Abstract
We performed an enzymatic screening of synthetic peptides based on β-amyloid precursor protein substrates. The template peptide sequence was a decapeptide derived from our previous screening study, which determined several effective unnatural amino acids. In this study, new libraries containing some unnatural amino acid compounds were prepared in the solid phase and digested with the β-site amyloid precursor protein-cleaving enzyme. The reaction mixture was analyzed using high-performance liquid chromatography combined with mass spectrometry. The peptides that showed a higher cleavage than the template sequence were determined and reported.
1. Introduction
Screening substrate peptides to find superior cleavage sequences is effective for designing substrate-based protease inhibitors [1,2,3]. We previously reported the screening of the substrate peptides of the β-site amyloid precursor protein-cleaving enzyme 1 (BACE1) [4]. BACE1 has been a target enzyme for developing drugs against Alzheimer’s disease because it triggers amyloid β-peptide (Aβ) production by cleaving amyloid precursor protein (APP) at the N-terminus of the Aβ domain [5,6,7,8,9,10]. The oligomerization and fibrillization of Aβ are considered to play important roles in the pathogenesis of Alzheimer’s disease. As Aβs are quite aggregative, toxic, and known to cause Alzheimer’s-type dementia, BACE1 has been targeted to develop drugs against Alzheimer’s disease. Many peptidic inhibitors against BACE1 are prepared based on substrate sequences [7,8,9,10,11,12,13,14]. In a previous screening study, we replaced the amino acids in Swedish (SW) mutant APP-based dodecapeptides and digested them to find superior cleavage sequences [4]. In the SW peptide, the amino acids at the P4 to P1′ positions were independently replaced by several nonproteinogenic amino acid compounds, and the substrates were digested by recombinant BACE1. At the P2 to P1′ positions, new amino acids suitable for drug design were determined. In addition, combining selected amino acids showed enhanced efficiency against BACE1 (Figure 1). However, no significant unnatural amino acid was identified at the P4 and P3 positions.

Figure 1.
(A) The sequences of the Swedish (SW) mutant amyloid precursor protein (APP) and our previous study. (B) The structures and abbreviations of amino acids in the sequence.
Considering the design of potent BACE1 inhibitors, discovering new P3 and P4 amino acids is interesting because BACE1 inhibitors are sometimes prepared as pentapeptides incorporating the P4 to P1′ positions [11,12,13,14]. In this study, we prepared new peptidic compounds as decapeptides and digested them with BACE1 to find new effective amino acids at the P3 and P4 positions.
2. Materials and Methods
2.1. General Methods
All reagents were purchased and used as received. Fmoc-protected unnatural amino acids were purchased from Watanabe Chemical Industries (Hiroshima, Japan). Human recombinant BACE1 protein was purchased from R & D Systems (Minneapolis, MN, USA, 931-AS-050). High-performance liquid chromatography (HPLC) was performed using a JASCO PU-2028 Plus pump system (JASCO, Tokyo, Japan). Preparative HPLC was performed with a Cosmosil 5C18-AR-II column (20 × 250 mm, Nacalai Tesque, Kyoto, Japan) using a linear gradient of 0.05% trifluoroacetic acid (TFA) in CH3CN and 0.05% aqueous TFA at a flow rate of 5.0 mL min−1, and detection was at 220 nm. Analytical HPLC was performed with a JASCO PU-2028 Plus pump system and a Cosmosil 5C18-AR-II column (4.6 × 150 mm, Nacalai Tesque) using a linear gradient of 0.05% TFA in CH3CN and 0.05% TFA in H2O (1.0 mL min−1, 220 nm). Peptides were characterized using the LCMS-2020 (SHIMADZU, Kyoto, Japan) or matrix-assisted laser desorption/ionization time-of-flight mass spectrometry using an AXIMA-CFR plus (SHIMADZU, Kyoto, Japan) with α-cyano-4-hydroxycinnamic acid as the matrix.
2.2. Peptide Synthesis
Peptidic compounds were prepared in the solid phase using a standard Fmoc-based method. Peptide chain elongation was performed on 2-chlorotrityl chloride resin using Fmoc-amino acid, DIPCDI, HOBt∙H2O, and DIPEA. The coupling reactions were conducted in DMF for 2 h, and the deprotection of Fmoc was performed with 20% piperidine in DMF. At the P3 position, the resin was split into eight, and the corresponding amino acids for the library were introduced for each part of the resin. Peptide chain elongation was independently performed afterward. Total cleavage of the peptide from the resin was performed with TFA in the presence of thioanisole and H2O. Each peptide was purified through HPLC and lyophilized to give puffy powders. A similar procedure was conducted at the P4 position to prepare P4-modified peptidic compounds. The template sequence and P3, P4-modified substrate were also synthesized in a similar protocol. The characterization of the products is shown in Tables S1–S3.
2.3. Peptide Library Preparation
Each obtained peptide was independently adjusted to 250 μM solutions in H2O:DMSO (1:1). The library solution was prepared by mixing the single-peptide solutions at the same volumes as the peptide libraries.
2.4. Digestion of the Peptide Libraries with BACE1
Digestion of the libraries with recombinant BACE1 was performed based on a slightly modified procedure in previous research [4,11]. The enzyme assay was conducted by mixing the components to set the final concentrations at 20 ng/μL (280 nM) BACE1 and a 25 μM substrate peptide mix in 0.1 M sodium acetate buffer (pH = 4.0), including 7% DMSO. After incubation for 3 h at 37 °C, the reaction was stopped by adding 7 M guanidine hydrochloride solution. The solution mixture was analyzed through HPLC detection at 220 nm. The peptides were assigned using mass spectrometry. Compound processing was evaluated based on the HPLC peak areas with a percentage decrease in the substrate cleaved by the enzyme.
3. Results
We previously performed and reported a peptide library screening study to find superior cleavage sequences against BACE1 [4]. In a previous study, favorable unnatural amino acids were determined at the P2 to P1′ positions in the SW mutant. However, effective amino acids were not identified at the P3 and P4 positions. Thus, it is of interest to find the effective amino acids at the P3 and P4 positions because several BACE1 inhibitors are prepared in the P4 to P1′ positions [11,12,13,14]. In this study, new libraries were prepared based on the template sequence H-Ser-Glu-Ile-Thi-Thi↓Nva-Ala-Glu-Phe-Arg-OH (1), which was confirmed in our previous study as a superior substrate for BACE1 [4]. Figure 2 shows the new amino acids in this study that incorporate several types of side chains, such as linear or cyclic structures, as well as the main chain of a cyclic structure. Substrate peptides were prepared using standard solid-phase peptide synthesis, and the new amino acids (Figure 2) were introduced at the P3 or P4 positions to produce new libraries. Library peptides were incubated for 3 h with recombinant BACE1 and analyzed with HPLC.
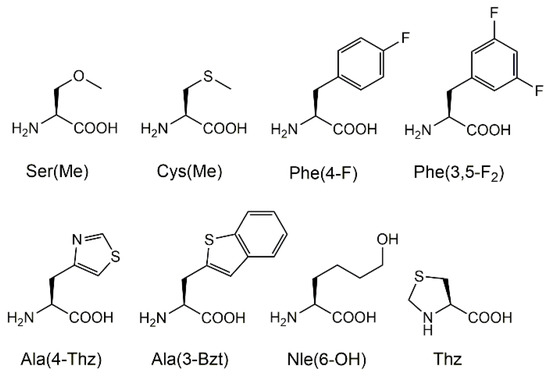
Figure 2.
The structures and abbreviations of the amino acids used in this study. These amino acids are incorporated into the P3 or P4 positions of the template sequence.
3.1. Digestion of P3 Library
Synthetic P3-modified peptides, H-Ser-Glu-Xaa3-Thi-Thi↓Nva-Ala-Glu-Phe-Arg-OH, were prepared as a library and digested with recombinant BACE1. For comparison, the original template 1 (Xaa3 = Ile) was added to the library. The reaction mixture was analyzed using HPLC, and the peaks were characterized using mass spectrometry. The HPLC profiles before and after the incubation of the P3 peptides are shown in Figure 3. Compared with the 0 h incubation, all the peptides showed peak area reductions of approximately 12.5–39.4% after the 3 h incubation. The Ser(Me)-containing sequence (P3-Ser(Me)) showed the maximum preference against BACE1. Furthermore, the cleaved substrate fragments, H-Ser-Glu-Xaa3-Thi-Thi-OH and H-Nva-Ala-Glu-Phe-Arg-OH, were observed in the elution profile after incubation. The N-terminal peptide of cleaved P3-Ser(Me) was assigned through mass analysis.
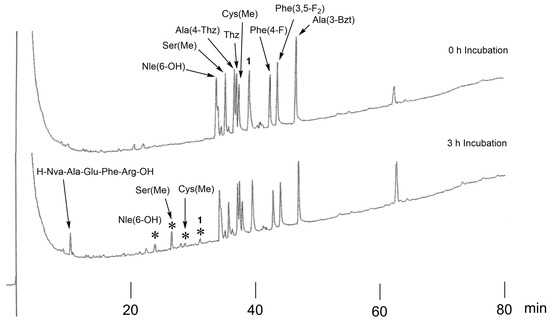
Figure 3.
The HPLC profiles of P3-modified peptides (H-Ser-Glu-Xaa3-Thi-Thi↓Nva-Ala-Glu-Phe-Arg-OH) before and after incubation. Asterisks (*) indicate the N-terminal region of the cleaved peptide, H-Ser-Glu-Xaa3-Thi-Thi-OH (the Xaa3 residue is shown beside *).
3.2. Digestion of P4 Library
The synthetic library of P4-modified compounds H-Ser-Xaa4-Ile-Thi-Thi↓Nva-Ala-Glu-Phe-Arg-OH, where Xaa4 is replaced by the unnatural amino acids listed in Figure 2, was digested with BACE1 in the presence of peptide 1 (Xaa4 = Glu). The reaction mixture was incubated for 3 h at 37 °C and analyzed using HPLC. The HPLC results before and after incubation are shown in Figure 4. The substrate containing Thz residue (P4-Thz) showed remarkable peak reduction (−50.6%) in HPLC. Moreover, the corresponding cleaved fragments of the N- and C-terminal regions were clearly observed. Sequences of other amino acids showed no notable peak reductions in HPLC.
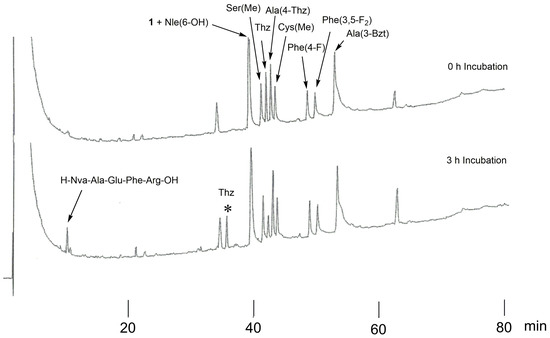
Figure 4.
The HPLC profiles of P4-modified peptides (H-Ser-Xaa4-Ile-Thi-Thi↓Nva-Ala-Glu-Phe-Arg-OH) before and after incubation. The asterisk (*) indicates the N-terminal region of the cleaved peptide, H-Ser-Thz-Ile-Thi-Thi-OH.
3.3. Digestion of P3, P4-Modified Peptide
Based on our results, we prepared a new P3, P4-modified substrate, H-Ser-Thz-Ser(Me)-Thi-Thi-Nva-Ala-Glu-Phe-Arg-OH (2), using solid-phase peptide synthesis and digested it with BACE1. For comparison, original template 1, P3-Ser(Me), and P4-Thz were mixed with 2 in a buffer and incubated. The elution profiles of before and after incubation are shown in Figure 5, and the results in the peak areas are summarized in Table 1. After incubation, the P4-Thz sequence showed a remarkable peak reduction (Figure 5), and the peak areas of peptides 1 and 2 were moderately reduced. In contrast, the P3-Ser(Me) substrate showed an increase in peak area, although its cleaved fragment appeared. MS analysis proved that the P3-Ser(Me) peak overlapped with the cleaved N-terminal fragment of the P4-Thz substrate (see Figure S1).
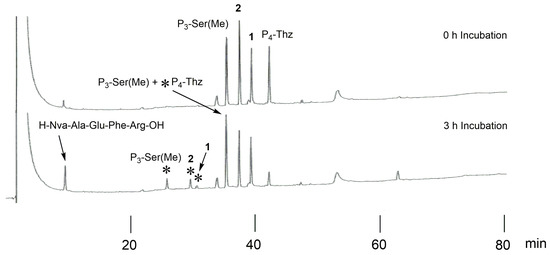
Figure 5.
HPLC profiles of 1, P3-Ser(Me), P4-Thz, and 2 before and after incubation. Asterisks (*) indicate the N-terminal region of the cleaved peptide (the corresponding sequences are shown beside *).

Table 1.
The sequences of the peptides and their peak area reductions in HPLC.
To estimate the peak reduction in P3-Ser(Me), the peptides were independently compared with the original compound 1. P3-Ser(Me), P4-Thz, and the P3, P4-modified peptide 2 were incubated with BACE1 in the presence of peptide 1. The reaction mixtures were adjusted for the HPLC analysis, and the obtained results are shown in Figure 6. In all cases, the peptides showed peak reductions in the HPLC along with corresponding cleaved fragments. The cleavage of the P4-Thz peptide was the highest, with −76.4% of peak reduction after incubation, whereas only −34.2% of peptide 1 decreased in the peak area under the same conditions. P3-Ser(Me) and peptide 2 showed −53.5% and −46.9% reductions, respectively. The modified peptides showed better cleavage in every case than peptide 1. The order of peptide processing with the enzyme was estimated as P4-Thz > P3-Ser(Me) > peptide 2 > peptide 1.
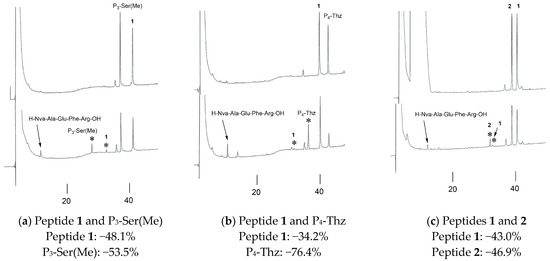
Figure 6.
The HPLC profiles of the 0 h (above) and 3 h (below) incubations with BACE1 in the presence of peptide 1. The peak reduction percentages of (a) P3-Ser(Me), (b) P4-Thz, and (c) peptide 2. Asterisks (*) indicate the N-terminal region of the cleaved peptide.
4. Discussion
Enzyme inhibitors are sometimes designed from their substrates. Several synthetic peptides based on substrate structures, which showed inhibitory activity against BACE1, have been reported [11,12,13,14]. BACE1 is known to cleave APP at the N-terminus, producing Aβs. As Aβs are quite aggregative and cause Alzheimer’s-type dementia, BACE1 has been a target enzyme to develop drugs against Alzheimer’s disease. Screening a library of substrates to find superior cleavage sequences is quite useful for future drug development [15,16,17]. The digestion of the P3 library suggested that the Ser(Me) residue is a candidate for future drug design. The P4 library assay proved that Thz is a suitable residue at the P4 position, as it showed a remarkable increase in cleaving efficiency. However, P3 and P4 residues were replaced with Ser(Me) and Thz, and substrate recognition by BACE1 decreased, probably due to the steric effect of each amino acid in the active site of BACE1. The substrate specificity against BACE1 was the highest at the P4-Thz residue in our template peptide.
5. Conclusions
Using the peptidic compounds, we performed a screening study to find new amino acids for the future design of substrate-based BACE1 inhibitors. The compound processing was evaluated with peak areas in HPLC. At the P4 position, the new amino acid, Thz, was confirmed. The screening of the P3 position suggested that the Ser(Me) residue with a linear side-chain showed superior efficiency against BACE1. Although P3-Ser(Me) and P4-Thz independently showed superior cleavage efficiency, their combination showed an inferior efficiency. Since several amino acids at P3 exhibited enhanced efficiency, testing different amino acids alongside P4-Thz remains our future research plan. In addition, theoretical calculations, such as docking studies, are also of interest because they will help understand the decrease in activity when combining the selected residues for P3 and P4. In addition to drug design, superior cleavage sequences are useful for establishing a fast screening system [17]. In this study, several improved substrates for BACE1 were obtained. Further investigations are currently underway to find improved sequences.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/compounds3020026/s1, Table S1: Obtained data of P3 library; Table S2: Obtained data of P4 library; Table S3: Obtained data for Figure 5; Figure S1: Mass spectra of overlapping peaks in HPLC, including P3-Ser(Me) and cleaved N-terminal region of P4-Thz.
Author Contributions
T.K. conceived, designed, and supervised the experiments; T.K. synthesized the P3 and P4 library peptides; R.Y. and N.T. performed P3 library assay; M.M. and R.F. performed P4 library assay, R.Y. prepared P3, P4-modified substrate; A.A. performed P3, P4-modified substrate enzymatic assay and assigned the fragments of the cleaved peptides. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Acknowledgments
We thank Soichiro Wada for his technical support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Tomasselli, A.G.; Oahwash, I.; Emmons, T.L.; Lu, Y.; Leone, J.W.; Lull, J.M.; Fok, K.F.; Bannow, C.A.; Smith, C.W.; Bienkowski, M.J.; et al. Employing a superior BACE1 cleavage sequence to probe cellular APP processing. J. Neurochem. 2003, 84, 1006–1017. [Google Scholar] [CrossRef] [PubMed]
- Grüninger-Leitch, F.; Schlatter, D.; Küng, E.; Nelböck, P.; Döbeli, H. Substrate and inhibitor profile of BACE (beta-secretase) and comparison with other mammalian aspartic proteases. J. Biol. Chem. 2002, 277, 4687–4693. [Google Scholar] [CrossRef] [PubMed]
- Turner, R.T., III; Koelsch, G.; Hong, L.; Castanheira, P.; Ghosh, A.; Tang, J. Subsite specificity of memapsin 2 (β-secretase): Implications for inhibitor design. Biochemistry 2001, 40, 10001–10006. [Google Scholar] [CrossRef] [PubMed]
- Kakizawa, T.; Sanjoh, A.; Kobayashi, A.; Hattori, Y.; Teruya, K.; Akaji, K. Evaluation of superior BACE1 cleavage sequences containing unnatural amino acids. Bioorg. Med. Chem. 2011, 19, 2785–2789. [Google Scholar] [CrossRef] [PubMed]
- Selkoe, D.J. Translating cell biology into therapeutic advances in Alzheimer’s disease. Nature 1999, 399, A23–A31. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.; Lieberburg, I. Cellular mechanisms of β-amyloid production and secretion. Proc. Natl. Acad. Sci. USA 1999, 96, 11049–11053. [Google Scholar] [CrossRef] [PubMed]
- Stachel, S.J. Progress toward the development of a viable BACE-1 inhibitor. Drug Dev. Res. 2009, 70, 101–110. [Google Scholar] [CrossRef]
- Ziora, Z.; Kimura, T.; Kiso, Y. Small-sized BACE1 inhibitors. Drugs Future 2006, 31, 53–63. [Google Scholar] [CrossRef]
- Hills, I.D.; Vacca, J.P. Progress toward a practical BACE inhibitor. Curr. Opin. Drug Discov. Dev. 2007, 10, 383–391. [Google Scholar]
- Fisha, P.V.; Steadmana, D.; Baylea, E.D.; Whiting, P. New approaches for the treatment of Alzheimer’s disease. Bioorg. Med. Chem. Lett. 2019, 29, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Kakizawa, T.; Hidaka, K.; Hamada, D.; Yamaguchi, R.; Uemura, T.; Kitamura, H.; Tagad, H.D.; Hamada, T.; Ziora, Z.; Hamada, Y.; et al. Tetrapeptides, as small-sized peptidic inhibitors; synthesis and their inhibitory activity against BACE1. J. Pept. Sci. 2010, 16, 257–262. [Google Scholar] [CrossRef] [PubMed]
- Kimura, T.; Shuto, D.; Hamada, Y.; Igawa, N.; Kasai, S.; Liu, P.; Hidaka, K.; Hamada, T.; Hayashi, Y.; Kiso, Y. Design and synthesis of highly active Alzheimer’s β-secretase (BACE1) inhibitors, KMI-420 and KMI-429, with enhanced chemical stability. Bioorg. Med. Chem. Lett. 2005, 15, 211–215. [Google Scholar] [CrossRef] [PubMed]
- Kimura, T.; Shuto, D.; Kasai, S.; Liu, P.; Hidaka, K.; Hamada, T.; Hayashi, Y.; Hattori, C.; Asai, M.; Kitazume, S.; et al. KMI-358 andKMI-370, highly potent and small-sized BACE1 inhibitors containing phenylnorstatine. Bioorg. Med. Chem. Lett. 2004, 14, 1527–1531. [Google Scholar] [CrossRef] [PubMed]
- Shuto, D.; Kasai, S.; Kimura, T.; Liu, P.; Hidaka, K.; Hamada, T.; Shibakawa, S.; Hayashi, Y.; Hattori, C.; Szabo, B.; et al. KMI-008, a novel β-secretase inhibitor containing a hydroxymethylcarbonyl isostere asa transition-state mimic: Design and synthesis of substrate-based octapeptides. Bioorg. Med. Chem. Lett. 2003, 13, 4273–4276. [Google Scholar] [CrossRef] [PubMed]
- Rut, W.; Groborz, K.; Zhang, L.; Sun, X.; Zmudzinski, M.; Pawlik, B.; Wang, X.; Jochmans, D.; Neyts, J.; Młynarski, W.; et al. SARS-CoV-2 Mpro inhibitors and activity-based probes for patient-sample imaging. Nat. Chem. Biol. 2021, 17, 222–228. [Google Scholar] [CrossRef] [PubMed]
- Uddin, M.J.; Akhter, H.; Chowdhury, U.; Mawah, J.; Karim, S.T.; Jomel, M.; Islam, M.S.; Islam, M.R.; Onin, L.A.B.; Ali, M.A.; et al. Large scale peptide screening against main protease of SARS CoV-2. J. Comput. Chem. 2023, 44, 887–901. [Google Scholar] [CrossRef] [PubMed]
- Meng, J.; Lai, M.-T.; Munshi, V.; Grobler, J.; McCauley, J.; Zuck, P.; Johnson, E.N.; Uebele, V.N.; Hermes, J.D.; Adam, G.C. Screening of HIV-1 protease using a combination of an ultra-high-throughput fluorescent-based assay and RapidFire mass spectrometry. J. Biomol. Screen. 2015, 20, 606–615. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).