Abstract
An overview of the biophysics activity at the Department of Physics and Chemistry Emilio Segrè of the University of Palermo is given. For forty years, the focus of the research has been on the protein structure–dynamics–function paradigm, with the aim of understanding the molecular basis of the relevant mechanisms and the key role of solvent. At least three research lines are identified; the main results obtained in collaboration with other groups in Italy and abroad are presented. This review is dedicated to the memory of Professors Massimo Ugo Palma, Maria Beatrice Palma Vittorelli, and Lorenzo Cordone, which were the founders of the Palermo School of Biophysics. We all have been, directly or indirectly, their pupils; we miss their enthusiasm for scientific research, their deep physical insights, their suggestions, their strict but always constructive criticisms, and, most of all, their friendship. This paper is dedicated also to the memory of Prof. Hans Frauenfelder, whose pioneering works on nonexponential rebinding kinetics, protein substates, and energy landscape have inspired a large part of our work in the field of protein dynamics.
1. Protein Dynamics: Equilibrium Fluctuations, Dynamics in Confinement, and Structural Relaxation
The wording “protein dynamics” may cover a wide ensemble of biophysical effects ranging from rheology and transport phenomena to angstrom-sized atomic movements in the active site of proteins. The scientific activity of the Biophysics Groups of the Department of Physics and Chemistry-Emilio Segrè at the University of Palermo has focused on the study of protein equilibrium fluctuations and substates interconversions occurring mainly in the picoseconds/nanoseconds time range and in the angstroms space range, both in solution and in confinement. Studies have been extended also to tertiary and quaternary structural relaxations that occur in wide time (from picoseconds to seconds or more, depending on temperature, solvent, confinement, etc.) and space (from tenths of an angstrom to nanometers, depending on the relaxation studied) ranges.
1.1. Protein Equilibrium Fluctuations
Our interest in this research field originated in the late 1970s from the suggestion by M.U. Palma to look at the temperature-induced difference spectra in hemoglobin (Hb). We analyzed the difference spectra using Gaussian or skewed Gaussian components; this enabled us to suggest the relevance of the effects of internal molecular dynamics and anharmonicity [1]. Correctly, it was noted that the absorption bands of both Oxy-and Deoxy-Hemoglobin show a composite structure, disproving interpretations in terms of single-skewed Gaussians. However, the rather limited temperature range investigated (5–35 °C) did not allow more quantitative conclusions to be drawn.
In the following years, thanks also to the leading efforts of L. Cordone, we developed a home-made optical dewar for the measurement of high-quality optical absorption spectra in the temperature interval of 10–320 °K; we also developed a protein-friendly protocol of sample preparation and handling to obtain homogeneous and transparent samples in the whole temperature interval. The temperature dependence of the hemoglobin and myoglobin (Mb) absorption spectra was first analyzed [2,3]. We observed a narrowing and a peak shift of all the bands with decreasing temperature, paralleled by a relevant increase in the integrated intensities of the charge-transfer bands of deoxyhemoglobin. Analysis of the first and second moment of the bands (modeled by Gaussian profiles), using the harmonic Franck–Condon approximation, gave information on the dynamic properties of the heme pocket. This approach was applied also to study the temperature dependence of the optical absorption spectra of blue-copper proteins [4], superoxide dismutase [5], and hemocyanin [6], also in relation with different solvent compositions.
We soon realized the intrinsic limits of the above “band moments approach” and that to obtain more in-depth information on structure–dynamics–function relationships in heme proteins, a more analytical approach to the band profile was needed. For this purpose, we used the theory of Franck–Condon linear coupling between an allowed electronic transition of a chromophore and the collective normal modes of the surrounding atoms; this approach was applied to obtain an analytical expression suitable for fitting the Soret band profile of hemeproteins at various temperatures [7]. The “0–0” transition was therefore modelled as a convolution of Lorentzian and Gaussian functions (Voigt functions) to account for the coupling of the Lorentzian band profile with a “bath” of low-frequency vibrational modes giving rise to a Gaussian broadening; the vibronic structure of the spectra was attributed to the coupling of the 0–0 transition with high-frequency modes. Spectral analysis was usually performed in the frequency domain; essentially analogous results were also obtained by using the full correlator approach in the time domain [8].
Using this approach, we were able to separate the contributions to the overall Soret band width arising from vibrational coupling with the high-frequency modes of the heme (as already shown by resonance Raman spectroscopy) from those arising from the coupling with the protein active site “soft” modes. The temperature dependence of the above quantities gives quantitative information on the dynamic properties of the protein active site and on their dependence upon protein ligation state and tertiary and/or quaternary conformation. It should also be mentioned that, for the deoxy derivatives, conformational heterogeneity gives rise to inhomogeneous broadening and contributes substantially to the linewidth [9,10]. One of the most relevant results obtained was to put in evidence the onset, at temperatures near 170 K, of large deviations of the Gaussian linewidth from the predictions of the harmonic model. These deviations are due to the onset of large-scale anharmonic motions that can be attributed to jumping among different conformational substates of the protein [11]. The remarkable agreement of our temperature-dependent optical absorption data with analogous data obtained with Mössbauer spectroscopy, neutron, and X-ray scattering (i.e., with techniques that monitor the motions of different atoms in different parts of the proteins) and with computer simulations adds to the generality and relevance of the phenomenon [12].
Relevant advancements in the understanding of the structure–dynamics–function relationships in heme proteins were obtained by combining the above low-temperature optical absorption spectroscopy approach with several “state of the art” experimental (such as, e.g., low-temperature flash photolysis, Raman, and Infra-Red spectroscopies) and computer simulation (such as, e.g., normal modes analysis and MD simulations) techniques. Experiments were extended to proteins with different functional roles (such as hemoglobins from different animal species [13,14,15,16] and mutant and/or crosslinked human hemoglobins [17,18,19,20] but also to cytochrome c and to various metal-substituted model porphyrins [21,22,23,24]. In this last case, by exploiting optical absorption spectroscopy in combination with resonance Raman spectroscopy, we were able to fully characterize the vibrational coupling of the Soret and to put in evidence the coupling of the out-of-plane motions of the porphyrin-central metal system with solvent motions [25].
1.2. Dynamics of Proteins in Confinement
Due to the high extinction coefficient, the optical absorption experiments described in the preceding section were performed on rather diluted protein solutions (concentrations of the order of 0.1–1% by weight) far away from the conditions experienced by proteins in vivo. In fact, within the cells, proteins are in a confined and crowded environment provided by other proteins and/or cell walls or other supramolecular structures. People usually speak of “soft” and “hard” confinement.
Soft confinement of biomolecules can be obtained either by a concentration increase (crowding or even self-crowding) or by coating them with suitable cosolutes; hard confinement is usually obtained by trapping the biomolecule, and its hydration layer, within the pores of a solid, crystalline, or glassy matrix. In both cases, the possible available conformations are reduced due to either excluded volume effects or to hindering of conformational transitions.
In this section, we will describe our studies of dynamics on protein powders at various hydration levels (where soft self-confinement is provided by other proteins of the same type) and on proteins encapsulated, via the sol–gel method, within the pores of a silica matrix (where hard confinement is provided by the rigid, hydrophilic pore walls). Elastic, quasielastic, and inelastic neutron scattering has been used as the main experimental technique, together with dielectric spectroscopy and differential scanning calorimetry [26], to investigate the dynamics in wide temporal windows and the thermodynamics of the investigated systems.
We first studied the effect of hard confinement on the so-called Protein Dynamical Transition (PDT), i.e., the steep increase in protein nonexchangeable hydrogen atoms’ mean square displacements linked to the onset of anharmonic protein motions, occurring at temperatures around 180 K and first observed by Doster and collaborators [27] in myoglobin hydrated powders. We showed that geometrical confinement within the matrix plays a crucial role in protein dynamics and conformational stability, the effect being essentially a reduction of collective protein motions likely related to the slowing down of solvent diffusion [28,29,30].
It is highly debated whether the PDT is a real physical effect, or a mere instrumental resolution effect linked to the fact that the protein–solvent β relaxation time enters at this temperature into the time window of the neutron spectrometer used. The question is complicated by the fact that the rotational activation of methyl groups belonging to amino acids side chains (MGA) is also present in myoglobin samples. The two effects are superimposed, and this complicates the analysis and interpretation of experimental data. By using neutron spectrometers of different time resolutions and homomeric polypeptides containing or not methyl groups (e.g., polyalanine and polyglycine) at various hydration levels [31,32], we were able to disentangle MGA and PDT and clearly showed that MGA, being an activated process, depends on temperature and instrumental resolution, whereas PDT (i.e., the appearance of large-scale anharmonic motions) is a real physical effect whose onset, in myoglobin hydrated powders, does not depend upon instrumental resolution and occurs at about 220 K [33]. Complementary differential scanning calorimetry and dielectric spectroscopy studies enabled us to suggest that the PDT is linked to the fragile-to-strong liquid–liquid transition occurring in water confined at the protein surface (hydration water) at approximately the same temperature [34].
This fascinating suggestion confirmed the biophysical relevance of detailed investigations of the properties (structure, dynamics, and thermodynamics) of confined water [35,36,37,38]. Further experimental evidence for the presence of a fragile-to-strong liquid–liquid crossover in supercooled water confined in silica gels has been obtained by our group by studying the temperature dependence of the so-called boson peak [39] and of the infrared bending bands [40] and by investigating, with elastic neutron scattering, the pressure dependence of mean square displacements (MSD) of confined water hydrogen atoms [41].
In recent works, the fascinating field of water diffusional dynamics inside living cells, and in the brain, has also been investigated by our group with quasielastic neutron scattering and diffusion nuclear magnetic resonance experiments [42,43,44].
1.3. Structural Relaxations
The research activity in this field has been devoted initially to the study of the different tertiary relaxations that occur in myoglobin, the so-called “hydrogen atom” of biophysics (obviously lacking the quaternary structure) in solution [45], and in human hemoglobin locked in different quaternary structures by encapsulation in a silica hydrogel able to prevent any quaternary conformational transitions in the time window of the experiments [46]. Flash photolysis pump-probe experiments were performed by time-resolved optical absorption spectroscopy in the Soret spectral region in the nanoseconds to milliseconds time interval and at temperatures between 120 and 260 K. We were able to investigate the energy landscape of these proteins and to characterize the time course of the so-called Kinetic Hole Burning effect and of the structural relaxations linked to ligand migration.
We then focused on the study of the quaternary R↔T transition of hemoglobin. Initially, we used the approach, developed in our laboratory, of hemoglobin encapsulation in porous silica matrices using coencapsulated solvents of suitable viscosity to slow down the characteristic times of the transition and to bring them in the temporal range of minutes/hours, accessible with a conventional spectrophotometer. We were able to identify spectroscopic markers of this transition, monitoring conformational variations at the heme pocket and at the α1β2 subunit interface, and this enabled us to study their temporal evolution and their dependence upon the viscosity of the solvent coencapsulated within the silica matrix [47,48]. The multistep character of the R↔T transition was clearly evidenced; moreover, the characteristic times of the transition were shown to depend on the solvent viscosity according to a power law, which suggested that the conformational transitions of the protein may be “slaved” to the solvent.
Optical spectroscopy, however, does not give direct structural information. In fact, to study the proteins’ structural relaxations more in depth, it is necessary to develop an experimental approach able to directly monitor the structure of proteins in solution with suitable space and time resolutions. In this respect, the Palermo group, within a prestigious international collaboration at the ESRF, Grenoble, France, has shown that time-resolved wide-angle X-ray scattering (TR-WAXS) using synchrotron radiation can be used to accurately probe protein conformational changes in solution with angstrom and nanosecond space and time resolutions [49]. TR-WAXS combines high time resolution with high structural sensitivity and can monitor changes in the position of all atoms of the protein rather than only structural modifications around a given spectroscopic marker (either endogenous or exogenous); therefore, it is complementary to time-resolved optical spectroscopy and allows one to track the time evolution of tertiary and quaternary conformational transitions.
We have used TR-WAXS to study the time course of the tertiary and quaternary change that occurs after laser flash photolysis of human HbCO [50]. The main result obtained was the observation of a multistep R->T transition consisting of a fast (characteristic time <300 ns) tertiary relaxation involving the clamshell formed by the E and F helices, followed by a quaternary relaxation with characteristic time ~2 μs and involving the relative rotation of αβ dimers. This last time constant was clearly identified as the time constant of the R0->T0 transition of human hemoglobin in nearly physiological solution at pH = 7 and room temperature, a new and relevant result.
TR-WAXS experiments were extended to human hemoglobin in physiological conditions, i.e., directly inside the red blood cells [51] and to the HbYG recombinant mutant hemoglobin, to test the kinetic validity of the Monod–Wyman–Changeux allosteric model [52].
The intrinsic time resolution of TR-WAXS experiments (>100 ps) does not allow one to follow structural events occurring in hemeproteins (e.g., myoglobin) in the sub-picosecond–picosecond time scale following ligand photolysis. In fact, it has been suggested that the initial ultrafast protein response to the photolysis of the iron–ligand bond by an intense laser pulse can be described as a quake-like motion, since the strain release propagation through the protein is like the propagation of acoustic waves during an earthquake. We used the femtosecond X-ray pulses produced by the Linear Coherent Light Source (LCLS) X-ray free-electron laser (X-FEL) to visualize the structural response of Mb after photolysis and therefore obtain direct experimental evidence of the hypothesized protein quake [53]. Results have confirmed the protein quake hypothesis: in fact, the photolysis-induced structural perturbation of the active site propagates to the global protein structure with a timescale set by the acoustic speed of sound.
The data also show that the radius of gyration and the volume of Mb exhibit an underdamped oscillatory time dependence in the picosecond time range (see Figure 1), implying a collective behavior of the protein atoms in this time scale. This intrinsically ballistic protein motion is captured by ultrafast studies but is generally obscured in measurements on large protein ensembles at longer time scales, due to ensemble and time averages.
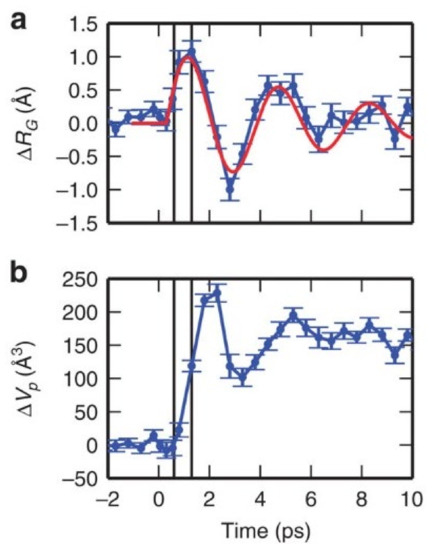
Figure 1.
Time-dependent changes of protein structural parameters. Panel (a): radius of gyration; the red curve represents a damped oscillation with a 3.6 ps time period and 6 ps decay time. Panel (b): protein volume. Reproduced with changes from Ref. [53], under the Creative Commons CC BY 4.0 license.
This is pictorially shown in Figure 2, where it is suggested that elementary single-protein motions have a quake-like ballistic nature, and therefore, the time evolution of an observable in an idealized single-molecule experiment has an oscillatory behavior, as reported in Figure 2a (red curve). Experiments on large protein ensembles (blue curve) can detect such quake-like oscillatory motions only at very short times (step I), where the initial synchronization imposed by the photoexcitation pulse is still preserved. At longer times (step II), the time evolution observed on a macroscopic protein sample is dominated by ensemble averaging and thermally activated transitions from one protein state to the other, which result in the ‘exponential-like’ kinetics typically observed in most time-resolved experiments at room temperature; indeed, in this case, different protein molecules will undergo the same structural transition but with a statistically distributed onset, resulting in exponential kinetics. This is shown in Figure 2b, in terms of a simplified free energy–conformational coordinate diagram.
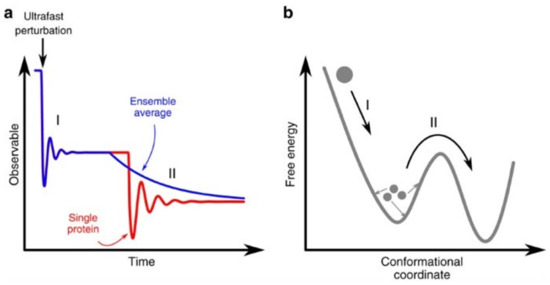
Figure 2.
Effect of ensemble average on an experimental observable after an ultrafast perturbation. (a) Time evolution of an observable in an idealized single-molecule experiment (red curve) and on large protein ensembles (blue curve). In the latter case, experiments can detect quake-like transitions only at very short times (step I), while at longer times (step II), the time evolution is dominated by thermally activated transitions among protein states, which result in an exponential-like kinetics. (b) Simplified free-energy diagram illustrating the difference between a downhill transition along the energy landscape (step I) and a thermally activated process (step II). Reproduced without changes from Ref. [53] under the Creative Commons CC BY 4.0 license.
Further experiments performed at the LCLS X-ray free electron laser with nearly 100 fs (FWHM) time resolution using time-resolved X-ray absorption measurements at selected energies near the Fe K-edge gave relevant information on more localized motions in the heme pocket of Mb after flash-photolysis, involving the doming of the heme and the displacement of the iron atom out of the heme plane [54]. Our data highlighted a two-step heme response whereby an initial fast (~70 fs) relaxation associated to a partial heme doming is followed by a slower (~400 fs) component that can be attributed to a residual iron out-of-plane motion coupled to the displacement of the Mb F-helix.
1.4. Myoglobin Structure–Dynamics–Function with Simulations
As already stated in the previous sections, Myoglobin is a test case for understanding how structure, dynamics, and function are related. Mb binds small ligands (O2, CO, and NO) at the iron atom in the heme. The active site is buried in the protein interior, and no pathway to it is visible. For long time results from experiments were interpreted, postulating the existence of only one pathway for ligand escape to the solvent, called histidine gate, very close to the heme. Molecular dynamics (MD) simulations, performed by artificially accelerating the ligand motion [55], showed that the ligand diffuses inside the protein interior and visits some hydrophobic cavities before escaping to the solvent. Years later, experiments determining the protein structure at different times after CO photolysis found the ligand in these cavities [56]. It was then proposed that multiple cavities in the protein interior play a role in the ligand-binding kinetics [57].
In principle, as stated, MD simulations could provide a more detailed description of the ligand pathways than the one provided by experiments. However, the time scales of ligand diffusion could be unattainable in full atomistic MD simulations. In an alternative to MD, one could perform free-energy calculations to calculate the potential of mean force (PMF) underlying the process. We investigated the pathways of CO diffusion inside myoglobin and to the solvent, in water at room temperature, by reconstructing the three-dimensional PMF of the CO position inside the protein. We assumed that the ligand diffusion could be described as a ligand surfing on the PMF, hopping between the PMF local minima, following the so-called minimum free-energy paths (MFEPs) [58]. Results showed that the local minima correspond to the known binding cavities in the protein matrix. Furthermore, the paths connecting the local minima form a complex network (Figure 3), starting from the primary docking site (or Distal Pocket, DP). In this network, the histidine gate was found to be the shortest and, most importantly, no intermediate minima were found along it.
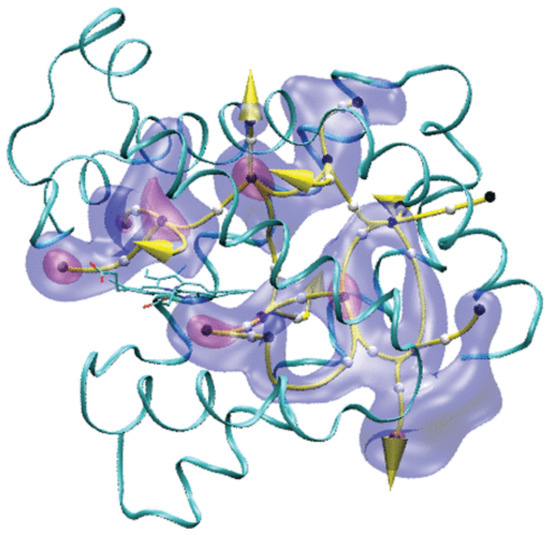
Figure 3.
CO migration pathways inside Mb. The yellow curves are MFEPs and identify possible CO routes. Two isosurfaces of the PMF map are shown (red, 2.0 kcal/mol; blue, 5.0 kcal/mol; with respect to the PFM at the Xenon 4 cavity, taken as zero). The white and black spheres represent, respectively, the locations of the energy barriers and the local minima along the pathways. The yellow arrows represent the locations of the CO exits to the solvent. The protein’s backbone is represented as ribbons and the heme as sticks. Reprinted with permission from Ref. [58]. Copyright 2010 American Chemical Society.
In another work, we modeled the processes of photolysis and geminate recombination of NO in myoglobin under the condition of continuous illumination [59]. We provided, for the first time, a single computational framework in which we could model all the events at the same time, e.g., photon absorption, ligand dissociation, diffusion in the protein matrix, and recombination back to the iron. We also monitored the relaxation of the photon energy into the protein matrix. The multiple-photons absorption put the system in a nonequilibrium state, and the following relaxation to equilibrium, after removing the illumination, was studied by exploiting the so-called D-NEMD approach [60].
Simulations were performed for the native myoglobin and for the mutants V68W and L29F. Results showed that the time and space scale of the geminate recombination and of the energy-relaxation processes are peculiar to each protein. Oscillations in the kinetic energy of the heme were observed on the picosecond time scale, i.e., the same time scale of the oscillation of Mb structural quantities observed in the X-FEL experiments, after the protein-quake [53]. We observed an energy funneling from the heme to the FG corner, a result that we interpreted in the frame of the allosteric transition in hemoglobin. Indeed, thermal excitation could induce structural changes of the FG corner of the β chain with respect to the C helix of the α chain, which are necessary to initiate the quaternary, large-scale motions, bringing the protein from the deoxy state to the oxy state.
2. Protein Aggregation Processes
The Biophysics Groups of the Department of Physics and Chemistry—Emilio Segrè at University of Palermo also addressed the molecular mechanisms underlying protein aggregation processes, because of their relevance in many fields of research, such as biophysics, biochemistry, and protein science. These processes play a critical role in many human diseases. Under specific destabilizing conditions, all proteins seem to have the ability to “misfold” and assemble themselves into supramolecular aggregates. For the research activities in this field at Palermo, the starting point was the pioneering scientific works of Profs. Massimo Ugo Palma and Beatrice Palma-Vittorelli, and their collaborators, who approached the problem from a point of view related to phase separation, from liquid–liquid demixing (LLD) to liquid–solid state transition [61,62,63,64].
The aggregation processes are the result of molecular mechanisms proceeding on different scales of time and space through different and interconnected phases [61,65,66]. The processes involve several steps as conformational and structural changes at the single-protein level, nucleation processes, oligomer formation, and protein–protein interactions of various nature with consequent formation of new supramolecular structures with different properties. General principles underlying aggregation and crystallization were suggested in Ref. [67].
2.1. Kinetics of Protein Aggregation
Since 2003, studies have focused on multiple aspects of aggregation kinetics, followed by complementary experimental techniques. This approach aimed to distinguish the hierarchies and the temporal evolution of the different paths that characterize aggregation processes studied through detailed kinetics of the aggregation of proteins such as Bovine Serum Albumin (BSA), a well-known globular protein formed by six helices essentially made of al α-helices. At room temperature, the tertiary structure of this model protein is well-defined and stable in its monomeric form [66]. As the temperature increases, results have shown that some molecular regions become available to new intermolecular interactions which produce soluble aggregates. In this study, BSA concentration was chosen at levels low enough to be far from the LLD instability region, thus allowing for the obtaining of information on tertiary and secondary structural changes which match with the initial intermolecular cross-linking processes.
The experimental approach consists of a kinetic study of steady-state fluorescence of intrinsic tryptophan of BSA. BSA has two tryptophans in two different domains: one, Trp-134, is buried in a hydrophobic pocket of domain I situated in the proximity of the protein surface, and the other, Trp-214, is in an internal portion of domain II [68]. Because their quantum yield depends on interaction with the neighbor environment and solvent exposure, the time evolution of tryptophan’s emission spectra allows us to obtain information on conformational changes in both these domains [69]. Most importantly, the extent of the overall aggregation process can be contemporary, followed by Rayleigh scattering of the excitation light (see Figure 4a) and circular dichroism, allowing for the highlighting of the occurrence of ordered β-sheet aggregates along the whole aggregation. The systematic experimental approach and the comparison of the result on BSA aggregation in different solution conditions allowed us to confirm that the progress of aggregates formation requires, as a minimum, the conformational changes at the tertiary structure level such as the partial unfolding of some domains. The role of electrostatic interaction was also highlighted. At pH close to the isoelectric point of BSA, the formation of intermolecular β-structures was not observed, and the protein was found to undergo a faster aggregation mainly toward hydrophobic interactions. At pH far from the isoelectric point, the formation of smaller aggregates was found to be pointed up by secondary structure changes probably arising in monomers which mutually interact to arrange an ordered structure. The onset of these changes is dependent on an opening of the tertiary structure, allowing the involvement of the free cysteine (Cys-34) in forming supramolecular assemblies [66,70,71].
Results obtained in the low-concentration regime are in line with the ones obtained by Militello et al., 2004 [72], where the initial steps of the BSA aggregation were followed by observing the infrared absorption bands of Amides to obtain complementary information on tertiary and secondary structure changes. The experimental approach consisted of studying the effects of pD changes on aggregation kinetics to understand if and how intra- and intermolecular interactions can depend on surface net charge. Time evolution of the Amide II band is a very strong “sensor” of the H–D exchange of the partially unfolded protein, so it allows for the obtaining of evidence of the conformational changes at the tertiary level. Analogously, changes at the level of the secondary structures are monitored by the time evolution of the Amide I’ band profile (see Figure 4b). This last band is a probe of α-helix, random coil, and β-sheet contents and gives information on protein–protein aggregation through the appearance of two new shoulders attributed to new β-sheet formation. The final picture proposed based on the overall experimental data allows us to sustain that the tertiary structures go toward partially unfolded conformations, and they are the first step that drives toward the macromolecular aggregation. Afterwards, the aggregation mechanism proceeds in an ordered way to form β-aggregates that prevalently originate from the a-helix changes for producing aggregates of small dimensions.
This was framed in the context of the emerging (at that time) evidence of the association of the deposits of aggregates and of amyloid aggregates with neurological pathologies such as Alzheimer’s, Parkinson’s, and Creutzfeldt–Jacob’s diseases [73,74,75]. Moreover, oligomers of misfolded proteins appear to be yet a further common denominator in these pathologies. Such structures are held to be intermediate forms of aggregation pathways, and they are believed to be key effecters of cytotoxicity and causes of a variety of amyloid-related diseases [76,77]. These ideas are nowadays well-assessed, and it is commonly accepted that amyloidogenic proteins or peptides, which do not share any sequence homology or common fold, form oligomer fibrils with relevant structural similarities, thus suggesting that common laws regulate both correct protein folding and aggregation. From a practical point of view, this means that a large variety of different aggregated protein structures can be achieved in vitro by selecting specific physicochemical conditions. Protein concentration, temperature, and presence of cosolvents, salts, or different thermal treatments may regulate the physicochemical properties of amyloid aggregate structures including size, molecular structure, and stability [78,79,80].
In the context of supramolecular assembly and of relevance for amyloid formation, nucleation mechanisms were also considered using different model systems: results pointed out that the destabilized protein molecules, after partial misfolding, interact with each other in forming a new high-energy species, the nucleus, from which, by subsequent addition of protein molecules, amyloid fibrils are formed [81,82,83]. In line with the literature, experimental results on the insulin model have shown that besides the tip-to-tip elongation of the fibrils, other highly cooperative growth mechanisms at the surface of already-formed fibrils in solution could happen, giving rise to spatially heterogeneous mechanisms such as branching, fragmentation, and secondary nucleation.
In the paper by Foderà et al., 2008 [82], fibril formation was followed through the fluorescence of Thioflavin T (ThT), a selective dye for the detection of amyloid fibrils [84]. With an excitation wavelength of 450 nm, this dye, in an aqueous solution, is characterized by a low emission quantum yield (in the 475–600 nm region). In contrast, in the presence of amyloid fibrils, it forms a highly fluorescent ThT-fibril complex, therefore allowing the use of ThT as a diagnostic dye for the detection of the presence and the rate of amyloid fibrils. The binding site for this dye is the cross-β-sheet structure in protofilaments, protofibrils, and fibrils. The experimental approach based on the analysis of ThT kinetics at different insulin concentrations revealed the intrinsic variability of the kinetic profile and its dependency on the concentration (see Figure 4, Panel c). Deeply looking at the data, it was evident that both the rate of fibrillation and the ThT fluorescence intensity reach saturation at high protein concentration. This brought to the hypothesis that, since ThT fluorescence depends on the accessibility on the surface of the fibrils, the overall fibrillation kinetics is mainly directed by this accessible surface through secondary nucleation mechanisms. Moreover, a statistical study of fibrillation has also revealed that the early stages of the process were affected by stochastic nucleation events.
In the same years, the systematic approach based on kinetics revealed technical aspects to be taken in account when using Thioflavin T, such as the possibility of the dye being chemical modified at basic pH [85] or triggering fibrils formation [86,87].
Based on the previous studies, with a similar kinetic approach and exploiting the power of quantitative fluorescence microscopy, the ongoing research is currently focused on the use of amyloid superstructures as new biocompatible biomaterials whose properties can be tailored modulating protein–protein and protein–solvent interactions [88]. The studies on protein aggregation were extended to the mutually disruptive interaction between cell membranes and aggregating proteins with the idea that in this case, common laws and interactions also regulate these phenomena [89,90,91]. Mutually disruptive mechanisms can be activated by membrane presence, promoting aggregation-prone conformations in proteins or favoring nucleation events and/or protein insertion in the membrane. This, in turn, may induce membrane destabilization, thinning, or breakage, and lipids can be incorporated into coaggregates [92,93].
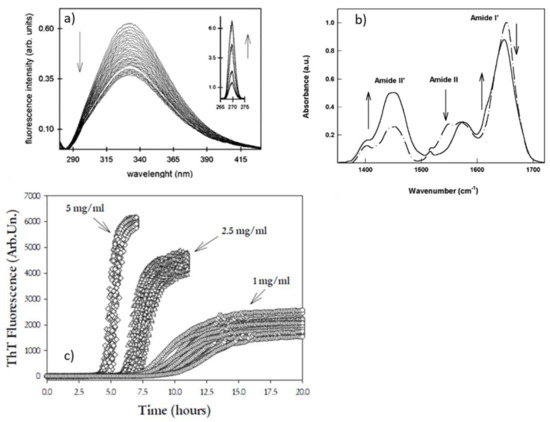
Figure 4.
Representative kinetics measurements on multiple observables that can be used to monitor the complexity of supramolecular assembly. (a) Time evolution of intrinsic fluorescence of BSA during incubation at high temperature; the inset shows Rayleigh scattering growth occurring simultaneously [66]. (b) FTIR spectra of BSA in its native form (dashed-dotted line) and in the aggregate aggregated form (solid line) [72]. Reprinted from Refs. [66,72] with permission from Elsevier (license number 5405291430367, 5405300216637). (c) ThT fluorescence kinetics for a human insulin sample at three different protein concentrations. Reprinted with permission from Ref. [82]. Copyright 2008 American Chemical Society.
2.2. The Role of Glycation in Protein Aggregation
In several papers [94,95,96], our group has studied the important role played by serum albumin in the transport of metabolites such as glucose and/or metals and how this binding influences the aggregation processes. In general, nonenzymatic glycation of circulating albumin can occur in case of hyperglycemia or diabetes, and a process, known also as the Maillard reaction, is a slow, nonenzymatic reaction that initially involves the attachment of glucose or derivatives with free amine groups of albumin (lysine or arginine) or free cysteine to reversibly form a Schiff base product, leading to the formation of stable Amadori products; several consequent rearrangements, oxidation, and polymerization of these early-stage glycation products give rise to irreversible conjugates, named advanced glycation end-products (AGE). This glycation process of albumin could generate thermodynamically stable aggregates with high content of β-sheet structures compared with its nonglycated form [97]. Our studies have demonstrated glycation’s protective role in the thermal aggregation of serum albumin. In particular, glycated albumin is characterized by aggregate formations but exhibits a reduced propensity for thermal aggregation.
On the other hand, metal ions such as copper or zinc are involved in the development of neurodegenerative pathologies and metabolic diseases such as diabetes mellitus, where, for example, albumin can be exposed to an increased level of glycation. In particular, both the Zn(II) and Cu(II) metabolisms and concentrations seem to have an injurious consequence on diabetes development [98]. Cu(II) concentrations have been found in the blood serum of diabetes mellitus patients [99], and treatment with a copper chelating agent seems to diminish insulin resistance and improve glucose intolerance. On the contrary, the Zn(II) plasma concentration is lower in diabetic patients than in healthy subjects, and patients treated with Zn(II) exhibited beneficial effects [100]. We have studied that both these metals promoted the inhibition of albumin glycation in a concentration-dependent manner. In particular, the copper ions appear to behave as a more efficient inhibitor in the glycation than zinc. Indeed, Cu(II) ions can reduce glycation (Amadori products) by a maximum of 94%, while Zn (II) ions can by 48%. In general, an increase in albumin glycation is coupled with a reduction of free amino groups such as lysine or arginine, the main accessible glycation sites [101]. For us, this attenuation in glycation due to the presence of metals is not automatically associated with the availability of these free primary amino groups of the protein, but with modification of these last groups for parallel processes including aggregation or oxidation initiated by the presence of both metal and glucose. First, as discussed in the previous paragraph, because of heating, BSA molecules undergo partial unfolding and conformational changes of their native structure, originating from a structure prevalently constituted by α-helices to more open β-rich structures. These changes are, overall, controlled by pH value, temperature, time of heating, and protein concentration. Secondly, “as concerns the aggregation, in the presence of copper/glucose mixture, the unfolding of BSA proceeds with a secondary structure evolution into intermolecular β-sheet structures, as observed by the fibrillar morphology and bigger size of aggregates studied with AFM microscopy. So, both metal ions, Cu(II) and Zn(II), during glycation, promoted aggregation and contemporary-induced aggregate formation with different morphologies in dependence of the nature and concentration of the metal. Therefore, our studies have shown that glycation and aggregation should not generally be considered two competitive processes. For example, Zn(II) ions promote the heat-induced aggregation of the protein without completely preventing the glycation process, while the presence of glucose enhances the albumin affinity for Cu(II), reducing the glycation extent. We attributed this increase in copper affinity to an advantageous conformational change in the tertiary structure of albumin induced by the presence of glucose. We have also established that generally, the glycated albumin, by following the thermal process, fails its ability to self-assemble into aggregates [102].
From another point of view, strictly binding to the spectroscopic methods applied through tryptophan fluorescence, the main intrinsic probe which monitors the denaturation and folding of proteins also discussed in the previous paragraph, we have found that the presence of metal ions showed a quenching of tryptophan fluorescence due to partial unfolding or protein oxidation induced by ions themselves. The improved oxidative state of the albumin has been estimated following the redox status of Cysteine 34 and the carbonyl levels. As expected, both the metals contribute to enhanced protein oxidation in addition to induce glycation and/or aggregation processes. The oxidation of some glycation sites by Cu(II) or Zn(II) ions has been explained by us with the “antiglycative” potential effect of these metals. For this purpose, we suggested schematic models of glycation and metal-induced changes in the structure and the properties of albumin (see in Figure 5). The different type of metal coordination explains why Zn(II) ions are more likely to promote the aggregation of BSA than Cu(II) ions in a heat-induced albumin treatment [103]. Then, the metal-binding mode involving Zn(II) ions can promote intermolecular cross-link, leading to the formation of large aggregates, whereas Cu(II) ions are more prone to induce intramolecular chelation [95].
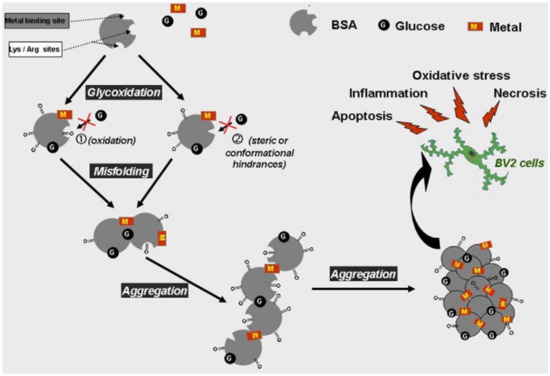
Figure 5.
Proposed schematic models for the glycation- and oxidation-induced changes in the structure and the properties of BSA molecule. Glycoxidation of albumin in the presence of metal and glucose induces protein misfolding and formation of high cytotoxic β-amyloid-like aggregates triggering microglia cell apoptosis and necrosis associated with oxidative stress. The inhibitory roles played by Zn(II) and especially Cu(II) ions in the glycation process could be attributed to potential oxidation at lysine and arginine residues (see (1)). We can hypothesize other contributing factors for “antiglycative” properties of Cu(II) and Zn(II): (i) The main lysine and arginine residues prone to glycation are not involved in specific metal binding, but some of these amino acids could be prevented from glycation because of metal coordination with adjacent or nearby histidine residues. Indeed, most histidine residues appear to be located near lysine or arginine, which could induce a steric hindrance (see (2)). (ii) Alteration in the tertiary and secondary structures of BSA induced by binding with metal ions could blunt the capacity of specific glycation sites to react with glucose residues (see (1)). Reproduced without changes from Ref. [94], Copyright 2014, with permission from Elsevier (license number 5405290900604).
The conformational changes involved in the process of glycoxidation, the different behavior of zinc and copper, and the specific inhibiting role played by copper in BSA glycation show that the glycation can protect the protein from thermal aggregation, that the metal ions are noncompletely chelated by the protein through the main binding sites, and that the role of nonspecific binding sites in glycation, aggregation, and oxidation is to make these processes not competitive.
3. Proteins in Extreme Conditions: Bioprotection by Saccharides and the Trehalose Peculiarity
Starting from 1998, part of the biophysics activity in Palermo was devoted to the investigation of saccharide amorphous systems containing carbon-monoxy myoglobin (MbCO), with complementary experimental techniques and theoretical modeling [104,105,106,107,108,109,110].
It was already known that saccharide glasses protect biomolecules from structural damages due to dehydration or high temperatures. Trehalose (α-d-glucopyranosyl-α-d-glucopyranoside), a sugar present in anhydrobiont organisms, was found to be the best stabilizer [111]. Different working hypotheses were already proposed in the literature for the trehalose bioprotective mechanism and its efficiency in terms of functional recovery of biomaterials, as, e.g., water substitution at very low hydration (water replacement hypothesis [112]) or water entrapment at the protein surface (sugar preferential exclusion hypothesis) [113,114]. Another hypothesis referred to the high viscosity of trehalose samples [115], which would reduce protein internal motions responsible for denaturation. Trehalose peculiarity was also attributed to its glass transition temperature, which is higher than in its homologue sucrose and maltose [116] at the same water/sugar ratio.
Data from the experimental activity in Palermo on saccharide–protein matrices, pioneered by Prof. Lorenzo Cordone in collaboration with other groups in Italy and abroad, provided a consistent, multiscale description of the properties of these systems, shedding light into the trehalose superior properties. Globular and membrane proteins as well as liposomes [117] were studied mostly in high-concentrated sugar matrices, but also in sugar solution [118].
First results from ENS [119], Mössbauer, and optical absorption spectroscopy [120] in trehalose/MbCO/water matrices pointed out a reduction of protein nonharmonic motions [27]. Flash photolysis experiments [121,122] pointed out how thermal fluctuations among A substates of MbCO [123] were hindered by dehydration. Protein atomic fluctuations were also analyzed by MD simulations performed, in parallel to experiments, on MbCO/trehalose systems and in aqueous solutions [124,125,126], and the results were found to be in agreement with experimental data on trehalose-coated hemeproteins [127]. Further simulations of MbCO in sucrose and maltose systems pointed out the superior ability of trehalose in hampering local fluctuations of protein residue stretches more prone to thermal-induced denaturation [128].
The analysis of the solvent distribution around the protein and of the hydrogen bonds (HB) patterns, provided by simulations, suggested an original model for the bioprotective mechanism by saccharides [125]. Even at very low water content, the sugar was found to lock a thin water layer at the protein surface, where water molecules bridge the protein and the sugar molecules. In this scenario, the formation of strong HB networks would impair the solvent rearrangements, allowing for large-scale protein motions, an interpretation then confirmed by vibrational echo experiments on MbCO embedded in trehalose glasses and silica gels [129,130]. Furthermore, the presence of interfacial tightly bound water could increase the water surface tension induced by cosolutes in ternary systems [113]. Trehalose was found to be the most efficient in setting up these peculiar protein–sugar–water structures with respect to maltose and sucrose, even at very low water content [128]. Noteworthily, this model was adopted in several computational studies of proteins embedded in saccharide systems [131,132,133,134,135] and recently confirmed by neutron scattering experiments of MbCO and other protein models in trehalose/water samples [136,137,138,139,140].
FTIR studies were performed at different water contents and in a wide range of temperatures, at a fixed sugar/protein ratio, using different sugars [108,141,142,143,144]. The results were interpreted in the light of a new hypothesis (or anchorage hypothesis, see in Refs. [145,146,147,148]) in which a protein–matrix dynamical coupling likely arise from the HB networks depicted by MD, anchoring the protein surface to the matrix, and whose stiffness is tuned by the residual water. In these studies, the bound CO stretching band (COB, ~1900–2000 cm−1) was used to study the protein properties [123]; the water association band (WAB, ~2000–2400 cm−1), adjacent to the COB and arising from a combination of bending and intermolecular modes, was monitored to study the matrix properties [149] and references therein [150]. This band was extensively studied in Palermo, and it was shown that it is a valuable spectroscopical probe for the solvent in low-water matrices, including saccharide samples both in the presence and in the absence of proteins. In particular, the analysis of the WAB subcomponents provided a new way to classify water molecules in saccharide systems. Different subsets characterized by strong or weak HB patterns were found, and their relative population was found to be strongly dependent on the type of sugar and on the sugar/protein ratio [151].
In FTIR studies, the comparison of ternary, protein-containing samples, with binary sugar–water samples, are allowed to extract information on the sugar–protein–water interplay (see Figure 6).
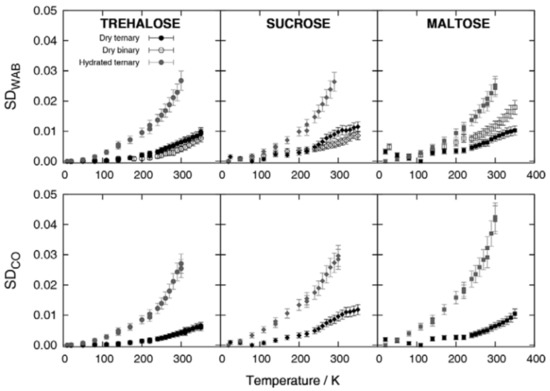
Figure 6.
Functional distance of the spectra (Spectra Distance, SD) as a function of temperature, from the spectrum measured at the lowest temperature investigated. Remarkably, in trehalose only, the SDWAB for binary samples has values comparable with dry ternary samples, a mark for a strong protein–matrix coupling. Reproduced without changes from Ref. [106], with permission from Springer Nature, Copyright 2013, license number 5405311084902.
In this respect, measurements pointed out the peculiarity of different disaccharides matrices toward the insertion of the protein component, especially at very low water content, where both protein and sugar compete for the few residual water molecules. Furthermore, both the COB and the WAB were largely exploited to study the thermal evolution of the matrix and the protein [107,108,152], providing results complementing information from neutron scattering and MD. Indeed, we could conceive that the PDT above a certain temperature (200–220 K), detected by ENS and simulations, arises from the exploration of the underlying equilibrium conformational landscape. In our model, this is a unique protein–solvent landscape; the COB and the WAB subcomponents map protein and solvent substates, respectively [107,152].
FTIR measurements were also performed on dry samples at different protein/sugar ratios [153]. An optimal sugar–protein formulation could be found, maximizing protein preservation at the minimal water content. Compared to other sugars, trehalose provided the most efficient mechanism, as trehalose matrices were found to better host the protein in a native conformation in a larger sugar-content range than other saccharides.
At high sugar content, the matrix becomes mostly like a sugar matrix that also contain domains without or with a small amount of protein. Such domains were also observed in small-angle X-ray scattering (SAXS) experiments [154,155], performed on the homologues’ binary and ternary disaccharide/water or disaccharide/water/protein samples, with the aim of investigating mutual protein–matrix effects on a large space scale (micro–nano). We proposed that the protein-poor domains could act as a water buffer against the effect of humidity variations by preferentially absorbing water molecules.
The thermal stability of MbCO in the homologues disaccharides was studied by differential scanning calorimetry (DSC) [156,157]. Results showed that the presence of the protein alter the glassy properties of the matrix, and, on the other hand, how protein stability is related to the glassy transition temperature. As for other measurements, the results in this case also showed a fairly different behavior of maltose, sucrose, and trehalose samples at very low water content. Trehalose was found able to form homogeneous matrices in a wide hydration range; in sucrose, we could observe phase separation and crystallization. The denaturation temperature (Tden) was found to increase upon dehydration in trehalose and sucrose, while in maltose, a reducing sugar, the Maillard reaction contributed to loss of regularity at low hydrations. In all the investigated systems, we observed a linear correlation between Tg and Tden; this was interpreted as a marker of protein–matrix coupling. It was found that trehalose is a better stabilizer than sucrose or maltose at equal Tg (Figure 7, upper panel).
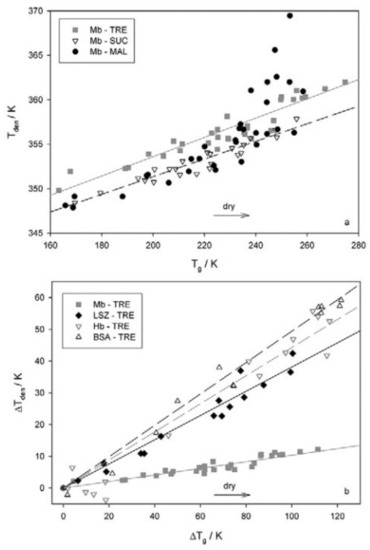
Figure 7.
(a): denaturation temperature (Tden) as a function of glassy transition temperature (Tg) for Mb-disaccharide-water systems. (b): Tden vs. Tg correlation for different proteins in trehalose samples. Reproduced without changes from Ref. [106], with permission from Springer Nature, Copyright 2013, license number 5405311084902.
Samples with proteins of different sizes and charges embedded in trehalose were also studied [157]. The Tg was found to increase at high to intermediate hydration, with a larger effect for a larger protein size, likely due to a confinement effect. At variance, at very low hydration, a Tg decrease was observed independent on the protein size. In this case, the underlying mechanism would be the onset of protein–sugar competition for residual water with a consequent weakening of sugar–water interactions. At low hydration, Tden was found to increase up to about 60–70 °C, depending on both protein size and charge. Furthermore, a protein-dependent linear correlation between Tg and Tden was observed (Figure 7, lower panel): specific protein–matrix interactions do influence the denaturation process, while confinement effects mostly regulate the glass transition temperature.
Protein thermal denaturation was studied in matrices containing other preserving agents [158,159] as gelatin. DSC and UV-Vis spectrophotometry measurements were performed on Mb in amorphous gelatin, in trehalose/gelatin matrices, and in only Mb matrices (self-crowding), at different hydrations. Our aim was at disentangling the specific (sugar–protein direct interactions) and aspecific (crowding, self-crowding, confinement) components of the bioprotection mechanism [159]. It was found that water plays a key role in switching from one to another mechanism in the same matrix. We could estimate the recovery fraction after thermal treatment at increasing heating times; this was considered to be a measure of massive protein denaturation. Results showed that at medium to high hydration (larger than 30 w/w%), trehalose stabilizes myoglobin, whereas gelatin destabilizes it. In this case, trehalose preservation occurs via water entrapment, thus avoiding interactions among proteins leading to protein denaturation. Spectroscopic measurements were exploited to probe the ability of the protein to retain the ligand (CO) after thermal treatments. It was found that, at low heating times, when the protein is likely still in native-like conformation, ligand retention is higher in trehalose than in gelatin. However, when massive denaturation takes place, the mixed trehalose/gelatine matrix was found to have a better performance.
Author Contributions
Writing—original draft preparation, G.C., A.C., M.L., V.V. and V.M.; writing—review and editing, all authors. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Acknowledgments
We thank all current and former coworkers in our laboratories, colleagues, postdocs, and students who contributed to the development of the research topics presented in this manuscript. We are also indebted to the many colleagues and friends, both in Italy and abroad, with whom we have collaborated during the years; we have learned much about biophysics from them and we have always enjoyed their collaboration and friendship.
Conflicts of Interest
The authors declare no conflict of interest.
References
- San Biagio, P.L.; Vitrano, E.; Cupane, A.; Madonia, F. Temperature induced difference spectra of Oxy and Deoxy hemoglobin in the near IR, visible and soret regions. Biochem. Biophys. Res. Commun. 1977, 77, 1158–1165. [Google Scholar] [CrossRef]
- Leone, M.; Cupane, A.; Vitrano, E.; Cordone, L. Dynamic properties of oxy-and carbonmonoxyhemoglobin probed by optical spectroscopy in the temperature range of 300–20 K. Biopolym. Orig. Res. Biomol. 1987, 26, 1769–1779. [Google Scholar] [CrossRef] [PubMed]
- Cordone, L.; Cupane, A.; Leone, M.; Vitrano, E.; Bulone, D. Interaction between external medium and haem pocket in myoglobin probed by low-temperature optical spectroscopy. J. Mol. Biol. 1988, 199, 213–218. [Google Scholar] [CrossRef]
- Cupane, A.; Leone, M.; Vitrano, E.; Cordone, L. Optical absorption spectra of azurin and stellacyanin in glycerol/water and ethylene glycol/water solutions in the temperature range 290–20 K. Biophys. Chem. 1990, 38, 213–224. [Google Scholar] [CrossRef]
- Cupane, A.; Leone, M.; Militello, V.; Stroppolo, M.E.; Polticelli, F.; Desideri, A. Low-temperature optical spectroscopy of native and azide-reacted bovine Cu, Zn superoxide dismutase. A structural dynamics study. Biochemistry 1994, 33, 15103–15109. [Google Scholar] [CrossRef]
- Vitrano, E.; Cupane, A.; Leone, M.; Militello, V.; Cordone, L.; Salvato, B.; Beltramini, M.; Bubacco, L.; Rocco, G.P. Low temperature optical spectroscopy of cobalt-substituted hemocyanin from Carcinus maenas. Eur. Biophys. J. 1993, 22, 157–167. [Google Scholar] [CrossRef]
- Di Pace, A.; Cupane, A.; Leone, M.; Vitrano, E.; Cordone, L. Protein dynamics. Vibrational coupling, spectral broadening mechanisms, and anharmonicity effects in carbonmonoxy heme proteins studied by the temperature dependence of the Soret band lineshape. Biophys. J. 1992, 63, 475–484. [Google Scholar] [CrossRef][Green Version]
- Cupane, A.; Cammarata, M.; Cordone, L.; Leone, M.; Vitrano, E.; Engler, N.; Parak, F. Spectral broadening of the Soret band in myoglobin: An interpretation by the full spectrum of low-frequency modes from a normal modes analysis. Eur. Biophys. J. 2005, 34, 881–889. [Google Scholar] [CrossRef]
- Cupane, A.; Leone, M.; Vitrano, E. Protein dynamics: Conformational disorder, vibrational coupling and anharmonicity in deoxy-hemoglobin and myoglobin. Eur. Biophys. J. 1993, 21, 385–391. [Google Scholar] [CrossRef]
- Cupane, A.; Leone, M.; Vitrano, E.; Cordone, L. Low temperature optical absorption spectroscopy: An approach to the study of stereodynamic properties of hemeproteins. Eur. Biophys. J. 1995, 23, 385–398. [Google Scholar] [CrossRef]
- Frauenfelder, H.; Petsko, G.A.; Tsernoglou, D. Temperature-dependent x-ray diffraction as a probe of protein structural dynamics. Nature 1979, 280, 558–563. [Google Scholar] [CrossRef] [PubMed]
- Melchers, B.; Knapp, E.W.; Parak, F.; Cordone, L.; Cupane, A.; Leone, M. Structural fluctuations of myoglobin from normal-modes, Mössbauer, Raman, and absorption spectroscopy. Biophys. J. 1996, 70, 2092–2099. [Google Scholar] [CrossRef][Green Version]
- Di Iorio, E.E.; Hiltpold, U.R.; Filipovic, D.; Winterhalter, K.H.; Gratton, E.; Vitrano, E.; Cupane, A.; Leone, M.; Cordone, L. Protein dynamics. Comparative investigation on heme-proteins with different physiological roles. Biophys. J. 1991, 59, 742–754. [Google Scholar] [CrossRef][Green Version]
- Cupane, A.; Leone, M.; Vitrano, E.; Cordone, L.; Hiltpold, U.R.; Winterhalter, K.H.; Yu, W.; Di Iorio, E.E. Structure-dynamics-function relationships in Asian elephant (Elephas maximus) myoglobin. An optical spectroscopy and flash photolysis study on functionally important motions. Biophys. J. 1993, 65, 2461–2472. [Google Scholar] [CrossRef][Green Version]
- Boffi, A.; Verzili, D.; Chiancone, E.; Leone, M.; Cupane, A.; Militello, V.; Vitrano, E.; Cordone, L.; Yu, W.; Di Iorio, E.E. Stereodynamic properties of the cooperative homodimeric Scapharca inaequivalvis hemoglobin studied through optical absorption spectroscopy and ligand rebinding kinetics. Biophys. J. 1994, 67, 1713–1723. [Google Scholar] [CrossRef]
- Falconi, M.; Desideri, A.; Cupane, A.; Leone, M.; Ciccotti, G.; Peterson, E.S.; Friedman, J.M.; Gambacurta, A.; Ascoli, F. Structural and Dynamic Properties of the Homodimeric Hemoglobin from Scapharca inaequivalvis Thr-72 → Ile Mutant: Molecular Dynamics Simulation, Low Temperature Visible Absorption Spectroscopy, and Resonance Raman Spectroscopy Studies. Biophys. J. 1998, 75, 2489–2503. [Google Scholar] [CrossRef]
- Militello, V.; Cupane, A.; Leone, M.; Brinigar, W.S.; Lu, A.L.; Fronticelli, C. Dynamic properties of some β-chain mutant hemoglobins. Proteins Struct. Funct. Bioinform. 1995, 22, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Militello, V.; Cupane, A.; Leone, M.; Vitrano, E. Dynamic and functional properties of a β-β crosslinked derivative of human hemoglobin. Phys. Med. 1993, 9, 43–46. [Google Scholar]
- Cupane, A.; Leone, M.; Militello, V.; Friedman, F.K.; Koley, A.P.; Vasquez, G.B.; Brinigar, W.S.; Karavitis, M.; Fronticelli, C. Modification of α-Chain or β-Chain Heme Pocket Polarity by Val (E11) → Thr Substitution Has Different Effects on the Steric, Dynamic, and Functional Properties of Human Recombinant Hemoglobin: Deoxy Derivatives. J. Biol. Chem. 1997, 272, 26271–26278. [Google Scholar] [CrossRef]
- Karavitis, M.; Fronticelli, C.; Brinigar, W.S.; Vasquez, G.B.; Militello, V.; Leone, M.; Cupane, A. Properties of Human Hemoglobins with Increased Polarity in the α-or β-Heme Pocket: Carbonmonoxy Derivatives. J. Biol. Chem. 1998, 273, 23740–23749. [Google Scholar] [CrossRef]
- Militello, V.; Leone, M.; Cupane, A.; Santucci, R.; Desideri, A. Local dynamic properties of the heme pocket in native and solvent-induced molten-globule-like states of cytochrome c. Biophys. Chem. 2002, 97, 121–128. [Google Scholar] [CrossRef]
- Levantino, M.; Huang, Q.; Cupane, A.; Laberge, M.; Hagarman, A.; Schweitzer-Stenner, R. The importance of vibronic perturbations in ferrocytochrome c spectra: A reevaluation of spectral properties based on low-temperature optical absorption, resonance Raman, and molecular-dynamics simulations. J. Chem. Phys. 2005, 123, 054508. [Google Scholar] [CrossRef] [PubMed]
- Schweitzer-Stenner, R.; Levantino, M.; Cupane, A.; Wallace, C.; Laberge, M.; Huang, Q. Functionally relevant electric-field induced perturbations of the prosthetic group of yeast ferrocytochrome c mutants obtained from a vibronic analysis of low-temperature absorption spectra. J. Phys. Chem. B 2006, 110, 12155–12161. [Google Scholar] [CrossRef]
- Schweitzer-Stenner, R.; Cupane, A.; Leone, M.; Lemke, C.; Schott, J.; Dreybrodt, W. Anharmonic protein motions and heme deformations in myoglobin cyanide probed by absorption and resonance Raman spectroscopy. J. Phys. Chem. B 2000, 104, 4754–4764. [Google Scholar] [CrossRef]
- Cupane, A.; Leone, M.; Unger, E.; Lemke, C.; Beck, M.; Dreybrodt, W.; Schweitzer-Stenner, R. Dynamics of various metal-octaethylporphyrins in solution studied by resonance Raman and low-temperature optical absorption spectroscopies. Role of the central metal. J. Phys. Chem. B 1998, 102, 6612–6620. [Google Scholar] [CrossRef]
- Fomina, M.; Schirò, G.; Cupane, A. Hydration dependence of myoglobin dynamics studied with elastic neutron scattering, differential scanning calorimetry and broadband dielectric spectroscopy. Biophys. Chem. 2014, 185, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Doster, W.; Cusack, S.; Petry, W. Dynamical transition of myoglobin revealed by inelastic neutron scattering. Nature 1989, 337, 754–756. [Google Scholar] [CrossRef]
- Schiro, G.; Sclafani, M.; Natali, F.; Cupane, A. Hydration dependent dynamics in sol–gel encapsulated myoglobin. Eur. Biophys. J. 2008, 37, 543–549. [Google Scholar] [CrossRef]
- Schiro, G.; Sclafani, M.; Caronna, C.; Natali, F.; Plazanet, M.; Cupane, A. Dynamics of myoglobin in confinement: An elastic and quasi-elastic neutron scattering study. Chem. Phys. 2008, 345, 259–266. [Google Scholar] [CrossRef]
- Schiro, G.; Cupane, A.; Vitrano, E.; Bruni, F. Dielectric relaxations in confined hydrated myoglobin. J. Phys. Chem. B 2009, 113, 9606–9613. [Google Scholar] [CrossRef]
- Schiró, G.; Caronna, C.; Natali, F.; Cupane, A. Direct evidence of the amino acid side chain and backbone contributions to protein anharmonicity. J. Am. Chem. Soc. 2010, 132, 1371–1376. [Google Scholar] [CrossRef] [PubMed]
- Schiró, G.; Caronna, C.; Natali, F.; Cupane, A. Molecular origin and hydration dependence of protein anharmonicity: An elastic neutron scattering study. Phys. Chem. Chem. Phys. 2010, 12, 10215–10220. [Google Scholar] [CrossRef] [PubMed]
- Schiró, G.; Natali, F.; Cupane, A. Physical origin of anharmonic dynamics in proteins: New insights from resolution-dependent neutron scattering on homomeric polypeptides. Phys. Rev. Lett. 2012, 109, 128102. [Google Scholar] [CrossRef] [PubMed]
- Schirò, G.; Fomina, M.; Cupane, A. Communication: Protein dynamical transition vs. liquid-liquid phase transition in protein hydration water. J. Chem. Phys. 2013, 139, 121102. [Google Scholar] [CrossRef]
- Cupane, A.; Levantino, M.; Santangelo, M.G. Near-infrared spectra of water confined in silica hydrogels in the temperature interval 365–5 K. J. Phys. Chem. B 2002, 106, 11323–11328. [Google Scholar] [CrossRef]
- Cammarata, M.; Levantino, M.; Cupane, A.; Longo, A.; Martorana, A.; Bruni, F. Structure and dynamics of water confined in silica hydrogels: X-ray scattering and dielectric spectroscopy studies. Eur. Phys. J. E 2003, 12, 63–66. [Google Scholar] [CrossRef]
- Santangelo, G.; Di Matteo, A.; Müller-Plathe, F.; Milano, G. From mesoscale back to atomistic models: A fast reverse-mapping procedure for vinyl polymer chains. J. Phys. Chem. B 2007, 111, 2765–2773. [Google Scholar] [CrossRef]
- De Michele, V.; Romanelli, G.; Cupane, A. Dynamics of supercooled confined water measured by deep inelastic neutron scattering. Front. Phys. 2018, 13, 138205. [Google Scholar] [CrossRef]
- Cupane, A.; Fomina, M.; Schirò, G. The boson peak of deeply cooled confined water reveals the existence of a low-temperature liquid-liquid crossover. J. Chem. Phys. 2014, 141, 18C510. [Google Scholar] [CrossRef]
- De Michele, V.; Levantino, M.; Cupane, A. Hysteresis in the temperature dependence of the IR bending vibration of deeply cooled confined water. J. Chem. Phys. 2019, 150, 224509. [Google Scholar] [CrossRef]
- Cupane, A.; Fomina, M.; Piazza, I.; Peters, J.; Schirò, G. Experimental evidence for a liquid-liquid crossover in deeply cooled confined water. Phys. Rev. Lett. 2014, 113, 215701. [Google Scholar] [CrossRef] [PubMed]
- Piazza, I.; Cupane, A.; Barbier, E.L.; Rome, C.; Collomb, N.; Ollivier, J.; Gonzalez, M.A.; Natali, F. Dynamical properties of water in living cells. Front. Phys. 2018, 13, 138301. [Google Scholar] [CrossRef]
- Natali, F.; Dolce, C.; Peters, J.; Stelletta, C.; Demé, B.; Ollivier, J.; Leduc, G.; Cupane, A.; Barbier, E.L. Brain lateralization probed by water diffusion at the atomic to micrometric scale. Sci. Rep. 2019, 9, 14694. [Google Scholar] [CrossRef]
- Natali, F.; Dolce, C.; Peters, J.; Stelletta, C.; Demé, B.; Ollivier, J.; Boehm, M.; Leduc, G.; Piazza, I.; Cupane, A.; et al. Anomalous water dynamics in brain: A combined diffusion magnetic resonance imaging and neutron scattering investigation. J. R. Soc. Interface 2019, 16, 20190186. [Google Scholar] [CrossRef] [PubMed]
- Levantino, M.; Cupane, A.; Zimányi, L.; Ormos, P. Different relaxations in myoglobin after photolysis. Proc. Natl. Acad. Sci. USA 2004, 101, 14402–14407. [Google Scholar] [CrossRef]
- Levantino, M.; Cupane, A.; Zimányi, L. Quaternary structure dependence of kinetic hole burning and conformational substates interconversion in hemoglobin. Biochemistry 2003, 42, 4499–4505. [Google Scholar] [CrossRef]
- Schiro, G.; Cammarata, M.; Levantino, M.; Cupane, A. Spectroscopic markers of the T↔R quaternary transition in human hemoglobin. Biophys. Chem. 2005, 114, 27–33. [Google Scholar] [CrossRef]
- Schirò, G.; Cupane, A. Quaternary Relaxations in Sol–Gel Encapsulated Hemoglobin Studied via NIR and UV Spectroscopy. Biochemistry 2007, 46, 11568–11576. [Google Scholar] [CrossRef]
- Cammarata, M.; Levantino, M.; Schotte, F.; Anfinrud, P.A.; Ewald, F.; Choi, J.; Cupane, A.; Wulff, M.; Ihee, H. Tracking the structural dynamics of proteins in solution using time-resolved wide-angle X-ray scattering. Nat. Methods 2008, 5, 881–886. [Google Scholar] [CrossRef]
- Cammarata, M.; Levantino, M.; Wulff, M.; Cupane, A. Unveiling the timescale of the R–T transition in human hemoglobin. J. Mol. Biol. 2010, 400, 951–962. [Google Scholar] [CrossRef]
- Spilotros, A.; Levantino, M.; Schiro, G.; Cammarata, M.; Wulff, M.; Cupane, A. Probing in cell protein structural changes with time-resolved X-ray scattering. Soft Matter 2012, 8, 6434–6437. [Google Scholar] [CrossRef][Green Version]
- Levantino, M.; Spilotros, A.; Cammarata, M.; Schirò, G.; Ardiccioni, C.; Vallone, B.; Brunori, M.; Cupane, A. The Monod-Wyman-Changeux allosteric model accounts for the quaternary transition dynamics in wild type and a recombinant mutant human hemoglobin. Proc. Natl. Acad. Sci. USA 2012, 109, 14894–14899. [Google Scholar] [CrossRef] [PubMed]
- Levantino, M.; Schiro, G.; Lemke, H.T.; Cottone, G.; Glownia, J.M.; Zhu, D.; Chollet, M.; Ihee, H.; Cupane, A.; Cammarata, M. Ultrafast myoglobin structural dynamics observed with an X-ray free-electron laser. Nat. Commun. 2015, 6, 6772. [Google Scholar] [CrossRef] [PubMed]
- Levantino, M.; Lemke, H.T.; Schirò, G.; Glownia, M.; Cupane, A.; Cammarata, M. Observing heme doming in myoglobin with femtosecond X-ray absorption spectroscopy. Struct. Dyn. 2015, 2, 041713. [Google Scholar] [CrossRef] [PubMed]
- Elber, R.; Karplus, M. Enhanced sampling in molecular dynamics: Use of the time-dependent Hartree approximation for a simulation of carbon monoxide diffusion through myoglobin. J. Am. Chem. Soc. 1990, 112, 9161–9175. [Google Scholar] [CrossRef]
- Schotte, F.; Lim, M.; Jackson, T.A.; Smirnov, A.V.; Soman, J.; Olson, J.S.; Phillips, G.N., Jr.; Wulff, M.; Anfinrud, P.A. Watching a protein as it functions with 150-ps time-resolved x-ray crystallography. Science 2003, 300, 1944–1947. [Google Scholar] [CrossRef]
- Brunori, M.; Gibson, Q.H. Cavities and packing defects in the structural dynamics of myoglobin. EMBO Rep. 2001, 2, 674–679. [Google Scholar] [CrossRef]
- Maragliano, L.; Cottone, G.; Ciccotti, G.; Vanden-Eijnden, E. Mapping the network of pathways of CO diffusion in myoglobin. J. Am. Chem. Soc. 2010, 132, 1010–1017. [Google Scholar] [CrossRef]
- Cottone, G.; Lattanzi, G.; Ciccotti, G.; Elber, R. Multiphoton absorption of myoglobin–nitric oxide complex: Relaxation by D-NEMD of a stationary state. J. Phys. Chem. B 2012, 116, 3397–3410. [Google Scholar] [CrossRef]
- Ciccotti, G.; Jacucci, G. Direct computation of dynamical response by molecular dynamics: The mobility of a charged Lennard-Jones particle. Phys. Rev. Lett. 1975, 357, 789. [Google Scholar] [CrossRef]
- San Biagio, P.L.; Martorana, V.; Emanuele, A.; Vaiana, S.M.; Manno, M.; Bulone, D.; Palma-Vittorelli, M.B.; Palma, M.U. Interacting processes in protein coagulation. Proteins Struct. Funct. Genet. 1999, 37, 116–120. [Google Scholar] [CrossRef]
- Manno, M.; Martorana, V.; Emanuele, A.; Bulone, D.; San Biagio, P.L.; Palma-Vittorelli, M.B.; Palma, M.U. Multiple-path interactions and fractal structure kinetics in supramolecular self-assembly. Biophys. J. 1998, 74, A282. [Google Scholar]
- Bulone, D.; Martorana, V.; San Biagio, P.L. Effects of intermediates on aggregation of native bovine serum albumin. Biophys. Chem. 2001, 91, 61–69. [Google Scholar] [CrossRef]
- Vaiana, S.M.; Emanuele, A.; Palma-Vittorelli, M.B.; Palma, M.U. Irreversible formation of intermediate BSA oligomers requires and induces conformational changes. Proteins 2004, 1, 1053–1062. [Google Scholar] [CrossRef] [PubMed]
- Manno, M.; Craparo, E.F.; Bulone, D.; Martorana, V.; San Biagio, P.L. Kinetics of Insulin Aggregation: Disentanglement of Amyloid Fibrillation from Large-Size Cluster Formation. Biophys. J. 2006, 90, 585–4591. [Google Scholar] [CrossRef] [PubMed]
- Militello, V.; Vetri, V.; Leone, M. Conformational changes involved in thermal aggregation processes of Bovine Serum Albumin. Biophys. Chem. 2003, 105, 133–141. [Google Scholar] [CrossRef]
- Pullara, F.; Emanuele, A.; Palma-Vittorelli, M.B.; Palma, M.U. Protein Aggregation/Crystallization and Minor Structural Changes: Universal versus Specific Aspects. Biophys. J. 2007, 93, 3271–3278. [Google Scholar] [CrossRef]
- Moriyama, Y.; Ohta, D.; Hachiya, K.; Mitsui, Y.; Takeda, K. Fluorescence behaviour of tryptophan residues of bovine and human serum albumins in ionic surfactant solutions: A comparative study of the two and one tryptophan(s) of bovine and human Albumins. J. Protein Chem. 1996, 15, 265–271. [Google Scholar] [CrossRef]
- Lakovicz, J.R. Principles of Fluorescence Spectroscopy; Plenum Press: New York, NY, USA, 1983. [Google Scholar]
- Vetri, V.; Librizzi, F.; Leone, M.; Militello, V. Thermal aggregation of bovine serum albumin at different pH: Comparison with human serum albumin. Eur. Biophys. J. 2007, 36, 717–725. [Google Scholar] [CrossRef]
- Vetri, V.; D’Amico, M.; Foderà, V.; Leone, M.; Ponzoni, A.; Sberveglieri, G.; Militello, V. Bovine Serum Albumin protofibril-like aggregates formation: Solo but not simple mechanism. Arch. Biochem. Biophys. 2011, 508, 13–24. [Google Scholar] [CrossRef]
- Militello, V.; Casarino, C.; Emanuele, A.; Giostra, A.; Pullara, F.; Leone, M. Aggregation kinetics of Bovine Serum Albumin studied by FTIR spectroscopy and light Scattering. Biophys. Chem. 2004, 107, 175–187. [Google Scholar] [CrossRef] [PubMed]
- Bucciantini, M.; Giannoni, E.; Chiti, F.; Baroni, F.; Formigli, L.; Zurdo, J.; Taddei, N.; Ramponi, G.; Dobson, C.M.; Stefani, M. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature 2002, 416, 507–511. [Google Scholar] [CrossRef] [PubMed]
- Walsh, D.M.; Selkoe, D.J. Oligomers on the brain: The emerging role of soluble protein aggregates in neurodegeneration. Protein Pept. Lett. 2004, 11, 213–228. [Google Scholar] [CrossRef] [PubMed]
- Hartl, F.U. Protein misfolding diseases. Annu. Rev. Biochem. 2017, 86, 21–26. [Google Scholar] [CrossRef]
- Choi, M.L.; Gandhi, S. Crucial role of protein oligomerization in the pathogenesis of Alzheimer’s and Parkinson’s diseases. FEBS J. 2018, 285, 3631–3644. [Google Scholar] [CrossRef]
- Kulenkampff, K.; Wolf Perez, A.M.; Sormanni, P.; Habchi, J.; Vendruscolo, M. Quantifying misfolded protein oligomers as drug targets and biomarkers in Alzheimer and Parkinson diseases. Nat. Rev. Chem. 2021, 5, 277–294. [Google Scholar] [CrossRef]
- Vetri, V.; Foderà, V. The route to protein aggregate superstructures: Particulates and amyloid-like spherulites. FEBS Lett. 2015, 589, 2448–2463. [Google Scholar] [CrossRef]
- Vetri, V.; Piccirilli, F.; Krausser, J.; Buscarino, G.; Łapińska, U.; Vestergaard, B.; Zaccon, A.; Foderà, V. Ethanol controls the self-assembly and mesoscopic properties of human insulin amyloid spherulites. J. Phys. Chem. B 2018, 122, 3101–3112. [Google Scholar] [CrossRef]
- Fennema Galparsoro, D.; Zhou, X.; Jaaloul, A.; Piccirilli, F.; Vetri, V.; Foderà, V. Conformational transitions upon maturation rule surface and pH-responsiveness of α-lactalbumin microparticulates. ACS Appl. Bio Mater. 2021, 4, 1876–1887. [Google Scholar] [CrossRef]
- Librizzi, F.; Rischel, C. The kinetic behavior of insulin fibrillation is determined by heterogeneous nucleation pathways. Protein Sci. 2005, 14, 3129–3134. [Google Scholar] [CrossRef]
- Fodera, V.; Librizzi, F.; Groenning, M.; van de Weert, M.; Leone, M. Secondary nucleation and accessible surface in insulin amyloid fibril formation. J. Phys. Chem. B 2008, 112, 3853–3858. [Google Scholar] [CrossRef] [PubMed]
- Fodera, V.; Cataldo, S.; Librizzi, F.; Pignataro, B.; Spiccia, P.; Leone, M. Self-organization pathways and spatial heterogeneity in insulin amyloid fibril formation. J. Phys. Chem. B 2009, 113, 10830–10837. [Google Scholar] [CrossRef] [PubMed]
- LeVine, H. Thioflavine T interaction with amyloid β-sheet structures. Amyloid 1995, 2, 1–6. [Google Scholar] [CrossRef]
- Foderà, V.; Groenning, M.; Vetri, V.; Librizzi, F.; Spagnolo, S.; Cornett, C.; Olsen, L.; Van De Weert, M.; Leone, M. Thioflavin T hydroxylation at basic pH and its effect on amyloid fibril detection. J. Phys. Chem. B 2008, 112, 15174–15181. [Google Scholar] [CrossRef] [PubMed]
- D’Amico, M.; Di Carlo, M.G.; Groenning, M.; Militello, V.; Vetri, V.; Leone, M. Thioflavin T promotes Aβ(1-40) amyloid fibrils formation. J. Phys. Chem. Lett. 2012, 3, 1596–1601. [Google Scholar] [CrossRef] [PubMed]
- Di Carlo, M.G.; Minicozzi, V.; Foderà, V.; Militello, V.; Vetri, V.; Morante, S.; Leone, M. Thioflavin T templates amyloid β(1-40) conformation and aggregation pathway. Biophys. Chem. 2015, 206, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Navarra, G.; Peres, C.; Contardi, M.; Picone, P.; San Biagio, P.L.; Di Carlo, M.; Giacomazza, D.; Militello, V. Heat- and pH-induced BSA conformational changes, hydrogels formation and their applications as 3D cell scaffold. Arch. Biochem. Biophys. 2016, 606, 134–142. [Google Scholar] [CrossRef] [PubMed]
- De Luca, G.; Galparsoro, D.F.; Sancataldo, G.; Leone, M.; Foderà, V.; Vetri, V. Probing ensemble polymorphism and single aggregate structural heterogeneity in insulin amyloid self-assembly. J. Colloid Interface Sci. 2020, 574, 229–240. [Google Scholar] [CrossRef]
- Zhou, X.; Galparsoro, D.F.; Madsen, A.Ø.; Vetri, V.; van de Weert, M.; Nielsen, H.M.; Foderà, V. Polysorbate 80 controls Morphology, structure and stability of human insulin Amyloid-Like spherulites. J. Colloid Interface Sci. 2022, 606, 1928–1939. [Google Scholar] [CrossRef]
- Van Maarschalkerweerd, A.; Vetri, V.; Langkilde, A.E.; Fodera, V.; Vestergaard, B. Protein/lipid coaggregates are formed during α-synuclein-induced disruption of lipid bilayers. Biomacromolecules 2014, 15, 3643–3654. [Google Scholar] [CrossRef]
- Rao, E.; Foderà, V.; Leone, M.; Vetri, V. Direct observation of alpha-lactalbumin, adsorption and incorporation into lipid membrane and formation of lipid/protein hybrid structures. Biochim. Biophys. Acta BBA Gen. Subj. 2019, 1863, 784–794. [Google Scholar] [CrossRef] [PubMed]
- Anselmo, S.; Sancataldo, G.; Mørck Nielsen, H.; Foderà, V.; Vetri, V. Peptide–membrane interactions monitored by fluorescence lifetime imaging: A study case of Transportan 10. Langmuir 2021, 37, 13148–13159. [Google Scholar] [CrossRef] [PubMed]
- Rondeau, P.; Navarra, G.; Militello, V.; Bourdon, E. Aggregation of albumin: Influence of the protein glycation. In Protein Aggregation; Series: Protein Science and Engineering Microbiology Research Advances; Stein, D.A., Ed.; Nova Science Publishers: Hauppauge, NY, USA, 2011; Chapter 5; pp. 139–159. [Google Scholar]
- Baraka-Vidot, J.; Navarra, G.; Leone, M.; Bourdon, E.; Militello, V.; Rondeau, P. Deciphering metal-induced oxidative damages on glycated albumin structure and function. Biochim. Biophys. Acta BBA Gen. Subj. 2014, 1840, 1712–1724. [Google Scholar] [CrossRef] [PubMed]
- Baraka-Vidot, J.; Planesse, C.; Meilhac, O.; Militello, V.; van den Elsen, J.M.H.; Bourdon, E.; Rondeau, P. Glycation alters ligand-binding, enzymatic and pharmacological properties of human albumin. Biochemistry 2015, 54, 3051–3062. [Google Scholar] [CrossRef]
- Khan, M.W.; Rasheed, Z.; Khan, W.A.; Ali, R. Biochemical, biophysical, and thermodynamic analysis of in vitro glycated human serum albumin. Biochemistry 2007, 72, 146–152. [Google Scholar]
- Cohen, M.P.; Ziyadeh, F.N.; Chen, S. Amadori-modified glycated serum proteins and accelerated atherosclerosis in diabetes: Pathogenic and therapeutic implications. J. Lab. Clin. Med. 2006, 147, 211–219. [Google Scholar] [CrossRef]
- Tanaka, A.; Kaneto, H.; Miyatsuka, T.; Yamamoto, K.; Yoshiuchi, K.; Yamasaki, Y.; Shimomura, I.; Matsuoka, T.; Matsuhisa, M. Role of copper ion in the pathogenesis of type 2 diabetes. Endocr. J. 2009, 56, 699–706. [Google Scholar] [CrossRef]
- Al-Maroof, R.A.; Al-Sharbatti, S.S. Serum zinc levels in diabetic patients and effect of zinc supplementation on glycemic control of type 2 diabetics. Saudi Med. J. 2006, 27, 344–350. [Google Scholar]
- Rondeau, P.; Bourdon, E. The glycation of albumin: Structural and functional impacts. Biochimie 2011, 93, 645–658. [Google Scholar] [CrossRef]
- Rondeau, P.; Navarra, G.; Cacciabaudo, F.; Leone, M.; Bourdon, E.; Militello, V. Thermal aggregation of glycated bovine serum albumin. Biochim. Biophys. Acta Protein Proteom. 2010, 1804, 789–798. [Google Scholar] [CrossRef]
- Navarra, G.; Tinti, A.; Leone, M.; Militello, V.; Torreggiani, A. Influence of metal ions on thermal aggregation of bovine serum albumin: Aggregation kinetics and structural changes. J. Inorg. Biochem. 2009, 103, 1729–1738. [Google Scholar] [CrossRef] [PubMed]
- Cordone, L.; Cottone, G.; Giuffrida, S.; Palazzo, G.; Venturoli, G.; Viappiani, C. Internal dynamics and protein–matrix coupling in trehalose-coated proteins. Biochim. Biophys. Acta Proteins Proteom. 2005, 1749, 252–281. [Google Scholar] [CrossRef] [PubMed]
- Cordone, L.; Cottone, G.; Cupane, A.; Emanuele, A.; Giuffrida, S.; Levantino, M. Proteins in Saccharides Matrices and the Trehalose Peculiarity: Biochemical and Biophysical Properties. Curr. Org. Chem. 2015, 19, 1684–1706. [Google Scholar] [CrossRef]
- Giuffrida, S.; Cottone, G.; Bellavia, G.; Cordone, L. Proteins in Amorphous Saccharide Matrices: Structural and Dynamical Insights on Bioprotection. Eur. Phys. J. E 2013, 36, 7. [Google Scholar] [CrossRef]
- Cordone, L.; Cottone, G.; Giuffrida, S.; Librizzi, F. Thermal evolution of the CO stretching band in carboxy-myoglobin in the light of neutron scattering and molecular dynamics simulations. Chem. Phys. 2008, 345, 275–282. [Google Scholar] [CrossRef]
- Giuffrida, S.; Cottone, G.; Cordone, L. Role of solvent on protein-matrix coupling in MbCO embedded in water-saccharide systems: A Fourier transform infrared spectroscopy study. Biophys. J. 2006, 91, 968–980. [Google Scholar] [CrossRef]
- Cottone, G.; Giuffrida, S.; Bettati, S.; Bruno, S.; Campanini, B.; Marchetti, M.; Abruzzetti, S.; Viappiani, C.; Cupane, A.; Mozzarelli, A.; et al. More than a Confinement: “Soft” and “Hard” Enzyme Entrapment Modulates Biological Catalyst Function. Catalysts 2019, 9, 1024. [Google Scholar] [CrossRef]
- Da Silva, F.L.B.; Carloni, P.; Cheung, D.; Cottone, G.; Donnini, S.; Foegeding, E.A.; Gulzar, M.; Jacquier, J.C.; Lobaskin, V.; MacKernan, D.; et al. Understanding and Controlling Food Protein Structure and Function in Foods: Perspectives from Experiments and Computer Simulations. Annu. Rev. Food Sci. Technol. 2020, 11, 365–387. [Google Scholar] [CrossRef]
- Crowe, J.H.; Crowe, L.M.; Wolkers, W.F.; Oliver, A.E.; Ma, X.; Auh, J.H.; Tang, M.; Zhu, S.; Norris, J.; Tablin, F. Stabilization of Dry Mammalian Cells: Lessons from Nature. Integr. Comp. Biol. 2005, 45, 810–820. [Google Scholar] [CrossRef]
- Carpenter, J.F.; Crowe, J.H. An infrared spectroscopic study of the interactions of carbohydrates with dried proteins. Biochemistry 1989, 28, 3916–3922. [Google Scholar] [CrossRef]
- Xie, G.; Timasheff, S.N. The thermodynamic mechanism of protein stabilization by trehalose. Biophys. Chem. 1997, 64, 25–43. [Google Scholar] [CrossRef]
- Belton, P.S.; Gil, A.M. IR and Raman spectroscopic studies of the interaction of trehalose with hen egg white lysozyme. Biopolymers 1994, 34, 957–961. [Google Scholar] [CrossRef] [PubMed]
- Sampedro, J.G.; Uribe, S. Trehalose-enzyme interactions result in structure stabilization and activity inhibition. The role of viscosity. Mol. Cell. Biochem. 2004, 256, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Green, J.L.; Angell, C.A. Phase relations and vitrification in saccharide-water solutions and the trehalose anomaly. J. Phys. Chem. 1989, 93, 2880–2882. [Google Scholar] [CrossRef]
- Chiantia, S.; Giannola, L.; Cordone, L. Lipid phase transition in saccharide-coated cholate-containing liposomes: Coupling to the surrounding matrix. Langmuir 2005, 122, 4309–4317. [Google Scholar] [CrossRef]
- Panzica, M.; Emanuele, A.; Cordone, L. Thermal Aggregation of Bovine Serum Albumin in Trehalose and Sucrose Aqueous Solutions. J. Phys. Chem. B 2012, 116, 11829–11836. [Google Scholar] [CrossRef]
- Cordone, L.; Ferrand, M.; Vitrano, E.; Zaccai, G. Harmonic behavior of trehalose-coated carbon-monoxy-myoglobin at high temperature. Biophys. J. 1999, 76, 1043–1047. [Google Scholar] [CrossRef][Green Version]
- Cordone, L.; Galajda, P.; Vitrano, E.; Gassmann, A.; Ostermann, A.; Parak, F. A reduction of protein specific motions in co-ligated myoglobin embedded in a trehalose glass. Eur. Biophys. J. 1998, 27, 173–176. [Google Scholar] [CrossRef]
- Librizzi, F.; Vitrano, E.; Cordone, L. Dehydration and crystallization of trehalose and sucrose glasses containing carbonmonoxy-myoglobin. Biophys. J. 1999, 76, 2727–2734. [Google Scholar] [CrossRef]
- Librizzi, F.; Viappiani, C.; Abbruzzetti, S.; Cordone, L. Residual water modulates the dynamics of the protein and of the external matrix in “trehalose coated” MbCO: An infrared and flash-photolysis study. J. Chem. Phys. 2002, 116, 1193–1200. [Google Scholar] [CrossRef]
- Vojtěchovský, J.; Chu, K.; Berendzen, J.; Sweet, R.M.; Schlichting, I. Crystal Structures of Myoglobin-Ligand Complexes at Near-Atomic Resolution. Biophys. J. 1999, 77, 2153–2174. [Google Scholar] [CrossRef]
- Cottone, G.; Cordone, L.; Ciccotti, G. Molecular dynamics simulation of carboxy-myoglobin embedded in a trehalose-water matrix. Biophys. J. 2001, 80, 931–938. [Google Scholar] [CrossRef]
- Cottone, G.; Ciccotti, G.; Cordone, L. Protein–trehalose–water structures in trehalose coated carboxy-myoglobin. J. Chem. Phys. 2022, 117, 9862–9866. [Google Scholar] [CrossRef]
- Cottone, G.; Giuffrida, S.; Ciccotti, G.; Cordone, L. Molecular dynamics simulation of sucrose-and trehalose-coated carboxy-myoglobin. Proteins 2005, 59, 291–302. [Google Scholar] [CrossRef]
- Giachini, L.; Francia, F.; Cordone, L.; Boscherini, F.; Venturoli, G. Cytochrome c in a dry trehalose matrix: Structural and dynamical effects probed by x-ray absorption spectroscopy. Biophys. J. 2006, 92, 1350–1360. [Google Scholar] [CrossRef]
- Cottone, G. A comparative study of carboxy myoglobin in saccharide-water systems by molecular dynamics simulation. J. Phys. Chem. B 2007, 111, 3563–3569. [Google Scholar] [CrossRef]
- Massari, A.M.; Finkestein, I.J.; McClain, B.L.; Goj, A.; Wen, X.; Bren, K.L.; Loring, R.F.; Fayer, M.D.J. The influence of aqueous versus glassy solvents on protein dynamics: Vibrational echo experiments and molecular dynamics simulations. J. Am. Chem. Soc. 2005, 127, 14279–14289. [Google Scholar] [CrossRef]
- Massari, A.M.; Finkelstein, I.J.; Fayer, M.D. Dynamics of proteins encapsulated in silica sol–gel glasses studied with IR vibrational echo spectroscopy. J. Am. Chem. Soc. 2006, 128, 3990–3997. [Google Scholar] [CrossRef]
- Katyal, N.; Deep, S. Revisiting the conundrum of trehalose stabilization. Phys. Chem. Chem. Phys. 2014, 16, 26746–26761. [Google Scholar] [CrossRef]
- Lerbret, A.; Bordat, P.; Affouard, F.; Hédoux, A.; Guinet, Y.; Descamps, M. How do trehalose, maltose, and sucrose influence some structural and dynamical properties of lysozyme? Insight from molecular dynamics simulations. J. Phys. Chem. B 2007, 111, 9410–9420. [Google Scholar] [CrossRef]
- Lerbret, A.; Affouard, F.; Hédoux, A.; Krenzlin, S.; Siepmann, J.; Bellissent-Funel, M.C.; Descamps, M. How Strongly Does Trehalose Interact with Lysozyme in the Solid State? Insights from Molecular Dynamics Simulation and Inelastic Neutron Scattering. J. Phys. Chem. B 2012, 116, 11103–11116. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Liu, F.; Dong, X.; Sun, Y. Molecular Insight into the Counteraction of Trehalose on Urea-Induced Protein Denaturation Using Molecular Dynamics Simulation. J. Phys. Chem. B 2012, 116, 7040–7047. [Google Scholar] [CrossRef] [PubMed]
- Weng, L.; Stott, S.L.; Toner, M. Exploring Dynamics and Structure of Biomolecules, Cryoprotectants, and Water Using Molecular Dynamics Simulations: Implications for Biostabilization and Biopreservation. Annu. Rev. Biomed. Eng. 2019, 21, 1–31. [Google Scholar] [CrossRef] [PubMed]
- Olsson, C.; Jansson, H.; Swenson, J. The Role of Trehalose for the Stabilization of Proteins. J. Phys. Chem. B 2016, 120, 4723–4731. [Google Scholar] [CrossRef]
- Olsson, C.; Genheden, S.; Sakai, V.G.; Swenson, J. Mechanism of Trehalose-Induced Protein Stabilization from Neutron Scattering and Modeling. Phys. Chem. Chem. Phys. 2019, 123, 3679–3697. [Google Scholar] [CrossRef] [PubMed]
- Olsson, C.; Zangana, R.; Swenson, J. Stabilization of proteins embedded in sugars and water as studied by dielectric spectroscopy. Phys. Chem. Chem. Phys. 2020, 22, 21197–21207. [Google Scholar] [CrossRef] [PubMed]
- Di Gioacchino, M.; Bruni, F.; Ricci, M.A. Protection against Dehydration: A Neutron Diffraction Study on Aqueous Solutions of a Model Peptide and Trehalose. J. Phys. Chem. B 2018, 122, 10291–10295. [Google Scholar] [CrossRef]
- Di Gioacchino, M.; Bruni, F.; Ricci, M.A. N-Methylacetamide Aqueous Solutions: A Neutron Diffraction Study. J. Phys. Chem. B 2019, 123, 1808–1814. [Google Scholar] [CrossRef]
- Giuffrida, S.; Cottone, G.; Librizzi, F.; Cordone, L. Coupling between the thermal evolution of the heme pocket and the external matrix structure in trehalose coated carboxymyoglobin. J. Phys. Chem. B 2003, 107, 13211–13217. [Google Scholar] [CrossRef]
- Giuffrida, S.; Cottone, G.; Cordone, L. Structure– Dynamics Coupling between Protein and External Matrix in Sucrose-Coated and in Trehalose-Coated MbCO: An FTIR Study. J. Phys. Chem. B 2004, 108, 15415–15421. [Google Scholar] [CrossRef]
- Cordone, L.; Cottone, G.; Giuffrida, S. Role of residual water hydrogen bonding in sugar/water/biomolecule systems: A possible explanation for trehalose peculiarity. J. Phys. Condens. Matter 2007, 19, 205110. [Google Scholar] [CrossRef]
- Giuffrida, S.; Cottone, G.; Vitrano, E.; Cordone, L. A FTIR study on low hydration saccharide amorphous matrices: Thermal behaviour of the Water Association Band. J. Non-Cryst. Solids 2011, 357, 677–682. [Google Scholar] [CrossRef]
- Francia, F.M.; Dezi, A.; Mallardi, G.; Palazzo, L.; Cordone, L.; Venturoli, G. Protein–matrix coupling/uncoupling in “Dry” systems of photosynthetic reaction center embedded in trehalose/sucrose: The origin of trehalose peculiarity. J. Am. Chem. Soc. 2008, 130, 10240–10246. [Google Scholar] [CrossRef] [PubMed]
- Savitsky, A.; Malferrari, M.; Francia, F.; Venturoli, G.; Möbius, K. Bacterial Photosynthetic Reaction Centers in Trehalose Glasses: Coupling between Protein Conformational Dynamics and Electron-Transfer Kinetics as Studied by Laser-Flash and High-Field EPR Spectroscopies. J. Phys. Chem. B 2010, 114, 12729–12743. [Google Scholar] [CrossRef]
- Malferrari, M.; Nalepa, A.; Venturoli, G.; Francia, F.; Lubitz, W.; Möbius, K.; Savitsky, A. Structural and Dynamical Characteristics of Trehalose and Sucrose Matrices at Different Hydration Levels as Probed by FTIR and High-Field EPR. Phys. Chem. Chem. Phys. 2014, 16, 9831–9848. [Google Scholar] [CrossRef]
- Nalepa, A.; Marco Malferrari, M.; Lubitz, W.; Venturoli, G.; Möbius, K.; Savitsky, A. Local water sensing: Water exchange in bacterial photosynthetic reaction centers embedded in a trehalose glass studied using multiresonance EPR. Phys. Chem. Chem. Phys. 2017, 19, 28388–28400. [Google Scholar] [CrossRef]
- Giuffrida, S.; Cottone, G.; Cordone, L. The Water Association Band as a Marker of Hydrogen Bonds in Trehalose Amorphous Matrices. Phys. Chem. Chem. Phys. 2017, 19, 4251–4265. [Google Scholar] [CrossRef]
- Verma, P.K.; Kundu, A.; Puretz, M.S.; Dhoonmoon, C.; Chegwidden, O.S.; Londergan, C.H.; Cho, M. The Bend + Libration Combination Band Is an Intrinsic, Collective, and Strongly Solute-Dependent Reporter on the Hydrogen Bonding Network of Liquid Water. J. Phys. Chem. B 2018, 122, 2587–2599. [Google Scholar] [CrossRef]
- Giuffrida, S.; Cupane, A.; Cottone, G. Water Association Band in Saccharide Amorphous Matrices: Role of Residual Water on Bioprotection. Int. J. Mol. Sci. 2021, 22, 2496. [Google Scholar] [CrossRef]
- Librizzi, F.; Vitrano, E.; Paciaroni, A.; Cordone, L. Elastic neutron scattering of dry and rehydrated trehalose coated carboxy-myoglobin. Chem. Phys. 2008, 345, 283–288. [Google Scholar] [CrossRef]
- Giuffrida, S.; Cordone, L.; Cottone, G. Bioprotection Can Be Tuned with a Proper Protein/Saccharide Ratio: The Case of Solid Amorphous Matrices. J. Phys. Chem. B 2018, 122, 8642–8653. [Google Scholar] [CrossRef] [PubMed]
- Longo, A.; Giuffrida, S.; Cottone, G.; Cordone, L. Myoglobin Embedded in Saccharide Amorphous Matrices: Water-Dependent Domains Evidenced by Small Angle X-ray Scattering. Phys. Chem. Chem. Phys. 2010, 12, 6852–6858. [Google Scholar] [CrossRef] [PubMed]
- Giuffrida, S.; Panzica, M.; Giordano, F.; Longo, A. SAXS Study on Myoglobin Embedded in Amorphous Saccharide Matrices. Eur. Phys. J. E 2011, 34, 87. [Google Scholar] [CrossRef]
- Bellavia, G.; Cottone, G.; Giuffrida, S.; Cupane, A.; Cordone, L. Thermal Denaturation of Myoglobin in Water-Disaccharide Matrixes: Relation with the Glass Transition of the System. J. Phys. Chem. B 2009, 113, 11543–11549. [Google Scholar] [CrossRef] [PubMed]
- Bellavia, G.; Giuffrida, S.; Cottone, G.; Cupane, A.; Cordone, L. Protein thermal denaturation and matrix glass transition in different protein–trehalose–water systems. J. Phys. Chem. B 2011, 115, 6340–6346. [Google Scholar] [CrossRef] [PubMed]
- Giuffrida, S.; Troia, R.; Schiraldi, C.; D’Agostino, A.; De Rosa, M.; Cordone, L. MbCO Embedded in Trehalosyldextrin Matrices: Thermal Effects and Protein–Matrix Coupling. Food Biophys. 2011, 6, 217–226. [Google Scholar] [CrossRef]
- Semeraro, E.F.; Giuffrida, S.; Cottone, G.; Cupane, A. Biopreservation of Myoglobin in Crowded Environment: A Comparison between Gelatin and Trehalose Matrixes. J. Phys. Chem. B 2017, 121, 8731–8741. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).