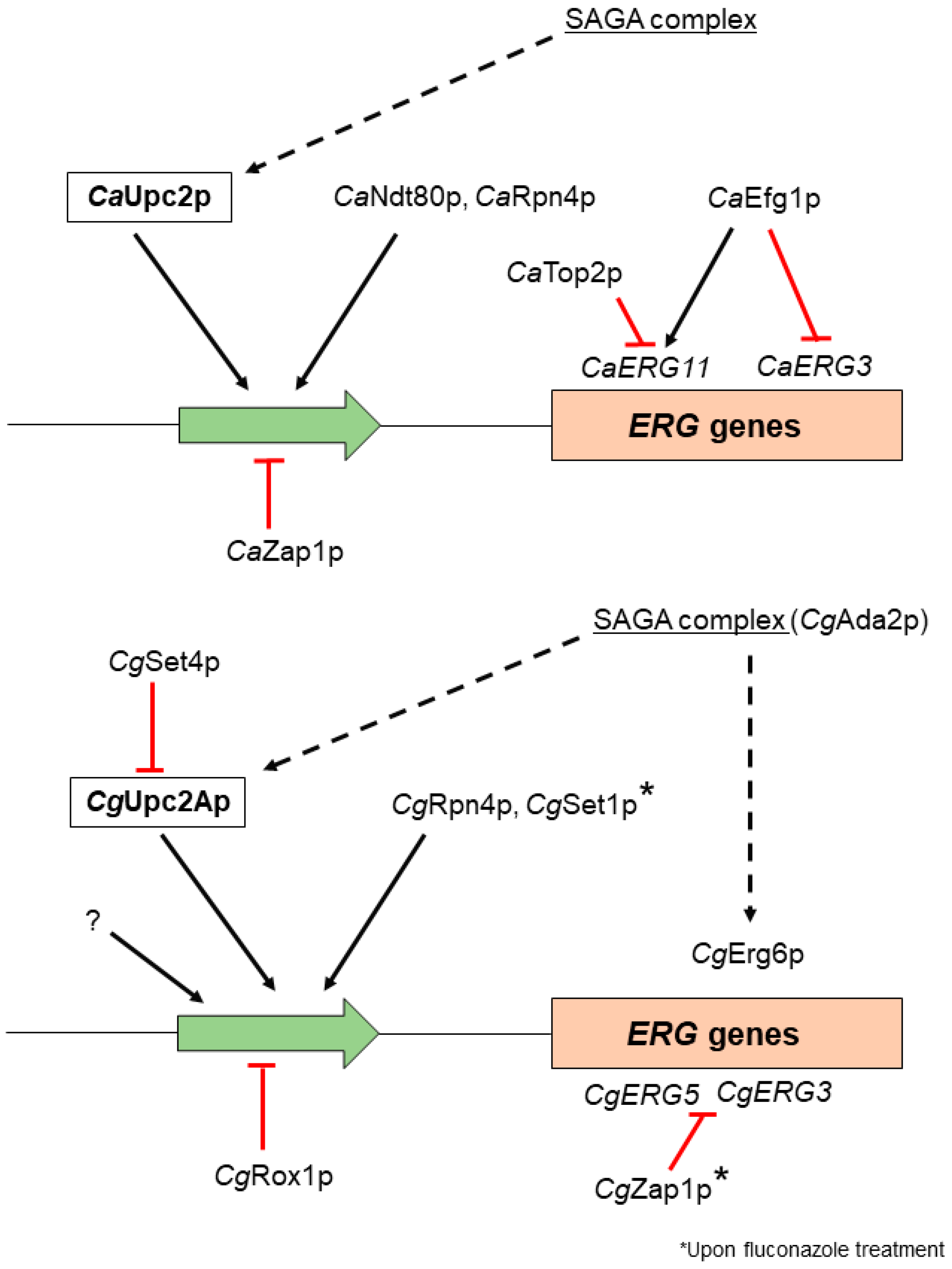
| Gene | C. albicans Gene ID | C. glabrata Gene ID | C. parapsilosis Gene ID | Predicted/Confirmed Function |
|---|---|---|---|---|
| ERG9 | C2_08610W_A [37] | CAGL0M07095g [38] | CPAR2_406760 * | squalene synthase |
| ERG1 | C1_08590C_A [39] | CAGL0D05940g [40] | CPAR2_210480 * | squalene epoxidase |
| ERG7 | C2_02460W_A [37] | CAGL0J10824g [41] | CPAR2_301800 * | lanosterol synthase |
| ERG11 | C5_00660C_A [32] | CAGL0E04334g [29] | CPAR2_303740 [42] | lanosterol 14α-demethylase |
| ERG24 | C2_09400C_A [43] | CAGL0I02970g * | CPAR2_405900 * | C-14 sterol reductase |
| ERG25 | CR_02370W_A [44] | CAGL0K04477g [41] | CPAR2_801410 * | C-4 methyl sterol oxidase |
| ERG26 | C4_06270C_A [45] | CAGL0G00594g [41] | CPAR2_302110 * | C-3 sterol dehydrogenase |
| ERG27 | CR_01140C_A [46] | CAGL0M11506g [41] | CPAR2_801560 * | 3-keto sterol reductase |
| ERG28 | C2_01090C_A * | CAGL0J02684g * | CPAR2_213750 * | scaffold activity |
| ERG6 | C3_02150C_A [30] | CAGL0H04653g [33] | CPAR2_405010 * | C-24 sterol methyltransferase |
| ERG2 | C1_00800C_A [47] | CAGL0L10714g [48] | CPAR2_201490 * | C-8 sterol isomerase |
| ERG3 | C1_04770C_A [31] | CAGL0F01793g [29] | CPAR2_105550 [49] | C-5 sterol desaturase |
| ERG5 | C7_02840C_A [36] | CAGL0M07656g * | CPAR2_703970 * | C-22 sterol desaturase |
| ERG4 | C3_00760W_A [50] | CAGL0A00429g * | CPAR2_502980 * | C-24 sterol reductase |
| Candida Species | Gene Involved in Ergosterol Biosynthesis | Gene Alteration | Alteration of Function | Reference |
|---|---|---|---|---|
| C. albicans | CaERG11 | deletion | Impaired biofilm formation/elevated CSH */impairment of filamentation Impairment of oral candidiasis Increased susceptibility to oxidative stress Inability to grow in the presence of L. fermentum | [116,147,166,167] |
| CaERG11 | overexpression | Elevated CSH * | [156] | |
| CaERG24 | deletion | Inability to form germ tubes | [43,47] | |
| CaERG6 | overexpression | Restoration of biofilm formation and virulence in G. mellonella | [148] | |
| CaERG3 | deletion | Impairment of filamentation and oral candidiasis | [27,34,55,166] | |
| CaERG3 | substitution mutation (L193R) or deletion mutation (Δ366–378) | Compromised virulence | [159] | |
| CaERG1 | deletion | Impairment of filamentation | [39] | |
| CaERG2 | deletion | Impairment of filamentation | [47] | |
| C. glabrata | CgERG11 | deletion | Increased susceptibility to oxidative stress and neutrophils | [168] |
| CgERG6 | deletion | Increased susceptibility to oxidative stress | [169] | |
| CgAUS1 | deletion | Compromised virulence | [113] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eliaš, D.; Tóth Hervay, N.; Gbelská, Y. Ergosterol Biosynthesis and Regulation Impact the Antifungal Resistance and Virulence of Candida spp. Stresses 2024, 4, 641-662. https://doi.org/10.3390/stresses4040041
Eliaš D, Tóth Hervay N, Gbelská Y. Ergosterol Biosynthesis and Regulation Impact the Antifungal Resistance and Virulence of Candida spp. Stresses. 2024; 4(4):641-662. https://doi.org/10.3390/stresses4040041
Chicago/Turabian StyleEliaš, Daniel, Nora Tóth Hervay, and Yvetta Gbelská. 2024. "Ergosterol Biosynthesis and Regulation Impact the Antifungal Resistance and Virulence of Candida spp." Stresses 4, no. 4: 641-662. https://doi.org/10.3390/stresses4040041
APA StyleEliaš, D., Tóth Hervay, N., & Gbelská, Y. (2024). Ergosterol Biosynthesis and Regulation Impact the Antifungal Resistance and Virulence of Candida spp. Stresses, 4(4), 641-662. https://doi.org/10.3390/stresses4040041






