Simple Summary
Our cellular DNA is packaged tightly into a structure called chromatin, where it is wrapped around specific proteins called histones. Histones can undergo modifications which alter the structure of chromatin and allow processes that promote cell growth and survival. One such modification of the histones, called acetylation, is controlled by a class of enzymes called histone deacetylases (HDACs). Altered levels of HDACs have has been implicated in the development of a variety of cancer types, but also HDACs have been suggested to play a role in the response of cancers to radiotherapy treatment. Therefore, inhibitors of HDAC enzymes have been investigated for their anti-cancer properties in combination with radiotherapy. In this review, we provide an up-to-date summary on HDACs and their roles in human cells, along with evidence suggesting that HDAC inhibitors can effectively increase the sensitivity of cancers to radiotherapy.
Abstract
In mammalian cells, genomic DNA is packaged with histone proteins and condensed into chromatin. To gain access to the DNA, chromatin remodelling is required that is enhanced through histone post-translational modifications, which subsequently stimulate processes including DNA repair and transcription. Histone acetylation is one of the most well understood modifications and is controlled by histone acetyltransferases (HATs) and histone deacetylases (HDACs). These enzymes play critical roles in normal cellular functioning, and the dysregulation of HDAC expression in particular has been linked with the development of a number of different cancer types. Conversely, tumour cell killing following radiotherapy is triggered through DNA damage and HDACs can help co-ordinate the cellular DNA damage response which promotes radioresistance. Consequently, HDAC inhibitors have been investigated as potential radiosensitizers in vitro and in vivo to improve the efficacy or radiotherapy in specific tumour types. In this review, we provide an up-to-date summary of HDACs and their cellular functions, including in DNA damage repair. We also review evidence demonstrating that HDAC inhibitors can effectively enhance tumour radiosensitisation, and which therefore show potential for translation into the clinic for cancer patient benefit.
1. Introduction
In eukaryotic cells, genomic DNA is packaged into condensed chromatin. Nucleosomes are the basic units of chromatin and are formed from ~145–147 base pairs of DNA wrapped around a histone octamer consisting of the core histone proteins H2A, H2B, H3 and H4 [1]. The nucleosomes are linked together by short (~20–90 base pair) segments of linker DNA to form the beads-on-a-string primary chromatin structure (euchromatin) [2], which folds into a higher-order structure of a 30-nanometre fibre (heterochromatin) [3]. This then undergoes supercoiling during cell division to form the metaphase chromosomes seen during mitosis and meiosis [4]. Pioneering research carried out in the 1960s identified that histone subunits can carry post-translational modifications (PTMs) [5], which can regulate and remodel the chromatin structure. Since then, several different types of PTMs have been identified, including histone acetylation, methylation, phosphorylation, SUMOylation and ubiquitylation. These PTMs occur on the histone N-terminal tails that protrude outward from the octamer structure [6] and are highly conserved in eukaryotes. The PTMs work alone, but also through the recruitment of ATP-dependent chromatin remodelling complexes, to influence the structure of chromatin [7]. Local changes in chromatin structure allow the cell to perform DNA-dependent processes, including regulating gene expression through transcription and stimulating DNA damage repair.
Histone acetylation is one of the most well understood histone PTMs. This modification is strictly regulated by both histone acetyltransferases (HATs) and histone deacetylases (HDACs). The HATs are divided into two main families based on sequence homology and shared functional roles. The Gcn5-related-acetyltransferase (GNAT) family of HATs acetylate the lysines of histones H2B, H3 and H4. They are distinguished from other HATs by the presence of a bromodomain, and members of this family include GCN5, PCAF, Hat1, and ATF-2. The MYST family of HATs is characterised by zinc finger and chromodomains. They acetylate histones H2A, H3 and H4, and include Tip60, MOZ, MORF and HBO1 [8]. The addition of acetyl groups to the N-terminal tails of the histones neutralises the positive charge of the octamer, leading to less interaction with the DNA [9], making the chromatin more accessible to the cellular transcription machinery to drive gene expression. Gene expression can be switched off by the activity of HDACs, which remove the acetyl groups and restore the strong interaction between the histone and DNA. In addition to gene expression, histone acetylation and deacetylation has been implicated in regulating DNA repair, in particular of DNA double strand breaks (DSBs) [10,11]. Indeed, a recent in-depth study using ChIP-sequencing revealed numerous histone modifications at sites of DSBs, but specifically that there were decreases in histone H4 lysine 12 (H4K12) acetylation and increases in histone H2B lysine 120 (H2BK120) acetylation that define DSB repair pathway choice [12]. Furthermore, the HAT Tip60 has been shown to bind to chromatin at sites of DSBs and that the HAT cofactor Trrap is essential for hyperacetylation of H4 and recruitment of repair proteins including 53BP1, BRCA1 and RAD51 [13]. Histone H4 lysine 16 (H4K16) acetylation catalysed by the HAT MOF has also been suggested to promote DSB repair [14].
A significant number of cancer patients (~50%) receive radiotherapy (ionising radiation; IR) during their treatment. Generally, conventional X-ray (photon) radiotherapy is used to promote tumour cell killing. However, there is increasing utilisation of particle ions, including proton beam therapy (PBT) that is a more targeted tumour treatment given the low entrance radiation dose which increases along the track, and that then peaks at a well-defined region known as the Bragg peak [15]. The Bragg peak therefore allows specific tumour targeting and thus minimising the radiation dose delivered to surrounding healthy tissues and organs at risk. Nevertheless, and despite the type of radiotherapy utilised, these largely cause tumour cell death through inducing sufficient DNA damage in the cells. Whilst DNA base damage and DNA single strand breaks (SSBs) predominate, particularly with sparsely ionising (low linear energy transfer, LET) radiation such as X-ray and γ-ray irradiation, the formation of DSBs but also complex DNA damage (CDD) containing multiple DNA lesions within close proximity are the major contributors to IR-induced cell death [15]. In fact, the higher the LET (such as with carbon ions), the greater the level and complexity of the DNA damage which drives cell killing. Following the generation of DNA damage, the cellular DNA damage response (DDR) is triggered which activates the base excision repair (BER) pathway for the repair of simple base damage and SSBs [16], whereas DSBs are repaired by either non-homologous end joining (NHEJ) or homologous recombination (HR) [17]. Interestingly, given the nature of the complexity within CDD, this is likely to require multiple DNA repair pathways to resolve the damage although we have recently demonstrated a critical role for proteins involved in SSB repair during resolving of CDD induced by proton irradiation [18,19]. Given that chromatin compaction is tightly regulated by histone PTMs, such as acetylation, there are subsequently opportunities to combine radiotherapy with drugs/inhibitors that target the enzymes controlling this phenotype. In this case, the combinatorial treatment can lead to altered DNA accessibility and impact on either DNA damage induction, DNA damage repair efficiency or the gene expression profile thus increasing radiotherapy efficacy.
Here, we provide an up-to-date review on HDACs and their cellular roles particularly in the DDR, along with reported evidence that HDAC inhibitors (HDACi) can enhance the effectiveness of radiotherapy in tumour models.
2. Histone Deacetylases (HDACs)
The HDACs are organised into classes I, II, III and IV based on function and sequence homology. Classes I, II and IV comprise the ‘classical’ Zn2+-dependent HDACs (Figure 1), while class III contains NAD+-dependent sirtuins [20]. Class I, II and IV HDACs share a Zn2+-dependent catalytic HDAC domain whereas Class II HDACs contain unique domains, including nuclear localisation and export signals, which allow for movement between the cytoplasm and nucleus [21]. Furthermore, class II HDACs also contain a binding domain specific to the myocyte enhancement factor 2 (MEF2) transcription factor [22]. HDAC6 contains a zinc finger binding domain, while HDAC10 contains a leucine-rich region. Sirtuins, the class III HDACs, are structurally distinct from classes I, II and IV. They are homologous to the yeast silent information regulator 2 (Sir2) protein [23], and are ubiquitously expressed in humans [24]. However, the sirtuins are not impacted by current HDAC inhibitors, and are therefore not covered in detail in this review. An understanding of their current cellular targets and roles of class I, II and IV HDACs as well as their associations with tumour development (also summarised in Table 1), are detailed below.
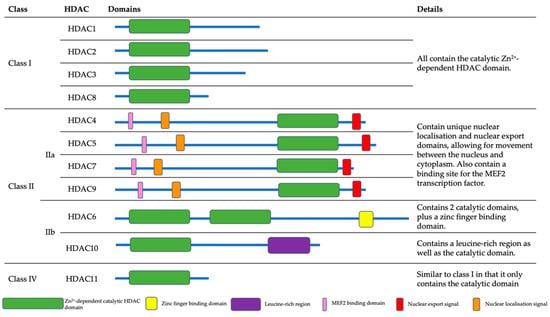
Figure 1.
The HDAC structures and domains. Class I, II and IV HDACs all contain a zinc-dependent HDAC domain. The class IIa HDACs also contain MEF2 binding domains, as well as nuclear localisation and export signals. HDAC6 alone has a zinc finger binding domain, whereas HDAC10 has a leucine-rich region.
2.1. Class I HDACs
Localised to the nucleus, class I HDACs (HDAC1, HDAC2, HDAC3 and HDAC8) have been shown to act as a catalytic subunit in protein complexes that are heavily involved in transcriptional repression [25]. They are important for maintaining correct gene expression in various tissues of the body.
2.1.1. HDAC1
HDAC1, along with HDAC2, is part of the CoREST complex which ensures that neuronal genes are transcriptionally silenced in non-neuronal tissues [26]. HDAC1 is also a component of the BRG1-RB1-HDAC1 complex, which negatively regulates transcription in resting neurons [27], and additionally targets the transcription factors SP1 and SP3 to regulate their function [28,29]. Furthermore, HDAC1 and HDAC2 can inhibit the transcriptional activity of NF-κB, which is associated with the development of specific cancers and autoimmune diseases [30]. In gastric cancer, HDAC1 overexpression has been suggested as a prognostic marker [31], and a study has shown that HDAC1 is upregulated in ~60% of cases [32]. In pancreatic cancer, HDAC1 appears to regulate the expression of the transcription factor hypoxia-inducible factor-1α (HIF-1α), which promotes tumour survival in reduced oxygen (hypoxic) conditions [33]. It was indicated that an increase in both HDAC1 and HIF-1α expression was associated with poor prognosis in pancreatic cancer patients. Upregulated HDAC1 has also been observed in colorectal cancer and prostate cancer [31,34]. Similarly, higher levels of HDAC1 expression are correlated with invasion, advanced stage disease and poor survival in hepatocellular carcinoma patients [35], while a meta-analysis of lung cancer patients showed that HDAC1 expression is negatively correlated with overall survival [36]. Interestingly, one study has shown that in breast cancer, increased HDAC1 was linked to better survival and smaller tumour sizes [37]. Collectively, these data show a strong correlation between HDAC1 expression and the development of specific tumour types.
2.1.2. HDAC2
HDAC2 forms a repressive transcription complex in association with DNA methyltransferase 1 (DNMT1) and the DNMT1-associated protein 1 (DMAP1). This complex functions during S-phase of the cell cycle to deacetylate histones following DNA replication [38]. It is also involved in metastasis tumour antigen 1 (MTA1)-mediated transcriptional co-repression of genes such as BRCA1, ESR1, TFF1 and CDKN1A, which are all linked to development of several cancer types [39]. Furthermore, HDAC2 has been shown to increase the DNA binding of the tumour suppressor protein p53, leading to increased inhibition of proliferation and the induction of senescence [40]. Like HDAC1, an increased expression of HDAC2 has been observed in multiple cancer types. In cervical cancers, HDAC2 is upregulated in the dysplasia transition region, and there is higher expression of HDAC2 during the polyp stage of carcinogenesis [41]. A retrospective study of gastric cancer patients showed that HDAC2 upregulation was associated with reduced 3-year survival rates [31]. In breast cancer, high HDAC2 expression is correlated with increased HER2 expression [42], and upregulation of HDAC1/2 has been linked to increased differentiation and proliferation of prostate cancers [43].
2.1.3. HDAC3
HDAC3 is thought to be involved in the functioning of the BCL6 transcriptional repressor through the deacetylation of histone H3 lysine 27 (H3K27) on gene enhancer elements. The changes in acetylation of enhancer elements allows B cells to undergo rapid changes in response to various cellular signals [44]. Moreover, HDAC3 is reported to interact with the X-box-binding protein 1 (XBP1) to protect endothelial cells from oxidative stress via the upregulation of AKT1 phosphorylation [45]. In acute promyelocytic leukaemia, HDAC3 has been shown to be involved in leukemogenesis through transcriptional repression in a complex with N-CoR [46], and a deficiency of HDAC3 in hepatocellular carcinoma is directly linked to the hyperacetylation of histone H3 lysine 9 (H3K9), leading to a loss of DNA damage control and the transcription of tumour-related genes [47]. Furthermore, HDAC3 has been associated with the proliferation and differentiation of colorectal cancer cells [48], and it has been suggested that downregulation of HDAC3 can reduce colorectal cancer proliferation [49]. In a study of 145 breast cancer patient samples, HDAC3 expression was correlated with negative expression of the oestrogen and progesterone receptors, and displayed a positive correlation with EGFR overexpression. High HDAC3 expression was also linked to poor overall survival [50].
2.1.4. HDAC8
HDAC8 has been implicated in the regulation of telomerase [51] and in tumour cell survival [52]. Furthermore, HDAC8 has been shown to modulate the transcriptional activity of oestrogen-related receptor α (ERRα), which is involved in processes such as glycolysis and oxidative phosphorylation [53]. In an expression study of HDAC1–11 in neuroblastoma, only HDAC8 expression was associated with tumour development. HDAC8 expression correlated with advanced disease and metastasis, as well as poor overall survival [54]. Furthermore, in breast cancer, HDAC8 expression has been shown to be upregulated, and associated with dissemination of breast cancer cells and epithelial-mesenchymal transition [55].

Table 1.
Cellular targets of HDACs and associations with tumour development.
Table 1.
Cellular targets of HDACs and associations with tumour development.
| HDAC Class | HDAC | Cellular Targets | Tumour Association | References |
|---|---|---|---|---|
| I | HDAC1 | CoREST transcriptional complex, BRG1-RB1-HDAC1 complex, SP1/3 transcription factors, NF-κB, HIF-1α | Gastric, pancreatic, colorectal, prostate, hepatocellular, lung, breast | [26,27,28,29,30,31,32,33,34,35,36,37,43,56] |
| HDAC2 | CoREST transcriptional complex, DNMT/DMAP1, MTA1, p53 | Cervical, gastric, breast, prostate | [26,31,38,39,40,41,42,43] | |
| HDAC3 | BCL6, XBP1, AKT1, N-CoR | Acute promyelocytic leukaemia, hepatocellular, colorectal, breast | [44,45,46,47,48,49,50] | |
| HDAC8 | Telomerase, ERRα | Neuroblastoma, breast | [51,53,54,55] | |
| II | HDAC4 | HIF-1α, CDK | Oesophageal, glioma, gastric, colorectal | [57,58,59,60,61,62] |
| HDAC5 | MEF2C, MTA1, ESR1, Rb, RARA | Hepatocellular, colorectal, breast, lung | [63,64,65,66,67,68,69,70] | |
| HDAC6 | ESR1, HIF-1α, Aggresome | Oral squamous cell carcinoma, ovarian, breast | [57,63,71,72,73,74] | |
| HDAC7 | RARA, FOXP3, STAT3 | Nasopharyngeal, lung, breast, gastric | [64,75,76,77,78,79] | |
| HDAC9 | MEF2, C-Jun, Aldehyde Dehydrogenase 1A3 | Oral squamous cell carcinoma, lung, hepatocellular, breast | [80,81,82,83,84,85] | |
| HDAC10 | PTPN22, AKT, DNA Repair | Lung, colorectal, ovarian | [86,87,88,89,90,91] | |
| IV | HDAC11 | p53, cell cycle progression, glycolysis | Pituitary, neuroblastoma, hepatocellular | [92,93,94] |
2.2. Class II HDACs
Class II HDACs are divided into class IIa (HDAC4, HDAC5, HDAC7 and HDAC9) and class IIb (HDAC6 and HDAC10). Class IIa HDACs are expressed in both the nucleus and cytoplasm and have been shown to play critical roles in the regulation of MEF2 expression, which is a vital protein in embryonic development [95,96]. Interestingly, the class IIa HDACs have minimal enzymatic activity, and are dependent on a multiprotein complex containing HDAC3 and a transcriptional corepressor, N-CoR/SMRT [97]. These proteins also appear to be tissue-specific, given that they are expressed in the skeletal, heart, and smooth muscle, as well as brain tissues [98].
2.2.1. HDAC4
Studies have shown that HDAC4 and HDAC6 are involved in the regulation of HIF-1α transcription and are therefore implicated in tumour angiogenesis [57]. Furthermore, HIF-1α undergoes acetylation at lysines 532 and 674 resulting in HIF-1α degradation or transcriptional activity, respectively [99,100]. It appears therefore that HDAC4 is involved largely in the hypoxic response, as further evidence demonstrates through a knockdown of HDAC4 that this leads to increases in the response and adaptation of cancer cells to hypoxia, and alters HIF-1-mediated transcription of genes involved in glycolysis and chemoresistance [101]. Overexpression of HDAC4 has been associated with higher tumour grade, advanced clinical stage and poor survival in oesophageal squamous cell carcinomas, and was found to promote proliferation via the upregulation of cyclin-dependent kinases (CDKs) [59]. The role of HDAC4 in proliferation has also been shown in glioblastoma [60], gastric cancer [61], and colorectal cancer where it promotes growth through p21 repression [62].
2.2.2. HDAC5
HDAC5 has been demonstrated to actively repress the transcription of the myocyte enhancer, MEF2C, which is involved in muscle maturation and vascular development [65]. Furthermore, HDAC5 is suggested to be involved in MTA1-mediated epigenetic regulation of the oestrogen receptor 1 (ESR1) gene, which encodes a nuclear hormone receptor, and is involved in cellular processes such as proliferation and differentiation [63]. HDAC5 has also been shown to serve as a co-repressor of the retinoic receptor alpha (RARA), which is involved in a variety of processes such as cell growth [64]. Studies have furthermore demonstrated that HDAC5 may interfere with the function of the Rb tumour suppressor gene to enhance tumourigenesis [66]. Overall, overexpression of HDAC5 has been observed in multiple cancer types, including hepatocellular carcinoma, colorectal, breast, and lung cancer, and is associated with tumour cell proliferation, invasion, and apoptosis inhibition [67,68,69,70].
2.2.3. HDAC6
Similar to HDAC5, HDAC6 has been shown to be involved in the MTA1-mediated regulation of ESR1 expression [63]. Another reported key role of HDAC6 is in the degradation of misfolded proteins. If misfolded proteins are too abundant to be degraded via the chaperone refolding system, HDAC6 mediates their transport to a cytoplasmic juxtanuclear structure called the aggresome [71]. HDAC6 has been shown to play a role in cancer cell metastasis [102], and in oral squamous cell carcinoma a higher level of HDAC6 has been linked to a higher primary tumour stage [74]. In ovarian cancer cell lines and tissues, levels of HDAC6 were found to be higher in both high-grade and low-grade samples compared to benign lesions and healthy ovarian cells [72]. Interestingly, HDAC6 mRNA levels have been shown to be higher in patients with small, low histologic grade, hormone positive breast cancers. HDAC6 levels were positively correlated with disease-free survival, and high HDAC6 levels were associated with a better response to endocrine therapy [73].
2.2.4. HDAC7
HDAC7 has been implicated in angiogenesis, as silencing of the protein lead to an alteration in morphology, migration and capillary forming abilities of endothelial cells in vitro [103]. Similarly to HDAC5, HDAC7 also acts as a corepressor of RARA, preventing its binding to DNA [64], and HDAC7 may play a role in transcriptional repression via complex formation with the FOXP3 regulator protein, which is involved in controlling the activity of regulatory T cells [76]. In nasopharyngeal carcinoma, lung cancer and breast cancer, an upregulation of HDAC7 is associated with higher levels of proliferation and thus disease progression [75,77,78]. Furthermore, HDAC7 has been shown to deacetylate and inhibit Stat3 expression in lung cancers to enhance tumourigenesis [75], and has been associated with poor prognosis and metastasis in gastric cancer [79].
2.2.5. HDAC9
Unsurprisingly, and like other members of the class II HDACs, it has been suggested that HDAC9 is involved in angiogenesis [85]. HDAC9 repressed MEF2-dependent transcription via the recruitment of HDAC1/3, which appears to inhibit skeletal muscle myogenesis and may be involved in heart development [80]. Furthermore, HDAC9 has been shown to repress c-Jun kinase activation, preventing downstream signalling through the MAPK pathway, which regulates a variety of processes in the cell, including proliferation, apoptosis and differentiation [82]. In oral squamous cell carcinoma, increased HDAC9 expression enhances growth and cell cycle progression [84], whereas in lung cancer, tumour cells had a decreased level of HDAC9 compared to healthy cells [83]. HDAC9 expression has also been positively correlated with the number of dedifferentiation markers in a database of hepatocellular carcinoma patients, and has been associated with aldehyde dehydrogenase 1A3 expression, a gene commonly mutated in hepatocellular carcinoma [81].
2.2.6. HDAC10
HDAC10 has been reported to play a role in DNA damage repair, through associations with DSB formation and controlling expression of the DNA repair mismatch genes mlh1 and msh2/6 [86,87], and promotes angiogenesis via the PTPN22/ERK signalling axis [88]. Furthermore, research suggests that HDAC10 regulates cell proliferation via the control of AKT signalling [89]. HDAC10 has been shown to have high expression levels in lung and colorectal cancer [87,89], and in ovarian cancer deletions of HDAC10 were linked to a sensitivity to platinum therapy [90].
2.3. Class IV HDACs
HDAC11 is the only class IV HDAC. It is mainly expressed in the nucleus, and is generally localised to the brain, heart, skeletal muscle and kidney tissues [104]. HDAC11 has been shown to display a negative correlation with p53 levels in pituitary tumours [94], and studies in neuroblastoma cells have identified a role for HDAC11 in cell cycle progression [93]. A study in hepatocellular carcinoma suggested that HDAC11 can also regulate cellular metabolic processes, such as glycolysis [92].
3. HDACs in the Cellular DDR
Histone acetylation has been associated with the cellular DDR. In response to IR induced DNA damage, the chromatin structure is altered to allow DNA repair factors access to the DNA and repair the damage, followed by chromatin structure and integrity being restored to its original state. HDACs have therefore been implicated in the maintenance of dynamic acetylation equilibrium of DDR proteins [105].
3.1. Class I HDACs
Class I HDACs, and particularly HDAC1 and HDAC2, have been associated with the control of several key DDR proteins, including ATM, ATR and BRCA1, involved in the signalling and repair of DSBs. Vorinostat, a pan-HDAC inhibitor, has been shown to reduce the activation of ATM after induction of DSBs in a dose- and time-dependent manner [106]. HDAC1 and HDAC2 have also been demonstrated to localise rapidly (within 5 min) to IR-induced DNA damage sites, then gradually dissociate until being no longer visible (30 min after treatment). At the damage sites, HDAC1/2 are reported to regulate the acetylation of histone H3 lysine 56 (H3K56), which decreases upon DNA damage. This process is also sensitive to class I/II HDAC inhibitors [107]. In this study, it was also noted that HDAC1/2 siRNA-mediated depletion led to hypersensitivity to IR and phleomycin. DNA damage induced CHK1/2 and p53 phosphorylation was also higher in cells depleted of HDAC1/2 compared to controls, and inhibition of HDAC1/2 enhanced the levels of the DSB marker γH2AX. These observations suggest that HDAC1/2 impair DSB repair, and to support this, it was shown that HDAC1/2-depleted cells have major defects in the NHEJ pathway. In particular, HDAC1/2 was demonstrated to influence the persistence of Ku70 and Artemis at the sites of DNA damage [107].
HDAC3 function has been shown to be vital for efficient DNA repair, specifically of DSBs. The inactivation of HDAC3 in mouse embryonic fibroblasts (MEFs) increased the sensitivity of the cells to doxorubicin, a topoisomerase II inhibitor, and to cisplatin that generates DNA crosslinks [108]. These results suggest that DNA repair pathways are inefficient when HDAC3 is absent. Furthermore, DNA damage sensitivity and repair data in this study suggested that the loss of HDAC3 impacts the DDR by targeting the chromatin structure, as HDAC3 is not recruited to DNA damage sites, and HDAC3 inactivation does not impact the localisation of other DDR proteins, such as RAD51, BRCA1, and MRE11 [109]. HDAC3 inhibition has been shown to decrease global heterochromatin levels in vivo and led to a 5–8 fold increase in the number of chromosomal breaks and gaps in metaphase chromosomes. Thus, HDAC3 is thought to be crucial for maintaining genome stability and contributes to efficient DNA repair.
3.2. Class II HDACs
It has been demonstrated that HDAC4 interacts with 53BP1 during the repair of DSBs [110]. After treatment with IR, HDAC4 localised to nuclear foci in a dose-dependent manner and this was evident early post-treatment (within 5 min) and peaked at 1 h. By 24 h post-IR, the number of HDAC4 foci had returned to background levels. The HDAC4 foci displayed similar kinetics to 53BP1 foci, and further investigation showed that HDAC4 and 53BP1 colocalise to sites of DSBs. Silencing of HDAC4 expression resulted in decreased levels of 53BP1, and the reverse was also shown, suggesting that HDAC4 and 53BP1 contribute to the maintenance of the stability of each other. As expected, silencing of either protein led to a reduction in 53BP1 foci following DNA damage, further suggesting that HDAC4 is involved in the DDR following DSB induction.
HDAC6 has also been shown to play a regulatory role in the DDR via its interactions with MutL homolog 1 (MLH1) and MutS protein homolog 2 (MSH2), two key DNA mismatch repair proteins. HDAC6 was demonstrated to deacetylate MLH1 on lysines 33, 241, 361 and 377, which prevents the formation of the MutL⍺-MutS⍺ complex, thus blocking the mismatch repair pathway [111]. HDAC6 has been reported to deacetylate and lead to ubiquitination of the MSH2 protein, which reduces its stability [112]. Nevertheless, this evidence pinpoints an important role for HDAC6 in regulating the repair of DNA mismatches.
HDAC9 and HDAC10 are thought to be important for the HR pathway of DSB repair. It has been demonstrated that depletion of HDAC9/10 using siRNA significantly decreased cellular HR activity, and sensitised cells to mitomycin C [113]. However, the mechanism by which HDAC9 and HDAC10 influence HR is currently unclear.
4. HDAC Inhibitors in Enhancing IR Sensitivity
Due to their role in the regulation of chromatin but also in the cellular DDR, several HDAC inhibitors (HDACi) have been investigated for their impact on the radiosensitivity of specific tumour models both in vitro and in vivo (also summarised in Table 2).
4.1. Valproic Acid (VPA)
Multiple studies have focused on the class I HDAC inhibitor valproic acid (VPA), a branched-chain saturated fatty acid, which has been shown to induce radiosensitisation in several cancer types. In colorectal cancer cells (LS174T and HCT116), p53-dependent radiosensitisation was observed following treatment with 500 μM VPA for 16 h prior to irradiation (up to 8 Gy γ-rays; dose enhancement ratios, DER = 1.3–1.4) [114]. The VPA treatment significantly increased the sub-G1 population indicative of apoptosis in the LS174T and HCT116/p53+/+ cell lines but not in the HCT116/p53−/− cells, further demonstrating the importance of p53 in radiation-induced cell killing. Immunoblotting analysis revealed that levels of γH2AX were increased from 2–24 h with the combination treatment in the p53-positive cell lines, suggesting that VPA treatment increases the levels of IR-induced DSBs. The effects of VPA were also studied in vivo using nude mice with HCT116-derived xenograft tumours treated with VPA (6 × 300 mg/kg) and IR (10 Gy), either alone or in combination. This revealed that the combination treatment reduced tumour growth of p53-containing xenografts but not those that were p53-negative. These data are similar to another study conducted in oesophageal squamous cell carcinoma cells (TE9, TE10, TE11 and TE14), where treatment with VPA (500 μM for 24 h) enhanced radiosensitivity (2–6 Gy X-rays) [115]. This was associated with increases in γH2AX levels in cells 2 h post-IR (6 Gy), and interestingly in downregulation of the expression levels of RAD51 involved in HR repair of DSBs. Analysis of annexin V staining revealed that the combined treatment of VPA and irradiation after 48 h led to increases in both early and late apoptotic cells. However, this evidence was not further explored in in vivo studies to further support that VPA acts to radiosensitise oesophageal tumours.
More recently, VPA (1 mM for 24 h) has been shown to increase the sensitivity of thyroid cancer cells to γ-radiation (1–8 Gy), although the relative effect was mild in TPC-1 cells (DER = 1.1) compared to WRO cells (DER = 1.3) [116]. Normal thyroid epithelial cells (Nthy-ori 3–1) also displayed apparent increases in radiosensitivity (DER = 1.3). The combination treatment did not appear to have a dramatic impact on cell cycle distribution of TPC-1 and WRO cells compared to the irradiation alone, although there was some evidence of increases of cell death (apoptosis and necrosis) with VPA pretreatment before γ-ray irradiation (3 Gy; at 24 h and 48 h in the WRO and TPC-1 cells, respectively). As to DSB damage and through γH2AX foci analysis, the combination treatment significantly increased γH2AX foci numbers at 30 min and 4 h post-IR in both cell lines. Furthermore, and similar to the study described above in oesophageal cancer cells, VPA treatment with γ-rays led to decreases in RAD51 protein levels post-IR (at 4 h).
4.2. Vorinostat
The non-selective HDACi vorinostat, which is a synthetic hydroxamic acid derivative (also known as MK0683 or suberoylanilide hydroxamic acid, SAHA), like VPA, has been shown to increase radiosensitivity of different cancers both in vitro and in vivo. Pretreatment with vorinostat (2.5 µM for 24 h) led to increases in radiosensitivity of melanoma (A375 and MeWo; DER = 1.4 and 1.3, respectively) and non-small cell lung cancer (A549; DER = 1.7) cells following γ-rays (2–6 Gy) [117]. In A375 cells, the combination of vorinostat (2.5–10 µM) and radiation (5 Gy) led to increased apoptosis compared to the HDACi alone, and which appeared to be associated with decreased protein levels of Ku70, Ku86 and RAD50. However, no quantitative analysis was completed to support the data significance. γH2AX foci analysis showed that vorinostat (2.5 µM) plus γ-irradiation (5 Gy) led to increased DSB levels and persistence 30 min-24 h post-treatment compared to the radiation alone, suggesting that this effect is responsible for the enhanced cellular radiosensitisation. In another study, vorinostat (0.5 µM for 16 h) caused enhanced sensitisation of breast tumour brain metastasis cells (MDA-MB-231-BR; DER = 1.3) as well as breast adenocarcinoma (T47D; DER = 1.2) and ovarian adenocarcinoma (NCI/ADR-RES; DER = 1.5) cells following X-ray irradiation [118]. In MDA-MB-231-BR cells, it was further shown that vorinostat (1 µM) in combination with X-rays (2 Gy) led to significant increases in γH2AX foci 1–24 h post-IR compared to irradiation alone suggesting defects in DSB repair. This was associated with significant increases in mitotic catastrophe in the cells post-IR (at 72 h). Finally, mice bearing MDA-MB-231-BR xenografts were found to exhibit significant tumour growth delay when treated with vorinostat (50 mg/kg) before irradiation (3 Gy) compared to the treatments alone. This provides evidence that vorinostat can act as a tumour radiosensitiser both in vitro and in vivo.
The impact of vorinostat in radiosensitising colorectal cancer cells (HCT116, HT29, KM20L2 and SW620) under conditions of both normoxia and hypoxic conditions has been studied. All cells treated with vorinostat (1–2 μM for 18 h) were significantly sensitised following a single dose of X-rays (5 Gy) in 1% hypoxia [119]. Similar observations were seen for all cell lines irradiated under conditions of normoxia, although data specifically using SW620 cells were not significant. Subsequently, in vivo experiments showed using SW620 and HCT116-derived xenografts that the addition of vorinostat (daily injection of 100 mg/kg for four days prior to irradiation) enhanced tumour growth delay compared to the treatments alone. Furthermore, addition of capecitabine (359 mg/kg) led to optimal tumour growth delay. Whilst these results support that vorinostat acts a radiosensitiser of colorectal cancer cells and xenograft tumours, no mechanistic insight was performed to understand the HDACi-induced radiosensitisation.
4.3. CUDC-101
CUDC-101 is a multitargeting small molecule inhibitor of HDAC1–10, but also epidermal growth factor receptor (EGFR) and the HER2 receptor. Using pancreatic cancer cell lines (MIA PaCa-2, Su.86.86 and T3M-4) treated with CUDC-101 (0.5 μM for 24 h), this led to significantly reduced cell survival following either a 2.5 or 5 Gy dose of X-rays compared to the treatment with CUDC-101 alone [120]. The radiosensitisation effect of CUDC-101 was also demonstrated on 3D spheroid cultures of Su.86.86 and T3M-4 cells treated with either 0.25 or 0.5 µM CUDC-101 with 5 Gy X-rays. To uncover the mechanism behind radiosensitisation, it was shown in all three pancreatic cancer cell lines treated with CUDC-101 (3 µM for 24 h) and X-rays (5 Gy) that this significantly reduced PARP-1 levels. However, no impact on DSB repair via γH2AX foci analysis was observed. Increases in the sub-G1 component of cells was demonstrated with the combination treatment (using doses of either 0.5 or 3 µM CUDC-101 with 5 Gy X-rays), suggesting increases in apoptosis and which was further supported by decreases in the anti-apoptotic proteins survivin and XIAP. Taken together, these results show that CUDC-101 is a potent radiosensitiser of pancreatic cancer cells in vitro, and interestingly this study also showed that CUDC-101 was a more effective radiosensitiser than vorinostat. There is also evidence from a scientific conference report in glioblastoma and breast cancer cell lines (U251 and MDA-MB-231, respectively) treated with CUDC-101 (either 0.5 μM or 1 μM), that this enhances radiosensitivity (DEF = 1.42) and is potentially related to delays in DSB repair and increases in mitotic catastrophe [121]. However, to our knowledge, the data from this abstract has not been reported in full.
4.4. Panobinostat
Panobinostat (also known as LBH-589 or Farydak) is a cinnamic hydroxamic acid analogue and another non-selective HDACi. Panobinostat (2.5–15 µM for 24 h, dependent on the cell line) has been shown to significantly reduce survival of prostate cancer cells (PC-3 and LNCaP; DEF = 1.3 and 1.8, respectively) in response to photon irradiation (2–8 Gy) [122]. Interestingly, increased radiosensitivity was not observed in normal prostate epithelial cells (RWPE-1). The combination treatment led to increased numbers of prostate cancer cells in subG1 phase (2–72 h post-irradiation with 2 Gy), indicative of apoptosis. This appeared to be associated with decreased activation of CHK1 and CHK2, suggesting a lack of cell cycle checkpoint activation. Additionally in the prostate cancer cells, there was an increase in the levels of γH2AX with panobinostat and radiation (at 24 and 72 h post-IR) compared to the radiation alone. Decreased BRCA1, BRCA2 and RAD51 foci formation were observed with the combination treatment suggesting a deficiency in HR repair of DSBs, although protein levels of Ku70 and Ku86 analysed by immunoblotting were also supressed suggesting that NHEJ activity may also be affected and responsible for the enhanced cellular radiosensitisation.
In a more recent study, panobinostat (10 mg/kg for 6 h) has been shown to enhance the response of X-ray irradiation (5 × 4 Gy) in bladder cancer (RT112-derived) xenografts compared to the HDACi or radiation alone [123]. When delivered intravenously, panobinostat was demonstrated to preferentially accumulate in the xenografts relative to the plasma, suggesting possible tumour cell specificity, although this was not observed where the drug was administered intraperitoneally. This study was then expanded in vitro to investigate the specificity of the HDAC. This revealed in bladder cancer cells (RT112, CAL29 and T24) that panobinostat alone (50 nM for 24 h) led to decreases in gene expression and protein levels of HDAC2 and HDAC7. However, variable decreases and increases in other HDACs were observed which was cell-line dependent, but also these effects were not studied in combination with IR. Nevertheless, an siRNA knockdown of HDAC1 and HDAC2 led to increased radiosensitivity of RT112 cells, suggesting these are the potentially important targets for panobinostat.
4.5. Romidepsin
Romidepsin (also known as FK288, FR901228 or Istodax) is a bicyclic depsipeptide antibiotic, and a class I-specific HDACi. There appears to be only a single study which has analysed this as a radiosensitiser. In bladder cancer cell lines (RT112, MBT2 and HT1376), romidepsin (0.4–2.5 nM for 24 h, dependent on the cell line) was effective at enhancing cellular sensitivity to γ-rays (2–6 Gy) [124]. Subsequently, the combination of romidepsin (4 mg/kg) with γ-radiation (6 Gy) was investigated in a RT112-derived xenograft model, which revealed that the combination treatment was significantly more effective in reducing tumour growth than the monotherapies alone. There was also some limited evidence in RT112 cells that romidepsin led to an increased level of γH2AX immediately and 4 h post-IR compared to the radiation only. Also, the efficiencies of HR and NHEJ activities were found to be reduced in romidepsin only treated cells using GFP reporter assays, but which indicates that the HDACi induces radiosensitivity through deficiencies in DSB repair.
4.6. Mocetinostat
A single study involving the benzamide class I HDACi mocetinostat (also known as MGCD0103) has shown that the survival of bladder cancer cells (RT112 and T24) were reduced by the HDACi (0.75–1.5 µM for 24 h, dependent on the cell line) in combination with X-ray irradiation (2–8 Gy) [123]. Whilst direct comparative analysis was not performed, it was suggested that radiosensitisation of both cell lines was less effective than with the HDACi panobinostat. However, similar to panobinostat, mocetinostat appeared to cause reductions in the protein levels of HDAC2, but also in the MRE11 exonuclease suggesting that impaired DSB repair could be the mechanism behind the observed radiosensitisation. Despite this, further evidence is required to support whether mocetinostat is an effective radiosensitiser in bladder cancer and other tumour models.
4.7. Belinostat
Belinostat (also known as PXD101) is a hydroxamic acid-type non-selective HDACi, and similar to romidepsin and mocetinostat described above, there is only a single report describing this as a radiosensitiser. Belinostat (0.23 or 0.41 µM for 24 h) was demonstrated to enhance the radiosensitivity of rhabdomyosarcoma cells (RD and RH3) to a single dose of photon radiation (4 Gy) compared to the HDACi or IR alone [125]. Mechanistically, the combination of belinostat with IR appeared to cause increases in γH2AX formation 24 h post-IR compared to the radiation alone, although this was only observed in one of the cell lines (RD). However, there were significant elevations in reactive oxygen species formation in both rhabdomyosarcoma cell lines with the combination treatment. Additionally, there was some data to suggest that belinostat in combination with IR supresses DNA-Pk and BRCA1 phosphorylation (3 and 24 h post-IR) compared to radiation alone. Finally, in vivo experiments using RD and RH30-derived xenografts treated with belinostat (40 mg/kg daily for 12 days) followed by photon irradiation (6 × 2 Gy) led to significant tumour growth delay compared to either treatment alone. Collectively, these data confirmed that belinostat is an effective radiosensitiser of rhabdomyosarcoma in vitro and in vivo, although additional evidence is necessary to expand on these observations, particularly using a broader range of tumour models.
4.8. Abexinostat
Abexinostat (also known as PCI-24781 or CRA-024781) is a hydroxamate-based pan-HDACi, which has been demonstrated to induce radiosensitivity in cervical, colorectal and non-small cell lung cancer cell lines. In the first study, SiHa cervical cancer (SiHa) and colorectal cancer (WiDr) cells were treated with abexinostat (0.3 or 3 μM for 20 h before and 4 h post-IR) in combination with X-rays (2–8 Gy), and which led to significantly reduced cell survival (DER = 1.5 and 1.2 for SiHa and WiDr, respectively) [126]. No impact of the combination was apparent on normal fibroblasts. Interestingly, abexinostat appeared to have no influence on the induction or rate of repair of DSBs through γH2AX and comet assay analysis. In the second study, significantly enhanced radiosensitisation of non-small cell lung cancer cell lines (A549 and H460) was observed with abexinostat (0.4 and 0.7 μM in A549, 0.1 and 0.2 μM in H460 treated for 24 h) following γ-radiation (2–6 Gy) [127]. The degree of radiosensitisation was dependent on the dose of the HDACi (DER = 1.3–1.7 and 1.2–1.9 for A549 and H460, respectively). Interestingly, significantly enhanced radiosensitivity of the combination treatment was also observed under conditions of severe hypoxia (0.1% oxygen), and where the impact of the HDACi was more pronounced (DER = 2.3–2.4 and 1.7–3.2 for A549 and H460, respectively). Abexinostat alone (0.2 µM for 24 or 48 h) in H460 cells was demonstrated to cause an increase in cellular apoptosis, although this was not further elevated in combination with IR. In contrast to the previous study, the combination treatment of abexinostat (0.2 µM) and γ-radiation (4 Gy) in non-small cell lung cancer cell lines was shown to cause delays in DSB repair as shown through γH2AX and 53BP1 foci formation (24 h post-IR), compared to the treatments alone. There were also indications that this led to significant decreases in the protein levels of MRE11, RAD51 and NBS1. Finally, and using A549 and H460-derived xenografts, abexinostat (25 mg/kg twice daily) and X-ray irradiation (3 × 2 Gy) caused a significant tumour growth delay compared to the treatments alone. This provided evidence of abexinostat’s function as an effective radiosensitiser both in vitro and in vivo for non-small cell lung cancer.
4.9. HDACi in Response to PBT
Currently, very little research has been performed examining the impact of HDACi in response to PBT, which whilst being a more precision-targeted radiotherapy treatment, displays significant biological uncertainties due to increases in LET at and around the Bragg peak where the radiation dose is delivered. This can therefore alter the DNA damage spectrum, particularly with increases in CDD formation, as well as the triggering of alternate cell death mechanisms [15]. To date, only two studies have been reported investigating the combination of either VPA or panobinostat with PBT on tumour models. Firstly, in hepatocellular carcinoma cells (Hep3B) treated with VPA (1 mM for 3 h), this was shown to enhance the sensitivity of cells to both photon and proton (230 MeV) irradiation (2–6 Gy) [128]. The impact of protons alone was shown to be more effective than photon irradiation (relative biological effectiveness, RBE = 1.08) which is close to that used in clinical treatment. There were indications that PBT in combination with VPA enhanced the numbers of cells arrested in G2/M phase of the cell cycle 24 h post-IR (6 Gy) compared to photon irradiation, although this appeared to be alleviated at 72 h post-IR. However, it was shown that the combination of PBT and VPA led to persistence of γH2AX levels and foci 24 h post-IR, compared to photons and VPA, and was also associated with an increase in reactive oxygen species and apoptotic cells. Furthermore, using Hep3B-derived xenografts, PBT (3 × 3 Gy) with VPA (3 × 300 mg/kg) was shown to be the most effective treatment at supressing tumour growth compared to photons and VPA, or the treatments alone.
In another more recent study conducted by the same authors and using hepatocellular carcinoma cells (Hep3B and Huh7), panobinostat (5 nM for 3 h) was shown to only moderately enhance the radiosensitivity of cells in response to PBT (DER = 1.2–1.3) and photon (DER = 1.1–1.2) irradiation (2–6 Gy) [129]. As for the previous study, PBT alone was more effective than photon irradiation in these cells (RBE = 1.2–1.3). Significant increases in reactive oxygen species with PBT or photons (6 Gy) in combination with panobinostat were observed compared to the radiation or HDACi alone. However, there was a suggestion that the PBT-HDACi combination led to persistent levels of γH2AX foci (24 h post-IR) and increases in apoptotic cells (72 h post-IR), compared to the photon-HDACi combination. These results are interesting in the context of potentially enhanced effectiveness of HDACi in combination with PBT on hepatocellular carcinoma cell models, although more expansive data is required, particularly expanding into other cancer types but also to explore the potential contribution of LET and CDD to the phenotypic response.

Table 2.
Evidence of enhanced radiosensitisation of tumour cells with HDAC inhibitors.
Table 2.
Evidence of enhanced radiosensitisation of tumour cells with HDAC inhibitors.
| HDAC Inhibitor | Target | Radiosensitivity Impact | Reference |
|---|---|---|---|
| Valproic Acid | Non-Selective | Effectively radiosensitised colorectal cancer, oesophageal cancer and thyroid cancer cells to photons, and hepatocellular carcinoma cells to PBT | [114,115,116,128] |
| Vorinostat | Non-Selective | Increased sensitivity of melanoma, lung cancer, breast cancer, and colorectal cancer cells to photons | [117,118,119] |
| CUDC-101 | HDAC1–10, EGFR, HER2 | Radiosensitised pancreatic cancer, breast cancer, and glioblastoma cells to photons | [120,121] |
| Panobinostat | Non-Selective | Sensitised bladder cancer, hepatocellular carcinoma, prostate cancer cells to photons, and hepatocellular carcinoma cells to PBT | [122,123,129] |
| Romidepsin | Class I | Increases the sensitivity of bladder cancer cells to photons | [124] |
| Mocetinostat | Class I | Radiosensitised bladder cancer cells to photons | [123] |
| Belinostat | Non-Selective | Sensitises rhabdomyosarcoma, cervical cancer and colorectal cancer cell lines to photons | [125,126] |
5. Discussion
The HDACs are well known and characterised to play important roles in controlling vital DNA-dependent processes, including transcription and DNA damage repair. Consequently, evidence determined to date has suggested that HDACs may be important therapeutic targets for cancer therapy and particularly that HDACi’s can be used in combination with radiotherapy to exert a radiosensitising effect on a variety of cancer types to suppress tumour growth. Indeed, there is promising evidence that non-selective HDACi’s have the potential to be most effective due to their multi-targeting effects. For example, VPA, vorinostat and panobinostat have been shown to be effective tumour radiosensitisers both in vitro and in vivo. However, there is only limited evidence using the more selective inhibitors, such as romidepsin and moectinostat, as radiosensitisers and therefore it is clear that further experimentation is required with these inhibitors specifically to determine their potential for eventual translation through to the clinic. Furthermore, it is evident that most experimental data surrounding the mechanism of action of most HDACi’s in combination with IR appears to centre on their impact relating to DNA damage repair, particularly the efficiency of repair of DSBs through either NHEJ or HR. This is understandable given that the major critical target for IR leading to cell death is DNA. However, it is likely that HDACi have more wide-ranging effects on DNA and chromatin structure, leading to impacts on other cellular processes, including gene regulation, cell signalling and proliferation. It is therefore important to gain a more comprehensive understanding of the way that potent HDACi’s work at the molecular level in both tumour but also the associated normal cell models to fully exploit their future therapeutic potential.
It is also apparent from the studies highlighted in this review that much of the research to date has utilised immortalised tumour cell lines as models to study the effects of HDAC inhibition on radiosensitivity. Whilst this is an appropriate starting point, expansion of these data in the appropriate and relevant 3D tumour models, such as spheroids and particularly patient-derived organoids that are a better representation of a tumour structure and its response to treatment, are required. This can provide a more accurate insight into the potential benefits of HDACi’s in radiosensitising tumours and preventing their growth or ultimately in driving tumour cell death. Additionally, these experiments should be complemented with the relevant preclinical in vivo models (e.g., tumour xenografts) to provide further evidence of their applicability for translation. Another important aspect, particularly in relation to solid tumours, is the impact of HDACi with radiotherapy on tumour hypoxia, which is well established to play a significant role in driving radioresistance. Only two studies to date, presented in the previous sections, have examined the impact of specific HDACi’s under different hypoxic conditions and tumour cell models. Therefore, it is clear from these studies that there is also scope for significant expansion, ideally directing comparing the radiosensitising effect of HDACi under conditions of hypoxia (0.1–1%) versus normoxia. This preclinical research must then work towards the ultimate goal of providing optimal treatment strategies using radiotherapy for cancer treatment.
Another noticeable point to consider is that the majority of the studies investigating HDACi’s as tumour radiosensitisers have only been investigated for their effect in combination with conventional photon (X-rays or γ-rays) radiation. However, with the increasing use of particle beam therapy, such as PBT, which provide more targeted delivery of the radiation dose to the tumour, this mode of radiotherapy should be examined for its ability to radiosensitise the cancer cells in combination with HDACi’s. Ideally, these experiments should be run in parallel with photon experiments to directly compare the efficacy but also define the precise mechanism of action. Given that PBT, but more so heavy ions including carbon, are associated with different biological effects due to the increased LET leading to elevations in the induction and complexity of CDD, more research must be carried out to fully understand the impact of HDACi with these different radiation types to appreciate their potential benefit translationally.
6. Conclusions
In summary, research carried out to date both in vitro and in vivo have shown the promising radiosensitising potential of HDACi’s in the treatment of a variety of cancer types, particularly the non-selective HDACi’s such as VPA, vorinostat and panobinostat, due to their multi-targeting effects. Evidence for the radiosensitising effects of the more selective inhibitors, such as romidepsin and mocetinostat, is much more limited and where further experimentation is required to determine their potential. Importantly, more research must be carried out using the appropriate and relevant 3D tumour models and environments (e.g. hypoxia) to investigate the ability of HDACi’s to radiosensitise the tumour cells. Additionally, the impact of the HDACi’s should be explored using alternative sources of ionising radiation other than conventional X-rays or γ-rays, such as PBT and heavy ions, which are increasingly being used in a clinical setting.
Author Contributions
Conceptualization, J.L.P.; writing—original draft preparation, J.A.; writing—review and editing, J.A. and J.L.P.; supervision, J.L.P.; funding acquisition, J.L.P. All authors have read and agreed to the published version of the manuscript.
Funding
J.A. is supported by the Bark Medal Studentship from the University of Liverpool. J.L.P. is supported by grants from North West Cancer Research (CR1197), the Medical Research Council (MR/V028944/1), and the National Institutes of Health (R01CA256854).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Luger, K.; Mäder, A.W.; Richmond, R.K.; Sargent, D.F.; Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 1997, 389, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Woodcock, C.L.; Dimitrov, S. Higher-order structure of chromatin and chromosomes. Curr. Opin. Genet. Dev. 2001, 11, 130–135. [Google Scholar] [CrossRef]
- Finch, J.T.; Klug, A. Solenoidal model for superstructure in chromatin. Proc. Nat. Acad. Sci. USA 1976, 73, 1897–1901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilbert, N.; Allan, J. Supercoiling in DNA and chromatin. Curr. Opin. Genet. Dev. 2014, 25, 15–21. [Google Scholar] [CrossRef] [Green Version]
- Allfrey, V.G.; Faulkner, R.; Mirskey, A. Acetylation and Methylation of Histones and Their Possible Role in thr Regulation of RNA Synthesis. Proc. Nat. Acad. Sci. USA 1964, 51, 786–794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morales, V.; Richard-Foy, H. Role of Histone N-Terminal Tails and Their Acetylation in Nucleosome Dynamics. Mol. Cell Biol. 2000, 20, 7230–7237. [Google Scholar] [CrossRef] [Green Version]
- Clapier, C.R.; Cairns, B.R. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 2009, 78, 273–304. [Google Scholar] [CrossRef]
- Lee, K.K.; Workman, J.L. Histone acetyltransferase complexes: One size doesn’t fit all. Nat. Rev. Mol. Cell Biol. 2007, 8, 284–295. [Google Scholar] [CrossRef]
- Garcia-Ramirez, M.; Rocchini, C.; Ausio, J. Modulation of chromatin folding by histone acetylation. J. Biol. Chem. 1995, 270, 17923–17928. [Google Scholar] [CrossRef] [Green Version]
- Tamburini, B.A.; Tyler, J.K. Localized histone acetylation and deacetylation triggered by the homologous recombination pathway of double-strand DNA repair. Mol. Cell Biol. 2005, 25, 4903–4913. [Google Scholar] [CrossRef] [Green Version]
- Robert, C.; Nagaria, P.K.; Pawar, N.; Adewuyi, A.; Gojo, I.; Meyers, D.J.; Cole, P.A.; Rassool, F.V. Histone deacetylase inhibitors decrease NHEJ both by acetylation of repair factors and trapping of PARP1 at DNA double-strand breaks in chromatin. Leuk. Res. 2016, 45, 14–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clouaire, T.; Rocher, V.; Lashgari, A.; Arnould, C.; Aguirrebengoa, M.; Biernacka, A.; Skrzypczak, M.; Aymard, F.; Fongang, B.; Dojer, N.; et al. Comprehensive Mapping of Histone Modifications at DNA Double-Strand Breaks Deciphers Repair Pathway Chromatin Signatures. Mol. Cell 2018, 72, 250–262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murr, R.; Loizou, J.I.; Yang, Y.G.; Cuenin, C.; Li, H.; Wang, Z.Q.; Herceg, Z. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat. Cell Biol. 2006, 8, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Sharma, G.G.; So, S.; Gupta, A.; Kumar, R.; Cayrou, C.; Avvakumov, N.; Bhadra, U.; Pandita, R.K.; Porteus, M.H.; Chen, D.J.; et al. MOF and histone H4 acetylation at lysine 16 are critical for DNA damage response and double-strand break repair. Mol. Cell Biol. 2010, 30, 3582–3595. [Google Scholar] [CrossRef] [Green Version]
- Vitti, E.T.; Parsons, J.L. The Radiobiological Effects of Proton Beam Therapy: Impact on DNA Damage and Repair. Cancers 2019, 11, 946. [Google Scholar] [CrossRef] [Green Version]
- Carter, R.J.; Parsons, J.L. Base Excision Repair, a Pathway Regulated by Posttranslational Modifications. Mol. Cell Biol. 2016, 36, 1426–1437. [Google Scholar] [CrossRef] [Green Version]
- Mao, Z.; Bozzella, M.; Seluanov, A.; Gorbunova, V. DNA repair by nonhomologous end joining and homologous recombination during cell cycle in human cells. Cell Cycle. 2008, 7, 2902–2906. [Google Scholar] [CrossRef]
- Carter, R.J.; Nickson, C.M.; Thompson, J.M.; Kacperek, A.; Hill, M.A.; Parsons, J.L. Complex DNA Damage Induced by High Linear Energy Transfer Alpha-Particles and Protons Triggers a Specific Cellular DNA Damage Response. Int. J. Radiat. Oncol. Biol. Phys. 2018, 100, 776–784. [Google Scholar] [CrossRef] [Green Version]
- Carter, R.J.; Nickson, C.M.; Thompson, J.M.; Kacperek, A.; Hill, M.A.; Parsons, J.L. Characterisation of Deubiquitylating Enzymes in the Cellular Response to High-LET Ionizing Radiation and Complex DNA Damage. Int. J. Radiat. Oncol. Biol. Phys. 2019, 104, 656–665. [Google Scholar] [CrossRef] [Green Version]
- de Ruijter, A.J.; van Gennip, A.H.; Caron, H.N.; Kemp, S.; van Kuilenburg, A.B. Histone deacetylases (HDACs): Characterization of the classical HDAC family. Biochem. J. 2003, 15, 737–749. [Google Scholar] [CrossRef]
- Muslin, A.J.; Xing, H. 14-3-3 proteins: Regulation of subcellular localization by molecular interference. Cell Signal. 2000, 12, 703–709. [Google Scholar] [CrossRef]
- Jayathilaka, N.; Han, A.; Gaffney, K.J.; Dey, R.; Jarusiewicz, J.A.; Noridomi, K.; Philips, M.A.; Lei, X.; He, J.; Ye, J.; et al. Inhibition of the function of class IIa HDACs by blocking their interaction with MEF2. Nucleic. Acids. Res. 2012, 40, 5378–5388. [Google Scholar] [CrossRef] [Green Version]
- Blander, G.; Guarente, L. The Sir2 family of protein deacetylases. Annu. Rev. Biochem. 2004, 73, 417–435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dai, Y.; Faller, D.V. Transcription Regulation by Class III Histone Deacetylases (HDACs)—Sirtuins. Transl. Oncogenomics 2008, 3, 53–65. [Google Scholar] [CrossRef] [PubMed]
- Watson, P.J.; Millard, C.J.; Riley, A.M.; Roberston, N.S.; Wright, L.C.; Godage, H.Y.; Cowley, S.M.; Jamieson, A.G.; Potter, B.V.L.; Schwabe, J.W.R. Insights into the activation mechanism of class I HDAC complexes by inositol phosphates. Nature Commun. 2016, 48, 829–834. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Myers, S.J.; Dingledine, R. Transcriptional repression by REST: Recruitment of Sin3A and histone deacetylase to neuronal genes. Nat. Neurosci. 1999, 2, 867–872. [Google Scholar] [CrossRef]
- Qiu, Z.; Ghosh, A. A calcium-dependent switch in a CREST-BRG1 complex regulates activity-dependent gene expression. Neuron 2008, 60, 775–787. [Google Scholar] [CrossRef] [Green Version]
- Ammanamanchi, S.; Freeman, J.W.; Brattain, M.G. Acetylated sp3 is a transcriptional activator. J. Biol. Chem. 2003, 278, 35775–35780. [Google Scholar] [CrossRef] [Green Version]
- Hung, J.J.; Wang, Y.T.; Chang, W.C. Sp1 deacetylation induced by phorbol ester recruits p300 to activate 12(S)-lipoxygenase gene transcription. Mol. Cell Biol. 2006, 26, 1770–1785. [Google Scholar] [CrossRef] [Green Version]
- Ashburner, B.P.; Westerheide, S.D.; Baldwin, A.S., Jr. The p65 (RelA) subunit of NF-kappaB interacts with the histone deacetylase (HDAC) corepressors HDAC1 and HDAC2 to negatively regulate gene expression. Mol. Cell Biol. 2001, 21, 7065–7077. [Google Scholar] [CrossRef] [Green Version]
- Weichert, W.; Röske, A.; Gekeler, V.; Beckers, T.; Ebert, M.P.A.; Pross, M.; Dietel, M.; Denkert, C.; Röcken, C. Association of patterns of class I histone deacetylase expression with patient prognosis in gastric cancer: A retrospective analysis. Lancet Oncol. 2008, 9, 139–148. [Google Scholar] [CrossRef]
- Choi, J.-H.; Kwon, H.J.; Yoon, B.-I.; Kim, J.-H.; Han, S.U.; Joo, H.J.; Kim, D.-Y. Expression Profile of Histone Deacetylase 1 in Gastric Cancer Tissues. Jpn. J. Cancer Res. 2001, 92, 1300–1304. [Google Scholar] [CrossRef] [PubMed]
- Miyake, K.; Yoshizumi, T.; Imura, S.; Sugimoto, K.; Batmunkh, E.; Kanemura, H.; Morine, Y.; Shimada, M. Expression of Hypoxia-Inducible Factor-1α, Histone Deacetylase 1, and Metastasis-Associated Protein 1 in Pancreatic Carcinoma.Correlation With Poor Prognosis With Possible Regulation. Pancreas 2008, 36, e1–e9. [Google Scholar] [CrossRef] [PubMed]
- Halkidou, K.; Gaughan, L.; Cook, S.; Leung, H.Y.; Neal, D.E.; Robson, C.N. Upregulation and nuclear recruitment of HDAC1 in hormone refractory prostate cancer. Prostate 2004, 59, 177–189. [Google Scholar] [CrossRef]
- Rikimaru, T.; Taketomi, A.; Yamashita, Y.; Shirabe, K.; Hamatsu, T.; Shimada, M.; Maehara, Y. Clinical significance of histone deacetylase 1 expression in patients with hepatocellular carcinoma. Oncology 2007, 72, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.L.; Song, X.; Pei, L.; Liu, L.; Wang, H.; Jia, M. Histone deacetylase HDAC1 expression correlates with the progression and prognosis of lung cancer: A meta-analysis. Medicine 2017, 96, e7663. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Yamashita, H.; Toyama, T.; Sugiura, H.; Ando, Y.; Mita, K.; Hamaguchi, M.; Hara, Y.; Kobayashi, S.; Iwase, H. Quantitation of HDAC1 mRNA expression in invasive carcinoma of the breast. Breast Cancer Res. Treat. 2005, 94, 11–16. [Google Scholar] [CrossRef]
- Rountree, M.R.; Bachman, K.E.; Baylin, S.B. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat. Genet. 2000, 25, 269–277. [Google Scholar] [CrossRef]
- Cong, L.; Pakala, S.B.; Ohshiro, K.; Li, D.Q.; Kumar, R. SUMOylation and SUMO-interacting motif (SIM) of metastasis tumor antigen 1 (MTA1) synergistically regulate its transcriptional repressor function. J. Biol. Chem. 2011, 286, 43793–43808. [Google Scholar] [CrossRef] [Green Version]
- Harms, K.L.; Chen, X. Histone deacetylase 2 modulates p53 transcriptional activities through regulation of p53-DNA binding activity. Cancer Res. 2007, 67, 3145–3152. [Google Scholar] [CrossRef] [Green Version]
- Huang, B.H.; Laban, M.; Leung, C.H.; Lee, L.; Lee, C.K.; Salto-Tellez, M.; Raju, G.C.; Hooi, S.C. Inhibition of histone deacetylase 2 increases apoptosis and p21Cip1/WAF1 expression, independent of histone deacetylase 1. Cell Death. Differ. 2005, 12, 395–404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Müller, B.M.; Jana, L.; Kasajima, A.; Lehmann, A.; Prinzler, J.; Budczies, J.; Winzer, K.; Dietel, M.; Weichert, W.; Denkert, C. Differential expression of histone deacetylases HDAC1, 2 and 3 in human breast cancer—Overexpression of HDAC2 and HDAC3 is associated with clinicopathological indicators of disease progression. BMC Cancer 2013, 13, 215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weichert, W.; Roske, A.; Gekeler, V.; Beckers, T.; Stephan, C.; Jung, K.; Fritzsche, F.R.; Niesporek, S.; Denkert, C.; Dietel, M.; et al. Histone deacetylases 1, 2 and 3 are highly expressed in prostate cancer and HDAC2 expression is associated with shorter PSA relapse time after radical prostatectomy. Br. J. Cancer 2008, 98, 604–610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hatzi, K.; Jiang, Y.; Huang, C.; Garrett-Bakelman, F.; Gearhart, M.D.; Giannopoulou, E.G.; Zumbo, P.; Kirouac, K.; Bhaskara, S.; Polo, J.M.; et al. A hybrid mechanism of action for BCL6 in B cells defined by formation of functionally distinct complexes at enhancers and promoters. Cell Rep. 2013, 4, 578–588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, D.; Li, Y.; Yang, J.; Wang, G.; Margariti, A.; Jiang, Z.; Yu, H.; Zampetaki, A.; Hu, Y.; Xu, Q.; et al. Unspliced X-box-binding protein 1 (XBP1) protects endothelial cells from oxidative stress through interaction with histone deacetylase 3. J. Biol. Chem. 2014, 289, 30625–30634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atsumi, A.; Tomita, A.; Kiyoi, H.; Naoe, T. Histone deacetylase 3 (HDAC3) is recruited to target promoters by PML-RARalpha as a component of the N-CoR co-repressor complex to repress transcription in vivo. Biochem. Biophys. Res. Commun. 2006, 345, 1471–1480. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.; Zhou, Y.; Zhuang, X.; Zhu, Y.; Wu, Z.; Lu, Y.; Li, S.; Zeng, Y.; Lu, Q.R.; Huo, Y.; et al. HDAC3 Deficiency Promotes Liver Cancer through a Defect in H3K9ac/H3K9me3 Transition. Cancer Res. 2019, 79, 3676–3688. [Google Scholar] [CrossRef]
- Spurling, C.C.; Godman, C.A.; Noonan, E.J.; Rasmussen, T.P.; Rosenberg, D.W.; Giardina, C. HDAC3 overexpression and colon cancer cell proliferation and differentiation. Mol. Carcinog. 2008, 47, 137–147. [Google Scholar] [CrossRef]
- Li, J.; Hu, M.; Liu, N.; Li, H.; Yu, Z.; Yan, Q.; Zhou, M.; Wang, Y.; Song, Y.; Pan, G.; et al. HDAC3 deteriorates colorectal cancer progression via microRNA-296-3p/TGIF1/TGFbeta axis. J. Exp. Clin Cancer Res. 2020, 39, 248. [Google Scholar] [CrossRef]
- Cui, Z.; Xie, M.; Wu, Z.; Shi, Y. Relationship Between Histone Deacetylase 3 (HDAC3) and Breast Cancer. Med. Sci. Monit. 2018, 24, 2456–2464. [Google Scholar] [CrossRef]
- Lee, H.; Sengupta, N.; Villagra, A.; Rezai-Zadeh, N.; Seto, E. Histone deacetylase 8 safeguards the human ever-shorter telomeres 1B (hEST1B) protein from ubiquitin-mediated degradation. Mol. Cell Biol. 2006, 26, 5259–5269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vannini, A.; Volpari, C.; Filocamo, G.; Casavola, E.C.; Brunetti, M.; Renzoni, D.; Chakravarty, P.; Paolini, C.; De Francesco, R.; Gallinari, P.; et al. Crystal structure of a eukaryotic zinc-dependent histone deacetylase, human HDAC8, complexed with a hydroxamic acid inhibitor. Proc. Natl. Acad. Sci. USA 2004, 101, 15064–15069. [Google Scholar] [CrossRef] [Green Version]
- Wilson, B.J.; Tremblay, A.M.; Deblois, G.; Sylvain-Drolet, G.; Giguere, V. An acetylation switch modulates the transcriptional activity of estrogen-related receptor alpha. Mol. Endocrinol. 2010, 24, 1349–1358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oehme, I.; Deubzer, H.E.; Wegener, D.; Pickert, D.; Linke, J.P.; Hero, B.; Kopp-Schneider, A.; Westermann, F.; Ulrich, S.M.; von Deimling, A.; et al. Histone deacetylase 8 in neuroblastoma tumorigenesis. Clin Cancer Res. 2009, 15, 91–99. [Google Scholar] [CrossRef] [Green Version]
- An, P.; Chen, F.; Li, Z.; Ling, Y.; Peng, Y.; Zhang, H.; Li, J.; Chen, Z.; Wang, H. HDAC8 promotes the dissemination of breast cancer cells via AKT/GSK-3beta/Snail signals. Oncogene 2020, 39, 4956–4969. [Google Scholar] [CrossRef] [PubMed]
- Weichert, W.; Roske, A.; Niesporek, S.; Noske, A.; Buckendahl, A.C.; Dietel, M.; Gekeler, V.; Boehm, M.; Beckers, T.; Denkert, C. Class I histone deacetylase expression has independent prognostic impact in human colorectal cancer: Specific role of class I histone deacetylases in vitro and in vivo. Clin Cancer Res. 2008, 14, 1669–1677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qian, D.Z.; Kachhap, S.K.; Collis, S.J.; Verheul, H.M.; Carducci, M.A.; Atadja, P.; Pili, R. Class II histone deacetylases are associated with VHL-independent regulation of hypoxia-inducible factor 1 alpha. Cancer Res. 2006, 66, 8814–8821. [Google Scholar] [CrossRef] [Green Version]
- Bryant, D.W.; Haynes, R.H. Endonuclease a from Saccharomyces cerevisiae shows increased activity on ultraviolet irradiated native DNA. Mol. Gen. Genet. 1978, 167, 139–145. [Google Scholar] [CrossRef]
- Zeng, L.-S.; Yang, X.-Z.; Wen, Y.-F.; Mai, S.-J.; Wang, M.-H.; Zhang, M.-Y.; Zheng, X.F.S.; Wang, H.-Y. Overexpressed HDAC4 is associated with poor survival and promotes tumor progression in esophageal carcinoma. Aging 2016, 8, 1236–1248. [Google Scholar] [CrossRef] [Green Version]
- Cai, J.Y.; Xu, T.T.; Wang, Y.; Chang, J.J.; Li, J.; Chen, X.Y.; Chen, X.; Yin, Y.F.; Ni, X.J. Histone deacetylase HDAC4 promotes the proliferation and invasion of glioma cells. Int. J. Oncol. 2018, 53, 2758–2768. [Google Scholar] [CrossRef] [Green Version]
- Kang, Z.H.; Wang, C.Y.; Zhang, W.L.; Zhang, J.T.; Yuan, C.H.; Zhao, P.W.; Lin, Y.Y.; Hong, S.; Li, C.Y.; Wang, L. Histone deacetylase HDAC4 promotes gastric cancer SGC-7901 cells progression via p21 repression. PLoS ONE 2014, 9, e98894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilson, A.J.; Byun, D.-S.; Nasser, S.; Murray, L.B.; Auyyanar, K.; Arango, D.; Figueroa, M.; Melnick, A.; Kao, G.D.; Augenlicht, L.H.; et al. HDAC4 Promotes Growth of Colon Cancer Cells via Repression of p21. Mol. Biol. Cell 2008, 19, 4062–4075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, H.J.; Lee, M.H.; Kang, H.L.; Kim, S.H.; Ahn, J.R.; Na, H.; Na, T.Y.; Kim, Y.N.; Seong, J.K.; Lee, M.O. Differential regulation of estrogen receptor alpha expression in breast cancer cells by metastasis-associated protein 1. Cancer Res. 2014, 74, 1484–1494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, D.Y.; Lin, T.E.; Lee, C.I.; Zhou, J.; Huang, Y.H.; Lee, P.L.; Shih, Y.T.; Chien, S.; Chiu, J.J. MicroRNA-10a is crucial for endothelial response to different flow patterns via interaction of retinoid acid receptors and histone deacetylases. Proc. Natl. Acad. Sci. USA 2017, 114, 2072–2077. [Google Scholar] [CrossRef] [Green Version]
- Wein, M.N.; Spatz, J.; Nishimori, S.; Doench, J.; Root, D.; Babij, P.; Nagano, K.; Baron, R.; Brooks, D.; Bouxsein, M.; et al. HDAC5 controls MEF2C-driven sclerostin expression in osteocytes. J. Bone. Min. Res. 2015, 30, 400–411. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Jin, X.; Ma, J.; Ding, D.; Huang, Z.; Sheng, H.; Yan, Y.; Pan, Y.; Wei, T.; Wang, L.; et al. HDAC5 Loss Impairs RB Repression of Pro-Oncogenic Genes and Confers CDK4/6 Inhibitor Resistance in Cancer. Cancer Res. 2021, 81, 1486–1499. [Google Scholar] [CrossRef]
- Feng, G.-W.; Dong, L.-D.; Shang, W.-J.; Pang, X.-L.; Li, J.F.; Liu, L.; Wang, Y. HDAC5 promotes cell proliferation in human hepatocellular carcinoma by up-regulating Six1 expression. Eur. Rev. Med. Pharmacol. Sci. 2014, 18, 811–816. [Google Scholar]
- He, P.; Liang, J.; Shau, T.; Guo, Y.; Hou, Y.; Li, Y. HDAC5 promotes colorectal cancer cell proliferation by up-regulating DLL4 expression. Int. J. Clin Exp. Med. 2015, 8, 6510–6516. [Google Scholar]
- Li, A.; Liu, Z.; Li, M.; Zhou, S.; Xu, Y.; Xiao, Y.; Yang, W. HDAC5, a potential therapeutic target and prognostic biomarker, promotes proliferation, invasion and migration in human breast cancer. Oncotarget 2016, 7, 37966–37978. [Google Scholar] [CrossRef]
- Zhong, L.; Sun, S.; Yao, S.; Han, X.; Gu, M.; Shi, J. Histone deacetylase 5 promotes the proliferation and invasion of lung cancer cells. Oncol. Rep. 2018, 40, 2224–2232. [Google Scholar] [CrossRef] [Green Version]
- Olzmann, J.A.; Li, L.; Chudaev, M.V.; Chen, J.; Perez, F.A.; Palmiter, R.D.; Chin, L.S. Parkin-mediated K63-linked polyubiquitination targets misfolded DJ-1 to aggresomes via binding to HDAC6. J. Cell Biol. 2007, 178, 1025–1038. [Google Scholar] [CrossRef] [Green Version]
- Bazzaro, M.; Lin, Z.; Santillan, A.; Lee, M.K.; Wang, M.C.; Chan, K.C.; Bristow, R.E.; Mazitschek, R.; Bradner, J.; Roden, R.B. Ubiquitin proteasome system stress underlies synergistic killing of ovarian cancer cells by bortezomib and a novel HDAC6 inhibitor. Clin. Cancer Res. 2008, 14, 7340–7347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Yamashita, H.; Toyama, T.; Sugiura, H.; Omoto, Y.; Ando, Y.; Mita, K.; Hamaguchi, M.; Hayashi, S.; Iwase, H. HDAC6 expression is correlated with better survival in breast cancer. Clin. Cancer Res. 2004, 10, 6962–6968. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakuma, T.; Uzawa, K.; Onda, T.; Shiiba, M.; Yokoe, H.; Shiabahara, T.; Tanzawa, H. Aberrant expression of histone deacetylase 6 in oral squamous cell carcinoma. Int. J. Oncol. 2006, 29, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Lei, Y.; Liu, L.; Zhang, S.; Guo, S.; Li, X.; Wang, J.; Su, B.; Fang, Y.; Chen, X.; Ke, H.; et al. Hdac7 promotes lung tumorigenesis by inhibiting Stat3 activation. Mol. Cancer 2017, 16, 170. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Samanta, A.; Song, X.; Iacono, K.T.; Bembas, K.; Tao, R.; Basu, S.; Riley, J.L.; Hancock, W.W.; Shen, Y.; et al. FOXP3 interactions with histone acetyltransferase and class II histone deacetylases are required for repression. Proc. Natl. Acad. Sci. USA 2007, 104, 4571–4576. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.G.; Xiao, T.; Zhu, W.; Yu, Z.Z.; Huang, X.P.; Yi, H.; Lu, S.S.; Tang, Y.Y.; Huang, W.; Xiao, Z.Q. HDAC7 promotes the oncogenicity of nasopharyngeal carcinoma cells by miR-4465-EphA2 signaling axis. Cell Death. Dis. 2020, 11, 322. [Google Scholar] [CrossRef]
- Uzelac, B.; Krivokuca, A.; Susnjar, S.; Milovanovic, Z.; Supic, G. Histone Deacetylase 7 Gene Overexpression Is Associated with Poor Prognosis of Triple-Negative Breast Cancer Patients. Genet. Test Mol. Biomark. 2021, 25, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Cao, F.; Yu, X.; Zhou, P.; Di, Q.; Lei, J.; Tai, Y.; Wu, H.; Li, X.; Wang, X.; et al. The expression of HDAC7 in cancerous gastric tissues is positively associated with distant metastasis and poor patient prognosis. Clin. Transl. Oncol. 2017, 19, 1045–1054. [Google Scholar] [CrossRef]
- Haberland, M.; Arnold, M.A.; McAnally, J.; Phan, D.; Kim, Y.; Olson, E.N. Regulation of HDAC9 gene expression by MEF2 establishes a negative-feedback loop in the transcriptional circuitry of muscle differentiation. Mol. Cell Biol. 2007, 27, 518–525. [Google Scholar] [CrossRef] [Green Version]
- Kanki, K.; Watanabe, R.; Nguyen Thai, L.; Zhao, C.H.; Naito, K. HDAC9 Is Preferentially Expressed in Dedifferentiated Hepatocellular Carcinoma Cells and Is Involved in an Anchorage-Independent Growth. Cancers 2020, 12, 2734. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Wu, Y.; Tang, X.; Xia, Y.; He, G.; Min, Z.; Li, C.; Xiong, S.; Shi, Z.; Lu, Y.; et al. HDAC inhibitors suppress c-Jun/Fra-1-mediated proliferation through transcriptionally downregulating MKK7 and Raf1 in neuroblastoma cells. Oncotarget 2016, 7, 6727–6747. [Google Scholar] [CrossRef] [PubMed]
- Okudela, K.; Mitsui, H.; Suzuki, T.; Woo, T.; Tateishi, Y.; Umeda, S.; Saito, Y.; Tajiri, M.; Masuda, M.; Ohashi, K. Expression of HDAC9 in lung cancer—Potential role in lung carcinogenesis. Int. J. Clin. Exp. Pathol. 2014, 7, 213–220. [Google Scholar] [PubMed]
- Rastogi, B.; Raut, S.K.; Panda, N.K.; Rattan, V.; Radotra, B.D.; Khullar, M. Overexpression of HDAC9 promotes oral squamous cell carcinoma growth, regulates cell cycle progression, and inhibits apoptosis. Mol. Cell Biochem. 2016, 415, 183–196. [Google Scholar] [CrossRef]
- Salgado, E.; Bian, X.; Feng, A.; Shim, H.; Liang, Z. HDAC9 overexpression confers invasive and angiogenic potential to triple negative breast cancer cells via modulating microRNA-206. Biochem. Biophys. Res. Commun. 2018, 503, 1087–1091. [Google Scholar] [CrossRef] [PubMed]
- Ridinger, J.; Koeneke, E.; Kolbinger, F.R.; Koerholz, K.; Mahboobi, S.; Hellweg, L.; Gunkel, N.; Miller, A.K.; Peterziel, H.; Schmezer, P.; et al. Dual role of HDAC10 in lysosomal exocytosis and DNA repair promotes neuroblastoma chemoresistance. Sci. Rep. 2018, 8, 10039. [Google Scholar] [CrossRef] [Green Version]
- Tao, X.; Yan, Y.; Lu, L.; Chen, B. HDAC10 expression is associated with DNA mismatch repair gene and is a predictor of good prognosis in colon carcinoma. Oncol. Lett. 2017, 14, 4923–4929. [Google Scholar] [CrossRef] [Green Version]
- Duan, B.; Ye, D.; Zhu, S.; Jia, W.; Lu, C.; Wang, G.; Guo, X.; Yu, Y.; Wu, C.; Kang, J. HDAC10 promotes angiogenesis in endothelial cells through the PTPN22/ERK axis. Oncotarget 2017, 8, 61338–61349. [Google Scholar] [CrossRef]
- Yang, Y.; Huang, Y.; Wang, Z.; Wang, H.-T.; Duan, B.; Ye, D.; Wang, C.; Jing, R.; Leng, Y.; Zi, J.; et al. HDAC10 promotes lung cancer proliferation via AKT phosphorylation. Oncotarget 2016, 7, 59388–59401. [Google Scholar] [CrossRef] [Green Version]
- Islam, M.M.; Banerjee, T.; Packard, C.Z.; Kotian, S.; Selvendiran, K.; Cohn, D.E.; Parvin, J.D. HDAC10 as a potential therapeutic target in ovarian cancer. Gynecol. Oncol. 2017, 144, 613–620. [Google Scholar] [CrossRef]
- Liu, X.; Wang, Y.; Zhang, R.; Jin, T.; Qu, L.; Jin, Q.; Zheng, J.; Sun, J.; Wu, Z.; Wang, L.; et al. HDAC10 Is Positively Associated With PD-L1 Expression and Poor Prognosis in Patients With NSCLC. Front. Oncol. 2020, 10, 485. [Google Scholar] [CrossRef] [PubMed]
- Bi, L.; Ren, Y.; Feng, M.; Meng, P.; Wang, Q.; Chen, W.; Jiao, Q.; Wang, Y.; Du, L.; Zhou, F.; et al. HDAC11 Regulates Glycolysis through the LKB1/AMPK Signaling Pathway to Maintain Hepatocellular Carcinoma Stemness. Cancer Res. 2021, 81, 2015–2028. [Google Scholar] [CrossRef] [PubMed]
- Thole, T.M.; Lodrini, M.; Fabian, J.; Wuenschel, J.; Pfeil, S.; Hielscher, T.; Kopp-Schneider, A.; Heinicke, U.; Fulda, S.; Witt, O.; et al. Neuroblastoma cells depend on HDAC11 for mitotic cell cycle progression and survival. Cell Death. Dis. 2017, 8, e2635. [Google Scholar] [CrossRef]
- Wang, W.; Fu, L.; Li, S.; Xu, Z.; Li, X. Histone deacetylase 11 suppresses p53 expression in pituitary tumor cells. Cell Biol. Int. 2017, 41, 1290–1295. [Google Scholar] [CrossRef] [PubMed]
- Lemercier, C.; Verdel, A.; Galloo, B.; Curtet, S.; Brocard, M.P.; Khochbin, S. mHDA1/HDAC5 histone deacetylase interacts with and represses MEF2A transcriptional activity. J. Biol. Chem. 2000, 275, 15594–15599. [Google Scholar] [CrossRef] [Green Version]
- Dressel, U.; Bailey, P.J.; Wang, S.C.; Downes, M.; Evans, R.M.; Muscat, G.E. A dynamic role for HDAC7 in MEF2-mediated muscle differentiation. J. Biol. Chem. 2001, 276, 17007–17013. [Google Scholar] [CrossRef] [Green Version]
- Fischle, W.; Dequiedt, F.; Hendzel, M.J.; Guenther, M.G.; Lazar, M.A.; Voelter, W.; Verdin, E. Enzymatic Activity Associated with Class IIHDACs Is Dependent on a Multiprotein ComplexContaining HDAC3 and SMRT/N-CoR. Mol. Cell 2002, 9, 45–47. [Google Scholar] [CrossRef] [Green Version]
- Parra, M. Class IIa HDACs—New insights into their functions in physiology and pathology. FEBS J. 2015, 282, 1736–1744. [Google Scholar] [CrossRef]
- Jeong, J.W.; Bae, M.K.; Ahn, M.Y.; Kim, S.H.; Sohn, T.K.; Bae, M.H.; Yoo, M.A.; Song, E.J.; Lee, K.J.; Kim, K.W. Regulation and destabilization of HIF-1a by ARD1-mediated acetylation. Cell 2002, 111, 709–720. [Google Scholar] [CrossRef] [Green Version]
- Lim, J.H.; Lee, Y.M.; Chun, Y.S.; Chen, J.; Kim, J.E.; Park, J.W. Sirtuin 1 modulates cellular responses to hypoxia by deacetylating hypoxia-inducible factor 1alpha. Mol. Cell 2010, 38, 864–878. [Google Scholar] [CrossRef]
- Geng, H.; Harvey, C.T.; Pittsenbarger, J.; Liu, Q.; Beer, T.M.; Xue, C.; Qian, D.Z. HDAC4 protein regulates HIF1alpha protein lysine acetylation and cancer cell response to hypoxia. J. Biol. Chem. 2011, 286, 38095–38102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aldana-Masangkay, G.I.; Sakamoto, K.M. The role of HDAC6 in cancer. J. Biomed. Biotechnol. 2011, 2011, 875824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mottet, D.; Bellahcene, A.; Pirotte, S.; Waltregny, D.; Deroanne, C.; Lamour, V.; Lidereau, R.; Castronovo, V. Histone deacetylase 7 silencing alters endothelial cell migration, a key step in angiogenesis. Circ. Res. 2007, 101, 1237–1246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, L.; Cueto, M.A.; Asselbergs, F.; Atadja, P. Cloning and functional characterization of HDAC11, a novel member of the human histone deacetylase family. J. Biol. Chem. 2002, 277, 25748–25755. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Zhu, W.G. Targeting histone deacetylases for cancer therapy: From molecular mechanisms to clinical implications. Int. J. Biol. Sci. 2014, 10, 757–770. [Google Scholar] [CrossRef] [Green Version]
- Thurn, K.T.; Thomas, S.; Raha, P.; Qureshi, I.; Munster, P.N. Histone deacetylase regulation of ATM-mediated DNA damage signaling. Mol. Cancer Ther. 2013, 12, 2078–2087. [Google Scholar] [CrossRef] [Green Version]
- Miller, K.M.; Tjeertes, J.V.; Coates, J.; Legube, G.; Polo, S.E.; Britton, S.; Jackson, S.P. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat. Struct. Mol. Biol. 2010, 17, 1144–1151. [Google Scholar] [CrossRef]
- Bhaskara, S.; Knutson, S.K.; Jiang, G.; Chandrasekharan, M.B.; Wilson, A.J.; Zheng, S.; Yenamandra, A.; Locke, K.; Yuan, J.L.; Bonine-Summers, A.R.; et al. Hdac3 is essential for the maintenance of chromatin structure and genome stability. Cancer Cell 2010, 18, 436–447. [Google Scholar] [CrossRef] [Green Version]
- Bhaskara, S.; Chyla, B.J.; Amann, J.M.; Knutson, S.K.; Cortez, D.; Sun, Z.-W.; Hiebert, S.W. Deletion of Histone Deacetylase 3 reveals critical roles in S-phase progression and DNA damage control. Mol. Cell 2008, 30, 61–72. [Google Scholar] [CrossRef] [Green Version]
- Kao, G.D.; McKenna, W.G.; Guenther, M.G.; Muschel, R.J.; Lazar, M.A.; Yen, T.J. Histone deacetylase 4 interacts with 53BP1 to mediate the DNA damage response. J. Cell Biol. 2003, 160, 1017–1027. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.; Hu, C.; Moses, N.; Haakenson, J.; Xiang, S.; Quan, D.; Fang, B.; Yang, Z.; Bai, W.; Bepler, G.; et al. HDAC6 regulates DNA damage response via deacetylating MLH1. J. Biol. Chem. 2019, 294, 5813–5826. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Xiang, S.; Joo, H.Y.; Wang, L.; Williams, K.A.; Liu, W.; Hu, C.; Tong, D.; Haakenson, J.; Wang, C.; et al. HDAC6 deacetylates and ubiquitinates MSH2 to maintain proper levels of MutSalpha. Mol. Cell 2014, 55, 31–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kotian, S.; Liyanarachchi, S.; Zelent, A.; Parvin, J.D. Histone deacetylases 9 and 10 are required for homologous recombination. J. Biol. Chem. 2011, 286, 7722–7726. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Wong, P.; Radany, E.; Wong, J.Y.C. HDAC Inhibitor, Valproic Acid, Induces p53-Dependent Radiosensitization of Colon Cancer Cells. Cancer Biother. 2009, 24, 689–699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shoji, M.; Ninomiya, I.; Makino, I.; Kinoshita, J.; Nakamura, K.; Oyama, K.; Nakagawara, H.; Fujita, H.; Tajima, H.; Takamura, H.; et al. Valproic acid, a histone deacetylase inhibitor, enhances radiosensitivity in esophageal squamous cell carcinoma. Int. J. Oncol. 2012, 40, 2140–2146. [Google Scholar] [CrossRef] [Green Version]
- Perona, M.; Thomasz, L.; Rossich, L.; Rodriguez, C.; Pisarev, M.A.; Rosemblit, C.; Cremaschi, G.A.; Dagrosa, M.A.; Juvenal, G.J. Radiosensitivity enhancement of human thyroid carcinoma cells by the inhibitors of histone deacetylase sodium butyrate and valproic acid. Mol. Cell Endocrinol. 2018, 478, 141–150. [Google Scholar] [CrossRef]
- Munshi, A.; Tanaka, T.; Hobbs, M.L.; Tucker, S.L.; Richon, V.M.; Meyn, R.E. Vorinostat, a histone deacetylase inhibitor, enhances the response of human tumor cells to ionizing radiation through prolongation of gamma-H2AX foci. Mol. Cancer Ther. 2006, 5, 1967–1974. [Google Scholar] [CrossRef] [Green Version]
- Baschnagel, A.; Russo, A.; Burgan, W.E.; Carter, D.; Beam, K.; Palmieri, D.; Steeg, P.S.; Tofilon, P.; Camphausen, K. Vorinostat enhances the radiosensitivity of a breast cancer brain metastatic cell line grown in vitro and as intracranial xenografts. Mol. Cancer Ther. 2009, 8, 1589–1595. [Google Scholar] [CrossRef] [Green Version]
- Saelen, M.G.; Ree, A.H.; Kristian, A.; Fleten, K.G.; Furee, T.; Hektoen, H.H.; Flatmark, K. Radiosensitization by the histone deacetylase inhibitor vorinostat under hypoxia and with capecitabine in experimental colorectal carcinoma. Radiat. Oncol. 2012, 7, 165. [Google Scholar] [CrossRef] [Green Version]
- Moertl, S.; Payer, S.; Kell, R.; Winkler, K.; Anastasov, N.; Atkinson, M.J. Comparison of Radiosensitization by HDAC Inhibitors CUDC-101 and SAHA in Pancreatic Cancer Cells. Int. J. Mol. Sci. 2019, 20, 3295. [Google Scholar] [CrossRef] [Green Version]
- Schlaff, C.D.; Arscott, W.T.; Gordon, I.; Tandle, A.; Tofilon, P.; Camphausen, K. Radiosensitization Effects of Novel Triple-Inhibitor CUDC-101 in Glioblastoma Multiforme and Breast Cancer Cells In Vitro. Int. J. Radiat. Oncol. Biol. Phys. 2013, 87, S650. [Google Scholar] [CrossRef]
- Xiao, W.; Graham, P.H.; Hao, J.; Chang, L.; Ni, J.; Power, C.A.; Dong, Q.; Kearsley, J.H.; Li, Y. Combination therapy with the histone deacetylase inhibitor LBH589 and radiation is an effective regimen for prostate cancer cells. PLoS ONE 2013, 8, e74253. [Google Scholar] [CrossRef] [Green Version]
- Groselj, B.; Ruan, J.L.; Scott, H.; Gorrill, J.; Nicholson, J.; Kelly, J.; Anbalagan, S.; Thompson, J.; Stratford, M.R.L.; Jevons, S.J.; et al. Radiosensitization In Vivo by Histone Deacetylase Inhibition with No Increase in Early Normal Tissue Radiation Toxicity. Mol. Cancer Ther. 2018, 17, 381–392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paillas, S.; Then, C.K.; Kilgas, S.; Ruan, J.L.; Thompson, J.; Elliott, A.; Smart, S.; Kiltie, A.E. The Histone Deacetylase Inhibitor Romidepsin Spares Normal Tissues While Acting as an Effective Radiosensitizer in Bladder Tumors In Vivo. Int. J. Radiat. Oncol. Biol. Phys. 2020, 107, 212–221. [Google Scholar] [CrossRef] [Green Version]
- Marampon, F.; Di Nisio, V.; Pietrantoni, I.; Petragnano, F.; Fasciani, I.; Scicchitano, B.M.; Ciccarelli, C.; Gravina, G.L.; Festuccia, C.; Del Fattore, A.; et al. Pro-differentiating and radiosensitizing effects of inhibiting HDACs by PXD-101 (Belinostat) in in vitro and in vivo models of human rhabdomyosarcoma cell lines. Cancer Lett. 2019, 461, 90–101. [Google Scholar] [CrossRef]
- Banuelos, C.A.; Banath, J.P.; MacPhail, S.H.; Zhao, J.; Reitsema, T.; Olive, P.L. Radiosensitization by the histone deacetylase inhibitor PCI-24781. Clin Cancer Res. 2007, 13, 6816–6826. [Google Scholar] [CrossRef] [Green Version]
- Rivera, S.; Leteur, C.; Mégnin, F.; Law, F.; Martins, I.; Kloos, I.; Depil, S.; Modjtahedi, N.; Perfettini, J.L.; Hennequin, C.; et al. Time dependent modulation of tumor radiosensitivity by a pan HDAC inhibitor: Abexinostat. Oncotarget 2017, 8, 56210–56227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, J.I.; Choi, C.; Shin, S.W.; Son, A.; Lee, G.H.; Kim, S.Y.; Park, H.C. Valproic Acid Sensitizes Hepatocellular Carcinoma Cells to Proton Therapy by Suppressing NRF2 Activation. Sci. Rep. 2017, 7, 14986. [Google Scholar] [CrossRef] [PubMed]
- Choi, C.; Lee, G.H.; Son, A.; Yoo, G.S.; Yu, J.I.; Park, H.C. Downregulation of Mcl-1 by Panobinostat Potentiates Proton Beam Therapy in Hepatocellular Carcinoma Cells. Cells 2021, 10, 554. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).