Artificial Intelligence in Glioma Diagnosis: A Narrative Review of Radiomics and Deep Learning for Tumor Classification and Molecular Profiling Across Positron Emission Tomography and Magnetic Resonance Imaging
Abstract
1. Introduction
2. Materials and Methods
3. Results
4. Discussion
4.1. Radiomics in Glioma Diagnosis and Molecular Profiling
| Authors | Imaging Modality | Application of Radiomics | Model Type | Performance Metrics | Clinical Utility | Reference |
|---|---|---|---|---|---|---|
| Sun X. et al. (2024) | Conventional MRI | Prediction of glioma subtype based on automatic segmentation | 3D U-Net based CNN | Accuracy: up to 0.909 | Non-invasive glioma molecular characterization | [39] |
| Nakase T. et al. (2025) | Conventional MRI | IDH mutation status prediction | Elastic Net Neural Network | Accuracy: 0.86 | Non-invasive glioma profiling | [41] |
| Mora N. et al. (2023) | T1w, T2w, FLAIR MRI | ATRX mutation status prediction | Lasso Regression | Accuracy: 0.746 | Non-invasive glioma molecular classification | [42] |
| Russo G. et al. (2021) | 11[C]-MET PET/CT | Prediction of tumor grading | Discriminant Analysis | Accuracy: 0.85 | Aid clinical decisions and non-invasive grading | [44] |
| Meng L. et al. (2022) | T1w, T2w, FLAIR, ADC MRI | ATRX mutation status prediction | LASSO + Support Vector Machine (SVM) | Accuracy: 0.88 | Non-invasive genetic profiling | [45] |
| Truong N. et al. (2024) | Preoperative MRI | IDH mutation status prediction | Random Forest, XGBoost ensemble | Accuracy: up to 0.95 | Non-invasive glioma molecular classification | [47] |
| Minh T. et al. (2023) | Conventional MRI, DTI | MGMT methylation status prediction | Multi-stage ML model | Accuracy: 0.80 | Non-invasive therapy stratification in GBM | [48] |
| Zhou W. et al. (2024) | 11[C]-MET PET/CT | IDH mutation status and WHO prediction | LASSO + ML (SelectKBest, Spearman) | AUC (IDH): 0.87 and (WHO): 0.77 | Non-invasive molecular and grade stratification | [49] |
| Zhang C. et al. (2024) | Diffusion MRI (DTI) | IDH mutation status and glioma grade prediction | GAN-based super resolution | AUC (IDH): 0.88 and (grade): 0.81 | Non-invasive molecular status and tumor grading | [50] |
| Du P. et al. (2023) | Preoperative T1w, T2w MRI | Non-invasive prediction of diffuse astrocytic glioma, IDH-wildtype with GBM features (DAG-G) | Multiple ML classifiers (RF, SVM, etc.) | AUC: 0.89–0.91 in external validation | Aid treatment planning by identifying aggressive gliomas preoperatively | [51] |
| Zaragori et al. (2022) | 18F-DOPA PET | Predict IDH mutation and 1p/19q co-deletion status | Logistic Regression (IDH), SVM with RBF kernel (1p/19q) | AUC (IDH): 0.831 (1p/19q): 0.724 | Non-invasive glioma molecular characterization | [53] |
| Ahrari S. et al. (2024) | L-[18F]-fluoro-phenylalanine PET | Predict progression-free survival using delta radiomics | SVM + Recursive Feature Elimination + ElasticNet + GB-Linear | C-Index: 0.783 (Accuracy or AUC not provided) | Prognosis prediction in rare HGG | [56] |
| Lohman P. et al. (2020) | O-2-[18F]-fluoroethyl-L-tyrosine (FET) PET | Discriminate pseudoprogression from early tumor progression | Random Forest classifier + RFE (4 features) | Accuracy: 0.70 | Differentiation of pseudoprogression from tumor progression | [58] |
| Zhang L. et al. (2023) | [18F]-FDG PET + Multi-modal MRI | Predict ATRX mutation status in IDH-mutant LGG | Random Forest integrated with clinical Radiomics | AUC: 0.975 in validation | Non-invasive ATRX mutation prediction in LGGs | [59] |
| Bai J. et al. (2025) | 18F-FET PET/MRI (FLAIR, T1, ADC) | Prediction of molecular genotypes (IDH, TERT, MGMT) | Naïve Bayesian classifier | AUC (IDH): 0.97, (MGMT): 0.86 | Preoperative molecular genotype prediction in adult-type diffuse gliomas | [60] |
4.2. Challenges and Limitations of Radiomics in Gliomas
4.3. Deep Learning Approaches in Gliomas
| Authors | Imaging Modality | Application of Deep Learning | Model Type | Performance Metrics | Clinical Utility | Reference |
|---|---|---|---|---|---|---|
| Iqbal MS. et al. (2025) | Multimodal MRI | Non-invasive MGMT promoter status classification in GBM | 3D Residual U-Net for segmentation + 3D ResNet10 for classification | AUC: 0.66 | Support treatment planning by predicting MGMT status | [65] |
| Koska I. et al. (2025) | Multiparametric MRI (T1w, T2w, FLAIR) | Non-invasive MGMT promoter status prediction in GBM | 3D-ROI-based custom CNN | Accuracy: up to 0.88 | Preoperative prognostic biomarker for treatment planning | [66] |
| Gutsche R. et al. (2023) | 18F-FET PET | Automated metabolic tumor volume segmentation in glioma patients | Artificial Neural Network | Sensitivity: 0.93 and F1 score: 0.92 | Evaluation and response assessment in glioma patients | [67] |
| Rahimpour M. et al. (2023) | 18F-FET PET | Automatic glioma detection, segmentation and Tumor-to-Background ratio estimation | Multi-label CNN and single-label CNN | Sensitivity: 0.89 | Tumor delineation and uptake quantification, reducing inter-reader variability | [68] |
| Waghmare P. et al. (2021) | Multiparametric MRI (T1w, T2w, FLAIR) | Prediction of multiple glioma molecular markers (IDH, MGMT, 1p/19q, grade) | Semi-supervised hierarchical multi-task CNN | Accuracy: 0.823 | Non-invasive genomic profiling for treatment planning | [69] |
| Decuyper M. et al. (2021) | Preoperative MRI (T1w, T2w, FLAIR) | Glioma segmentation and prediction of grade, IDH and 1p/19q co-deletion status | 3D U-Net + multi-task CNN | AUC (grade): 0.93, (IDH): 0.94, (1p/19q): 0.82 | Non-invasive preoperative molecular marker prediction for prognosis and therapy planning | [70] |
| Park J. et al. (2021) | T1w-FLAIR MRI | Synthetic image generation for data augmentation and IDH mutation prediction in GBM | GAN for synthetic generation + diagnostic CNN model | Diagnostic accuracy: 0.90–0.93 | Improved training data and diagnostic accuracy for IDH mutation | [71] |
| Napolitano A. et al. (2021) | Multiparametric MRI (T1w, T2w, FLAIR) | GBM-specific IDH mutation prediction | 4-block 2D CNN | Accuracy: up to 0.83 | Non-invasive IDH status prediction in GBM | [72] |
| Li J. et al. (2023) | T2w MRI data | Automatic segmentation and prediction of H3K27M in diffuse midline gliomas | nnU-Net architecture | Accuracy: 0.85–0.92 | Prediction of H3K27M status for prognosis and treatment stratification | [73] |
4.4. Model Selection in Deep Learning
4.5. Challenges and Limitations of Deep Learning in Gliomas
4.6. Integrating Imaging Modalities with Genomics, Biomechanical, and Clinical Data
4.7. Clinical Translation and Real-World Application
4.8. Future Directions
5. Limitations of the Study
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AI | Artificial Intelligence |
| MRI | Magnetic Resonance Imaging |
| PET | Positron Emission Tomography |
| CNN | Convolutional Neural Network |
| GAN | Generative Adversarial Network |
| IDH | Isocitrate Dehydrogenase |
| MGMT | O6-methylguanine-DNA methyltransferase |
| 1p/19q | 1p/19q Codeletion |
| WHO | World Health Organization |
| AUC | Area Under the Curve |
| FLAIR | Fluid Attenuated Inversion Recovery |
| DTI | Diffusion Tensor Imaging |
| DWI | Diffusion Weighted Imaging |
| PWI | Perfusion Weighted Imaging |
| ML | Machine Learning |
| SVM | Support Vector Machine |
| ADC | Apparent Diffusion Coefficient |
| CT | Computed Tomography |
| TBR | Tumor-to-Background Ratio |
| SHAP | SHapley Additive exPlanations |
| Grad-CAM | Gradient-weighted Class Activation Mapping |
| XAI | Explainable Artificial Intelligence |
| EHR | Electronic Health Record |
| PACS | Picture Archiving and Communication System |
| HIPAA | Health Insurance Portability and Accountability Act |
| GDPR | General Data Protection Regulation |
| SaMD | Software as a Medical Device |
| MTV | Metabolic Tumor Volume |
| ROI | Region of Interest |
| TPR | True Positive Rate |
| FET | Fluoroethyltyrosine |
| FDG | Fluorodeoxyglucose |
| MET | Methionine |
| CCL2 | C-C Motif Chemokine Ligand 2 |
| CDKN2A/B | Cyclin Dependent Kinase Inhibitor 2A and 2B |
| ATRX | Alpha Thalassemia/Mental Retardation Syndrome X-Linked |
| TERT | Telomerase Reverse Transcriptase |
References
- Ostrom, Q.T.; Bauchet, L.; Davis, F.G.; Deltour, I.; Fisher, J.L.; Langer, C.E.; Pekmezci, M.; Schwartzbaum, J.A.; Turner, M.C.; Walsh, K.M.; et al. The epidemiology of glioma in adults: A “state of the science” review. Neuro Oncol. 2014, 16, 896–913. [Google Scholar] [CrossRef]
- Park, Y.W.; Vollmuth, P.; Foltyn-Dumitru, M.; Sahm, F.; Ahn, S.S.; Chang, J.H.; Kim, S.H. The 2021 WHO Classification for Gliomas and Implications on Imaging Diagnosis: Part 1—Key Points of the Fifth Edition and Summary of Imaging Findings on Adult-Type Diffuse Gliomas. Magn. Reson. Imaging 2023, 58, 677–689. [Google Scholar] [CrossRef] [PubMed]
- Tasci, E.; Zhuge, Y.; Zhang, L.; Ning, H.; Cheng, J.Y.; Miller, R.W.; Camphausen, K.; Krauze, A.V. Radiomics and AI-based prediction of MGMT methylation status in glioblastoma using multiparametric MRI: A hybrid feature weighting approach. Diagnostics 2025, 15, 1292. [Google Scholar] [CrossRef] [PubMed]
- Lyoo, Y.W.; Lee, H.; Lee, J.; Park, J.H.; Hwang, I.; Chung, J.W.; Choi, S.H.; Yoo, J.; Choi, K.S. Deep learning enhances reliability of dynamic contrast-enhanced MRI in diffuse gliomas: Bypassing post-processing and providing uncertainty maps. Eur. Radiol. 2025, 35, 6229–6239. [Google Scholar] [CrossRef]
- Li, D.; Hu, W.; Ma, L.; Yang, W.; Liu, Y.; Zou, J.; Ge, X.; Han, Y.; Gan, T.; Cheng, D.; et al. Deep learning radiomics nomograms predict Isocitrate dehydrogenase (IDH) genotypes in brain glioma: A multicenter study. Magn. Reson. Imaging 2025, 117, 110314. [Google Scholar] [CrossRef]
- Yang, Z.; Zhang, P.; Ding, Y.; Deng, L.; Zhang, T.; Liu, Y. Magnetic resonance imaging-based deep learning for predicting subtypes of glioma. Front. Neurol. 2025, 16, 1518815. [Google Scholar] [CrossRef]
- Langen, A.-J.; Galldiks, N.; Mauler, J.; Kocher, M.; Filß, C.P.; Stoffels, G.; Brambilla, C.R.; Stegmayr, C.; Willuweit, A.; Worthoff, W.A.; et al. Hybrid PET/MRI in Cerebral Glioma: Current Status and Perspectives. Cancers 2023, 15, 3577. [Google Scholar] [CrossRef]
- Horowitz, T.; Tabouret, E.; Graillon, T.; Salgues, B.; Chinot, O.; Verger, A.; Guedj, E. Contribution of nuclear medicine to the diagnosis and management of primary brain tumours. Rev. Neurol. 2023, 179, 394–404. [Google Scholar] [CrossRef]
- Harbi, E.; Aschner, M. Correction: Nuclear Medicine Imaging Techniques in Glioblastomas. Neurochem. Res. 2024, 49, 3014. [Google Scholar] [CrossRef]
- Zhang, H.; Zhou, B.; Zhang, H.; Zhang, Y.; Ouyang, Y.; Su, R.; Tang, X.; Lei, Y.; Huang, B. MultiCubeNet: Multitask deep learning for molecular subtyping and prognostic prediction in gliomas. Neurooncol. Adv. 2025, 7, vdaf079. [Google Scholar] [CrossRef]
- Kikuchi, K.; Togao, O.; Yamashita, K.; Momosaka, D.; Kikuchi, Y.; Kuga, D.; Yuhei, S.; Fujioka, Y.; Narutomi, F.; Obara, M.; et al. Comparison of diagnostic performance of radiologist- and AI-based assessments of T2-FLAIR mismatch sign and quantitative assessment using synthetic MRI in the differential diagnosis between astrocytoma, IDH-mutant and oligodendroglioma, IDH-mutant and 1p/19q-codeleted. Neuroradiology 2024, 66, 333–341. [Google Scholar] [PubMed]
- Doniselli, F.M.; Pascuzzo, R.; Mazzi, F.; Padelli, F.; Moscatelli, M.; Akinci D’Antonoli, T.; Cuocolo, R.; Aquino, D.; Cuccarini, V.; Sconfinza, L.M.; et al. Quality assessment of the MRI-radiomics studies for MGMT promoter methylation prediction in glioma: A systematic review and meta-analysis. Eur. Radiol. 2024, 34, 5802–5815. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Liao, Y.; Li, P.; Jin, W.; Fang, J.; Huang, J.; Feng, Y.; Xie, C.; Li, R.; Jin, Q.; et al. A systematic review and meta-analysis of deep learning and radiomics in predicting MGMT promoter methylation status in glioblastoma: Efficacy, reliability, and clinical implications. Displays 2025, 89, 103072. [Google Scholar] [CrossRef]
- Liu, S.; Shah, Z.; Sav, A.; Russo, C.; Berkovsky, S.; Qian, Y.; Coiera, E.; Di Ieva, A. Isocitrate dehydrogenase (IDH) status prediction in histopathology images of gliomas using deep learning. Sci. Rep. 2020, 10, 7733. [Google Scholar] [CrossRef]
- Niu, W.; Yan, J.; Hao, M.; Zhang, Y.; Li, T.; Liu, C.; Li, Q.; Liu, Z.; Su, Y.; Peng, B.; et al. MRI transformer deep learning and radiomics for predicting IDH wild type TERT promoter mutant gliomas. NPJ Precis. Oncol. 2025, 9, 89. [Google Scholar] [CrossRef]
- Ayaz, H.; Oladimeji, O.; McLoughlin, I.; Tormey, D.; Booth, T.C.; Unnikrishnan, S. An eXplainable deep learning model for multi-modal MRI grading of IDH-mutant astrocytomas. Results Eng. 2024, 24, 103353. [Google Scholar] [CrossRef]
- Ye, M.; Cao, Z.; Zhu, Z.; Chen, S.; Zhou, J.; Yang, H.; Li, X.; Chen, X.; Luan, Y.; Li, M.; et al. Integrating quantitative DCE-MRI parameters and radiomic features for improved IDH mutation prediction in gliomas. Front. Oncol. 2025, 15, 1530144. [Google Scholar] [CrossRef]
- Tang, C.; Chen, L.; Xu, Y.; Huang, L.; Zeng, Z. Prediction of TERT mutation status in gliomas using conventional MRI radiogenomic features. Front. Neurol. 2024, 15, 1439598. [Google Scholar] [CrossRef]
- Yuan, J.; Siakallis, L.; Li, H.B.; Brandner, S.; Zhang, J.; Li, C.; Mancini, L.; Bisdas, S. Structural- and DTI-MRI enable automated prediction of IDH Mutation Status in CNS WHO Grade 2–4 glioma patients: A deep Radiomics Approach. BMC Med. Imaging 2024, 24, 104. [Google Scholar] [CrossRef]
- Contreras, K.; Gutierrez-Rengifo, J.; Casanova-Carvajal, O.; Alvarez, A.L.; Vélez-Varela, P.E.; Urbano-Bojorge, A.L. Deep Learning for Glioblastoma Multiforme Detection from MRI: A Statistical Analysis for Demographic Bias. Appl. Sci. 2025, 15, 6274. [Google Scholar] [CrossRef]
- Galldiks, N.; Kaufmann, T.J.; Vollmuth, P.; Lohmann, P.; Smits, M.; Veronesi, M.C.; Langen, K.-J.; Rudà, R.; Albert, N.L.; Hattingen, E.; et al. Challenges, limitations, and pitfalls of PET and advanced MRI in patients with brain tumors: A report of the PET/RANO group. Neuro Oncol. 2024, 26, 1181–1194. [Google Scholar] [CrossRef]
- Galldiks, N.; Lohmann, P.; Albert, N.L.; Tonn, J.C.; Langen, K.-J. Current status of PET imaging in neuro-oncology. Neurooncol. Adv. 2019, 1, vdz010. [Google Scholar] [CrossRef]
- Galldiks, N.; Lohmann, P.; Fink, G.R.; Langen, K.-J. Amino Acid PET in Neurooncology. J. Nucl. Med. 2023, 64, 693–700. [Google Scholar] [CrossRef]
- Liu, Y.; Zhu, Z.; Zhou, J.; Wang, H.; Yang, H.; Yin, J.; Wang, Y.; Li, X.; Chen, F.; Li, Q.; et al. Radiomics prediction of MGMT promoter methylation in adult diffuse gliomas: A combination of structural MRI, DCE, and DTI. Front. Neurol. 2025, 16, 1493666. [Google Scholar] [CrossRef] [PubMed]
- Alongi, P.; Arnone, A.; Vultaggio, V.; Fraternali, A.; Versari, A.; Casali, C.; Arnone, G.; DiMeco, F.; Vetrano, I.G. Artificial Intelligence Analysis Using MRI and PET Imaging in Gliomas: A Narrative Review. Cancers 2024, 16, 407. [Google Scholar] [CrossRef] [PubMed]
- Ni, J.; Zhang, H.; Yang, Q.; Fan, X.; Xu, J.; Sun, J.; Zhang, J.; Hu, Y.; Xiao, Z.; Zhao, Y.; et al. Machine-learning and radiomics-based preoperative prediction of Ki-67 expression in glioma using MRI data. Acad. Radiol. 2024, 31, 3397–3405. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Xiao, F.; Wang, S.; Kuang, S.; Li, Z.; Zhong, Y.; Xu, D.; Cai, Y.; Li, S.; Chen, J.; et al. Preoperative prediction of MGMT promoter methylation in glioblastoma based on multiregional and multi-sequence MRI radiomics analysis. Sci. Rep. 2024, 14, 16031. [Google Scholar] [CrossRef]
- Bjørkeli, E.B.; Esmaeili, M. Multi-task glioma segmentation and IDH mutation and 1p19q codeletion classification via a deep learning model on multimodal MRI. Meta-Radiol. 2025, 3, 100152. [Google Scholar] [CrossRef]
- Foltyn-Dumitru, M.; Rastogi, A.; Cho, J.; Schell, M.; Mahmutoglu, M.A.; Kessler, T.; Sahm, F.; Wick, W.; Bendszus, M.; Brugnara, G.; et al. The potential of GPT-4 advanced data analysis for radiomics-based machine learning models. Neurooncol. Adv. 2024, 7, vdae230. [Google Scholar] [CrossRef]
- Lin, D.; Liu, J.; Ke, C.; Chen, H.; Li, J.; Xie, Y.; Ma, J.; Lv, X.; Feng, Y. Radiomics analysis of quantitative maps from synthetic MRI for predicting grades and molecular subtypes of diffuse gliomas. Clin. Neuroradiol. 2024, 34, 817–826. [Google Scholar] [CrossRef]
- Chouleur, T.; Etchegaray, C.; Villain, L.; Lesur, A.; Ferté, T.; Rossi, M.; Andrique, L.; Simoncini, C.; Giacobbi, A.-S.; Gambaretti, M.; et al. A strategy for multimodal integration of transcriptomics, proteomics, and radiomics data for the prediction of recurrence in patients with IDH-mutant gliomas. Int. J. Cancer 2025, 157, 573–587. [Google Scholar] [CrossRef]
- Lv, Q.; Liu, Y.; Sun, Y.; Wu, M. Insight into deep learning for glioma IDH medical image analysis: A systematic review. Medicine 2024, 103, E37150. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, S.; Botta, F.; Raimondi, S.; Origgi, D.; Fanciullo, C.; Morganti, A.G.; Bellomi, M. Radiomics: The facts and the challenges of image analysis. Eur. Radiol. Exp. 2018, 2, 36. [Google Scholar] [CrossRef] [PubMed]
- Horvat, N.; Papanikolaou, N.; Koh, D.-M. Radiomics Beyond the Hype: A Critical Evaluation Toward Oncologic Clinical Use. Radiol. Artif. Intell. 2024, 6, e230437. [Google Scholar] [CrossRef] [PubMed]
- Parmar, C.; Rios Velazquez, E.; Leijenaar, R.; Jermoumi, M.; Carvalho, S.; Mak, R.H.; Mitra, S.; Shankar, B.U.; Kikinis, R.; Haibe-Kains, B.; et al. Robust radiomics feature quantification using semiautomatic volumetric segmentation. PLoS ONE 2014, 9, e102107. [Google Scholar] [CrossRef]
- Kasap, D.N.G.; Mora, N.G.N.; Blömer, D.A.; Akkurt, B.H.; Heindel, W.L.; Mannil, M.; Musigmann, M. Comparison of MRI sequences to predict IDH mutation status in gliomas using radiomics-based machine learning. Biomedicines 2024, 12, 725. [Google Scholar] [CrossRef]
- He, J.; Ren, J.; Niu, G.; Liu, A.; Wu, Q.; Xie, S.; Ma, X.; Li, B.; Wang, P.; Shen, J.; et al. Multiparametric MR radiomics in brain glioma: Models comparison to predict biomarker status. BMC Med. Imaging 2022, 22, 137. [Google Scholar] [CrossRef]
- Dedhia, M.; Germano, I.M. The Evolving Landscape of Radiomics in Gliomas: Insights into Diagnosis, Prognosis, and Research Trends. Cancers 2025, 17, 1582. [Google Scholar] [CrossRef]
- Sun, X.; Li, S.; Ma, C.; Fang, W.; Jing, X.; Yang, C.; Zhang, Y.; Liu, H.; Wang, Q.; Zhao, L.; et al. Glioma subtype prediction based on radiomics of tumor and peritumoral edema under automatic segmentation. Sci. Rep. 2024, 14, 27471. [Google Scholar] [CrossRef]
- Leone, A.; Di Napoli, V.; Fochi, N.P.; Di Perna, G.; Spetzger, U.; Filimonova, E.; Angileri, F.; Carbone, F.; Colamaria, A. Virtual Biopsy for the Prediction of MGMT Promoter Methylation in Gliomas: A Comprehensive Review of Radiomics and Deep Learning Approaches Applied to MRI. Diagnostics 2025, 15, 251. [Google Scholar] [CrossRef]
- Nakase, T.; Henderson, G.A.; Barba, T.; Bareja, R.; Guerra, G.; Zhao, Q.; Francis, S.S.; Gevaert, O.; Kachuri, L. Integration of MRI radiomics and germline genetics to predict the IDH mutation status of gliomas. NPJ Precis. Oncol. 2025, 9, 187. [Google Scholar] [CrossRef]
- Nacul Mora, N.G.; Akkurt, B.H.; Kasap, D.; Blömer, D.; Heindel, W.; Mannil, M.; Musigmann, M. Comparison of MRI Sequences to Predict ATRX Status Using Radiomics-Based Machine Learning. Diagnostics 2023, 13, 2216. [Google Scholar] [CrossRef] [PubMed]
- Kocher, M.; Ruge, M.I.; Galldiks, N.; Lohmann, P. Applications of radiomics and machine learning for radiotherapy of malignant brain tumors. Strahlenther. Onkol. 2020, 196, 856–867. [Google Scholar] [CrossRef] [PubMed]
- Russo, G.; Stefano, A.; Alongi, P.; Comelli, A.; Catalfamo, B.; Mantarro, C.; Longo, C.; Altieri, R.; Certo, F.; Cosentino, S.; et al. Feasibility on the Use of Radiomics Features of 11[C]-MET PET/CT in Central Nervous System Tumours: Preliminary Results on Potential Grading Discrimination Using a Machine Learning Model. Curr. Oncol. 2021, 28, 5318–5331. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.; Zhang, R.; Fa, L.; Zhang, L.; Wang, L.; Shao, G. ATRX status in patients with gliomas: Radiomics analysis. Medicine 2022, 101, E30189. [Google Scholar] [CrossRef] [PubMed]
- Di Salle, G.; Tumminello, L.; Laino, M.E.; Shalaby, S.; Aghakhanyan, G.; Fanni, S.C.; Febi, M.; Shortrede, J.E.; Miccoli, M.; Faggioni, L.; et al. Accuracy of Radiomics in Predicting IDH Mutation Status in Diffuse Gliomas: A Bivariate Meta-Analysis. Radiol. Artif. Intell. 2024, 6, e220257. [Google Scholar] [CrossRef]
- Truong, N.C.D.; Yogananda, C.G.B.; Wagner, B.C.; Holcomb, J.M.; Reddy, D.; Saadat, N.; Hatanpaa, K.J.; Patel, T.R.; Fei, B.; Lee, M.D.; et al. Two-Stage Training Framework Using Multicontrast MRI Radiomics for IDH Mutation Status Prediction in Glioma. Radiol. Artif. Intell. 2024, 6, e230218. [Google Scholar] [CrossRef]
- Minh, T.N.T.; Le, V.H.; Le, N.Q.K. Diffusion-tensor imaging and dynamic susceptibility contrast MRIs improve radiomics-based machine learning model of MGMT promoter methylation status in glioblastomas. Biomed. Signal Process. Control. 2023, 86, 105122. [Google Scholar] [CrossRef]
- Zhou, W.; Wen, J.; Huang, Q.; Zeng, Y.; Zhou, Z.; Zhu, Y.; Chen, L.; Guan, Y.; Xie, F.; Zhuang, D.; et al. Development and Validation of Clinical-Radiomics Analysis for Preoperative Prediction of IDH Mutation Status and WHO Grade in Diffuse Gliomas: A Consecutive L-[methyl-11C] Methionine Cohort Study with Two PET Scanners. Eur. J. Nucl. Med. Mol. Imaging 2024, 51, 1423–1435. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, P.; He, J.; Wu, Q.; Xie, S.; Li, B.; Hao, X.; Wang, S.; Zhang, H.; Hao, Z.; et al. Super-Resolution Reconstruction Improves Multishell Diffusion: Using Radiomics to Predict Adult-Type Diffuse Glioma IDH and Grade. Front. Oncol. 2024, 14, 1435204. [Google Scholar] [CrossRef]
- Du, P.; Wu, X.; Liu, X.; Chen, J.; Cao, A.; Geng, D. Establishment of a Prediction Model Based on Preoperative MRI Radiomics for Diffuse Astrocytic Glioma, IDH-Wildtype, with Molecular Features of Glioblastoma. Cancers 2023, 15, 5094. [Google Scholar] [CrossRef]
- Kihira, S.; Derakhshani, A.; Leung, M.; Mahmoudi, K.; Bauer, A.; Zhang, H.; Polson, J.; Arnold, C.; Tsankova, N.M.; Hormigo, A.; et al. Multi-Parametric Radiomic Model to Predict 1p/19q Co-Deletion in Patients with IDH-1 Mutant Glioma: Added Value to the T2-FLAIR Mismatch Sign. Cancers 2023, 15, 1037. [Google Scholar] [CrossRef] [PubMed]
- Zaragori, T.; Oster, J.; Roch, V.; Hossu, G.; Chawki, M.B.; Grignon, R.; Pouget, C.; Gauchotte, G.; Rech, F.; Blonski, M.; et al. 18F-FDOPA PET for the Noninvasive Prediction of Glioma Molecular Parameters: A Radiomics Study. J. Nucl. Med. 2022, 63, 147–157. [Google Scholar] [CrossRef] [PubMed]
- Elsevier Adds Half a Million Records from ClinicalTrials.gov to Embase, Enabling a Seamless Search Experience in the World’s Most Comprehensive Biomedical Database. Available online: https://www.elsevier.com/about/press-releases/elsevier-adds-half-a-million-records-from-clinicaltrials-gov-to-embase (accessed on 30 July 2025).
- Tam, L.; Han, M.; Wright, J.; Toescu, S.; Campion, A.; Shpanskaya, K.; Mankad, K.; Ho, C.; Lober, R.; Cheshier, S.; et al. MRI-Based Radiomic Prognostic Markers of Diffuse Midline Glioma. Neuro-Oncol. 2020, 22, iii357. [Google Scholar] [CrossRef]
- Ahrari, S.; Zaragori, T.; Zinsz, A.; Oster, J.; Imbert, L.; Verger, A. Application of PET imaging delta radiomics for predicting progression-free survival in rare high-grade glioma. Sci. Rep. 2024, 14, 3256. [Google Scholar] [CrossRef]
- Taylor, C.; Ekert, J.O.; Sefcikova, V.; Fersht, N.; Samandouras, G. Discriminators of pseudoprogression and true progression in high-grade gliomas: A systematic review and meta-analysis. Sci. Rep. 2022, 12, 13258. [Google Scholar] [CrossRef]
- Lohmann, P.; Elahmadawy, M.A.; Gutsche, R.; Werner, J.-M.; Bauer, E.K.; Ceccon, G.; Kocher, M.; Lerche, C.W.; Rapp, M.; Fink, G.R.; et al. FET PET Radiomics for Differentiating Pseudoprogression from Early Tumor Progression in Glioma Patients Post-Chemoradiation. Cancers 2020, 12, 3835. [Google Scholar] [CrossRef]
- Zhang, L.; Pan, H.; Liu, Z.; Gao, J.; Xu, X.; Wang, L.; Wang, J.; Tang, Y.; Cao, X.; Kan, Y.; et al. Multicenter clinical radiomics-integrated model based on [18F]FDG PET and multi-modal MRI predict ATRX mutation status in IDH-mutant lower-grade gliomas. Eur. Radiol. 2023, 33, 872–883. [Google Scholar] [CrossRef]
- Bai, J.; Cui, B.; Li, F.; Han, X.; Yang, H.; Lu, J. Multiparametric radiomics signature for predicting molecular genotypes in adult-type diffuse gliomas utilizing 18F-FET PET/MRI. BMC Med. Imaging 2025, 25, 187. [Google Scholar] [CrossRef]
- Donelli, M.; Espa, G.; Feraco, P. A Semi-Unsupervised Segmentation Methodology Based on Texture Recognition for Radiomics: A Preliminary Study on Brain Tumours. Electronics 2022, 11, 1573. [Google Scholar] [CrossRef]
- Carré, A.; Klausner, G.; Edjlali, M.; Lerousseau, M.; Briend-Diop, J.; Sun, R.; Ammari, S.; Reuzé, S.; Alvarez Andres, E.; Estienne, T.; et al. Standardization of brain MR images across machines and protocols: Bridging the gap for MRI-based radiomics. Sci. Rep. 2020, 10, 12340. [Google Scholar] [CrossRef]
- Fukui, R.; Onishi, M.; Hasegawa, K.; Ohata, M.; Kida, K.; Goto, S. Effect of segmentation dimension on radiomics analysis for MGMT promoter methylation status in gliomas. Aktual. Neurol. 2024, 24, 8–14. [Google Scholar] [CrossRef]
- Wang, P.; Xie, S.; Wu, Q.; Weng, L.; Hao, Z.; Yuan, P.; Zhang, C.; Gao, W.; Wang, S.; Zhang, H.; et al. Model incorporating multiple diffusion MRI features: Development and validation of a radiomics-based model to predict adult-type diffuse gliomas grade. Eur. Radiol. 2023, 33, 8809–8820. [Google Scholar] [CrossRef]
- Iqbal, M.S.; Bajwa, U.I.; Raza, R.; Anwar, M.W. Integrated brain tumor segmentation and MGMT promoter methylation status classification from multimodal MRI data using deep learning. Digit. Health 2025, 11, 20552076251332018. [Google Scholar] [CrossRef]
- Koska, İ.Ö.; Koska, Ç. Deep learning classification of MGMT status of glioblastomas using multiparametric MRI with a novel domain knowledge augmented mask fusion approach. Sci. Rep. 2025, 15, 3273. [Google Scholar] [CrossRef]
- Gutsche, R.; Lowis, C.; Ziemons, K.; Kocher, M.; Ceccon, G.; Régio Brambilla, C.; Shah, N.J.; Langen, K.-J.; Galldiks, N.; Isensee, F.; et al. Automated Brain Tumor Detection and Segmentation for Treatment Response Assessment Using Amino Acid PET. J. Nucl. Med. 2023, 64, 1594–1602. [Google Scholar] [CrossRef] [PubMed]
- Rahimpour, M.; Boellaard, R.; Jentjens, S.; Deckers, W.; Goffin, K.; Koole, M. A multi-label CNN model for the automatic detection and segmentation of gliomas using [18F]FET PET imaging. Eur. J. Nucl. Med. Mol. Imaging 2023, 50, 2441–2452. [Google Scholar] [CrossRef] [PubMed]
- Tupe-Waghmare, P.; Malpure, P.; Kotecha, K.; Beniwal, M.; Santosh, V.; Saini, J.; Ingalhalikar, M. Comprehensive Genomic Subtyping of Glioma Using Semi-Supervised Multi-Task Deep Learning on Multimodal MRI. IEEE Access 2021, 9, 167900–167910. [Google Scholar] [CrossRef]
- Decuyper, M.; Bonte, S.; Deblaere, K.; Van Holen, R. Automated MRI based pipeline for segmentation and prediction of grade, IDH mutation and 1p19q co-deletion in glioma. Comput. Med. Imaging Graph. 2021, 88, 101831. [Google Scholar] [CrossRef]
- Park, J.E.; Eun, D.; Kim, H.S.; Lee, D.H.; Jang, R.W.; Kim, N. Generative adversarial network for glioblastoma ensures morphologic variations and improves diagnostic model for isocitrate dehydrogenase mutant type. Sci. Rep. 2021, 11, 9912. [Google Scholar] [CrossRef]
- Napolitano, A.; Pasquini, L.; Tagliente, E.; Dellepiane, F.; Lucignani, M.; Vidiri, A.; Ranazzi, G.; Stoppacciaro, A.; Moltoni, G.; Nicolai, M.; et al. A novel deep learning model to differentiate IDH-mutant from IDH-wild type glioblastoma multiforme on a cohort of GBM cases. Phys. Medica 2021, 92, S135. [Google Scholar] [CrossRef]
- Li, J.; Zhang, P.; Qu, L.; Sun, T.; Duan, Y.; Wu, M.; Weng, J.; Li, Z.; Gong, X.; Liu, X.; et al. Deep Learning for Noninvasive Assessment of H3 K27M Mutation Status in Diffuse Midline Gliomas Using MR Imaging. Magn. Reson. Imaging 2023, 58, 850–861. [Google Scholar] [CrossRef]
- Zhou, Q.; Wang, Y.; Zhang, Q.; Wei, X.; Yao, Y.; Xia, L. Noninvasive prediction of CCL2 expression level in high-grade glioma patients. Cancer Med. 2024, 13, e70016. [Google Scholar] [CrossRef]
- Gao, J.; Liu, Z.; Pan, H.; Cao, X.; Kan, Y.; Wen, Z.; Chen, S.; Wen, M.; Zhang, L. Preoperative Discrimination of CDKN2A/B Homozygous Deletion Status in Isocitrate Dehydrogenase-Mutant Astrocytoma: A Deep Learning-Based Radiomics Model Using MRI. J. Magn. Reson. Imaging 2024, 59, 1655–1664. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zeng, M.; Tong, Y.; Zhang, T.; Fu, Y.; Li, H.; Zhang, Z.; Cheng, Z.; Xu, X.; Yang, R.; et al. Automatic Prediction of MGMT Status in Glioblastoma via Deep Learning-Based MR Image Analysis. BioMed Res. Int. 2020, 2020, 9258649. [Google Scholar] [CrossRef] [PubMed]
- Conte, G.M.; Moassefi, M.; Faghani, S.; Sanvito, F.; Decker, P.; Kizilbash, S.H.; Eckel-Passow, J.; Ida, C.; Erickson, B. Performance of deep learning in mgmt promoter methylation status prediction using brain mri: Results from a large cohort of idh-wildtype gliomas tested by a single methylation assay. Neuro-Oncol. 2023, 25, v199. [Google Scholar] [CrossRef]
- Takahashi, S.; Takahashi, M.; Kinoshita, M.; Miyake, M.; Sese, J.; Kobayashi, K.; Tanaka, S.; Takayanagi, S.; Takami, H.; Yamazawa, E. RBIO-03. Initial result of develop robust deep learning model for detecting genomic status in gliomas against image differences among facilities. Neuro-Oncol. 2021, 23 (Suppl. S6), vi192. [Google Scholar] [CrossRef]
- Herr, J.; Stoyanova, R.; Mellon, E.A. Convolutional Neural Networks for Glioma Segmentation and Prognosis: A Systematic Review. Crit. Rev. Oncog. 2024, 29, 33–65. [Google Scholar] [CrossRef]
- Lost, J.; Verma, T.; Jekel, L.; von Reppert, M.; Tillmanns, N.; Merkaj, S.; Cassinelli Petersen, G.; Bahar, R.; Gordem, A.; Haider, M.A.; et al. Systematic literature review of artificial intelligence algorithms using pre-therapy MR imaging for GLIOMA molecular subtype classification. Neuro-Oncol. 2021, 23, vi139. [Google Scholar] [CrossRef]
- Hrapșa, I.; Florian, I.A.; Șușman, S.; Farcaș, M.; Beni, L.; Florian, I.S. External Validation of a Convolutional Neural Network for IDH Mutation Prediction. Medicina 2022, 58, 526. [Google Scholar] [CrossRef]
- Foltyn-Dumitru, M.; Schell, M.; Rastogi, A.; Sahm, F.; Kessler, T.; Wick, W.; Bendszus, M.; Brugnara, G.; Vollmuth, P. Impact of signal intensity normalization of MRI on the generalizability of radiomic-based prediction of molecular glioma subtypes. Eur. Radiol. 2024, 34, 2782–2790. [Google Scholar] [CrossRef]
- Farahani, S.; Hejazi, M.; Moradizeyveh, S.; Di Ieva, A.; Fatemizadeh, E.; Liu, S. Diagnostic Accuracy of Deep Learning Models in Predicting Glioma Molecular Markers: A Systematic Review and Meta-Analysis. Diagnostics 2025, 15, 797. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, Z.; Lin, J.; Wan, Z.; Weng, J.; Yuan, Z.; Xie, Y.; Liu, Z.; Xie, P.; Mao, S.; Wang, Z.; et al. Radiogenomic profiling of global DNA methylation associated with molecular phenotypes and immune features in glioma. BMC Med. 2024, 22, 352. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Liu, H.; Du, F.; Wang, D.; Huo, X.; Tian, J.; Song, L. MDPNet: A dual-path parallel fusion network for multi-modal MRI glioma genotyping. Front. Oncol. 2025, 15, 1574861. [Google Scholar] [CrossRef] [PubMed]
- Wen, S.; Yang, S.; Xu, L.; Yang, Y.; Qi, Y.; Hu, P.; Chen, Q.; Zhang, D. A one-stage multi-task network for molecular subtyping, grading, and segmentation of glioma. Biomed. Signal Process. Control 2025, 108, 107923. [Google Scholar] [CrossRef]
- Yang, X.C.; Gao, J. Advances in biomechanics: Exploring biophysical models in cellular mechanics. Comput. Mol. Biol. 2024, 14, 125–133. [Google Scholar] [CrossRef]
- Yan, J.; Zhang, S.; Sun, Q.; Wang, W.; Duan, W.; Wang, L.; Ding, T.; Pei, D.; Sun, C.; Wang, W.; et al. Predicting 1p/19q co-deletion status from magnetic resonance imaging using deep learning in adult-type diffuse lower-grade gliomas: A discovery and validation study. Lab. Investig. 2022, 102, 154–159. [Google Scholar] [CrossRef]
- Kim, B.-H.; Lee, H.; Choi, K.S.; Nam, J.G.; Park, C.-K.; Park, S.-H.; Chung, J.W.; Choi, S.H. Validation of MRI-Based Models to Predict MGMT Promoter Methylation in Gliomas: BraTS 2021 Radiogenomics Challenge. Cancers 2022, 14, 4827. [Google Scholar] [CrossRef]
- Park, C.J.; Park, Y.W.; Ahn, S.S.; Kim, D.; Kim, E.H.; Kang, S.-G.; Chang, J.H.; Kim, S.H.; Lee, S.-K. Quality of Radiomics Research on Brain Metastasis: A Roadmap to Promote Clinical Translation. Korean J. Radiol. 2022, 23, 77–88. [Google Scholar] [CrossRef]
- van Leeuwen, K.G.; Schalekamp, S.; Rutten, M.J.C.M.; van Ginneken, B.; de Rooij, M. Artificial intelligence in radiology: 100 commercially available products and their scientific evidence. Eur. Radiol. 2021, 31, 3797–3804. [Google Scholar] [CrossRef]
- Shin, H.; Park, J.E.; Jun, Y.; Eo, T.; Lee, J.; Kim, J.E.; Lee, D.H.; Moon, H.H.; Park, S.I.; Kim, S.; et al. Deep learning referral suggestion and tumour discrimination using explainable artificial intelligence applied to multiparametric MRI. Eur. Radiol. 2023, 33, 5859–5870. [Google Scholar] [CrossRef] [PubMed]
- Rosenbacke, R.; Melhus, Å.; McKee, M.; Stuckler, D. How Explainable Artificial Intelligence Can Increase or Decrease Clinicians’ Trust in AI Applications in Health Care: Systematic Review. JMIR AI 2024, 3, e53207. [Google Scholar] [CrossRef] [PubMed]
- Farhud, D.D.; Zokaei, S. Ethical Issues of Artificial Intelligence in Medicine and Healthcare. Iran. J. Public Health 2021, 50, I–V. [Google Scholar] [CrossRef] [PubMed]
- MacIntyre, M.R.; Cockerill, R.G.; Mirza, O.F.; Appel, J.M. Ethical considerations for the use of artificial intelligence in medical decision-making capacity assessments. Psychiatry Res. 2023, 328, 115466. [Google Scholar] [CrossRef]
- Casale, R.; Lavrova, E.; Sanduleanu, S.; Woodruff, H.C.; Lambin, P. Development and external validation of a non-invasive molecular status predictor of chromosome 1p/19q co-deletion based on MRI radiomics analysis of Low Grade Glioma patients. Eur. J. Radiol. 2021, 139, 109678. [Google Scholar] [CrossRef]
- Warraich, H.J.; Tazbaz, T.; Califf, R.M. FDA Perspective on the Regulation of Artificial Intelligence in Health Care and Biomedicine. JAMA 2025, 333, 241. [Google Scholar] [CrossRef]
- Alleman, K.; Knecht, E.; Huang, J.; Zhang, L.; Lam, S.; DeCuypere, M. Multimodal Deep Learning-Based Prognostication in Glioma Patients: A Systematic Review. Cancers 2023, 15, 545. [Google Scholar] [CrossRef]
- Miteva, M.; Nisheva-Pavlova, M. The power of integrating multiple data sources in medical imaging: A study of MGMT methylation status. Procedia Comput. Sci. 2024, 239, 1196–1203. [Google Scholar] [CrossRef]
- Xu, J.; Meng, Y.; Qiu, K.; Topatana, W.; Li, S.; Wei, C.; Chen, T.; Chen, M.; Ding, Z.; Niu, G. Applications of Artificial Intelligence Based on Medical Imaging in Glioma: Current State and Future Challenges. Front. Oncol. 2022, 12, 892056. [Google Scholar] [CrossRef]
- Pati, S.; Baid, U.; Edwards, B.; Sheller, M.; Wang, S.-H.; Reina, G.A.; Foley, P.; Gruzdev, A.; Karkada, D.; Davatzikos, C.; et al. Federated learning enables big data for rare cancer boundary detection. Nat. Commun. 2022, 13, 7346. [Google Scholar] [CrossRef]
- Ourotech, Inc. Prospective Evaluation of AI R&D Tool in Adult Glioma and Other Primary Brain Tumours (PEAR-GLIO); Report No.: NCT06038760; 2025 July. Available online: https://clinicaltrials.gov/study/NCT06038760 (accessed on 30 July 2025).
- Dasgupta, D.A. Spatial and Temporal Characterization of Gliomas Using Radiomic Analysis [Internet]; Report No.: NCT06036381; 2025 April. Available online: https://clinicaltrials.gov/study/NCT06036381 (accessed on 30 July 2025).
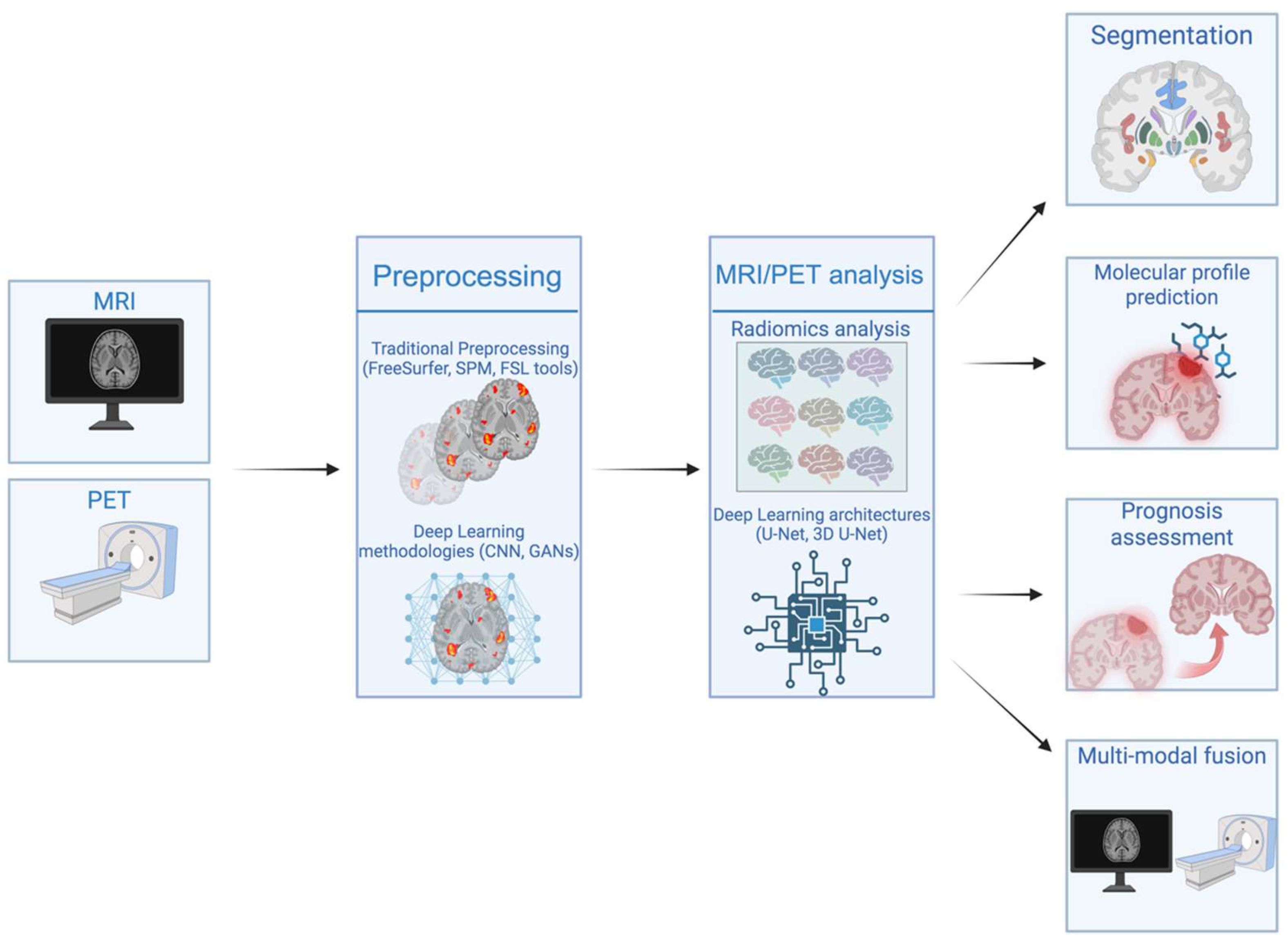
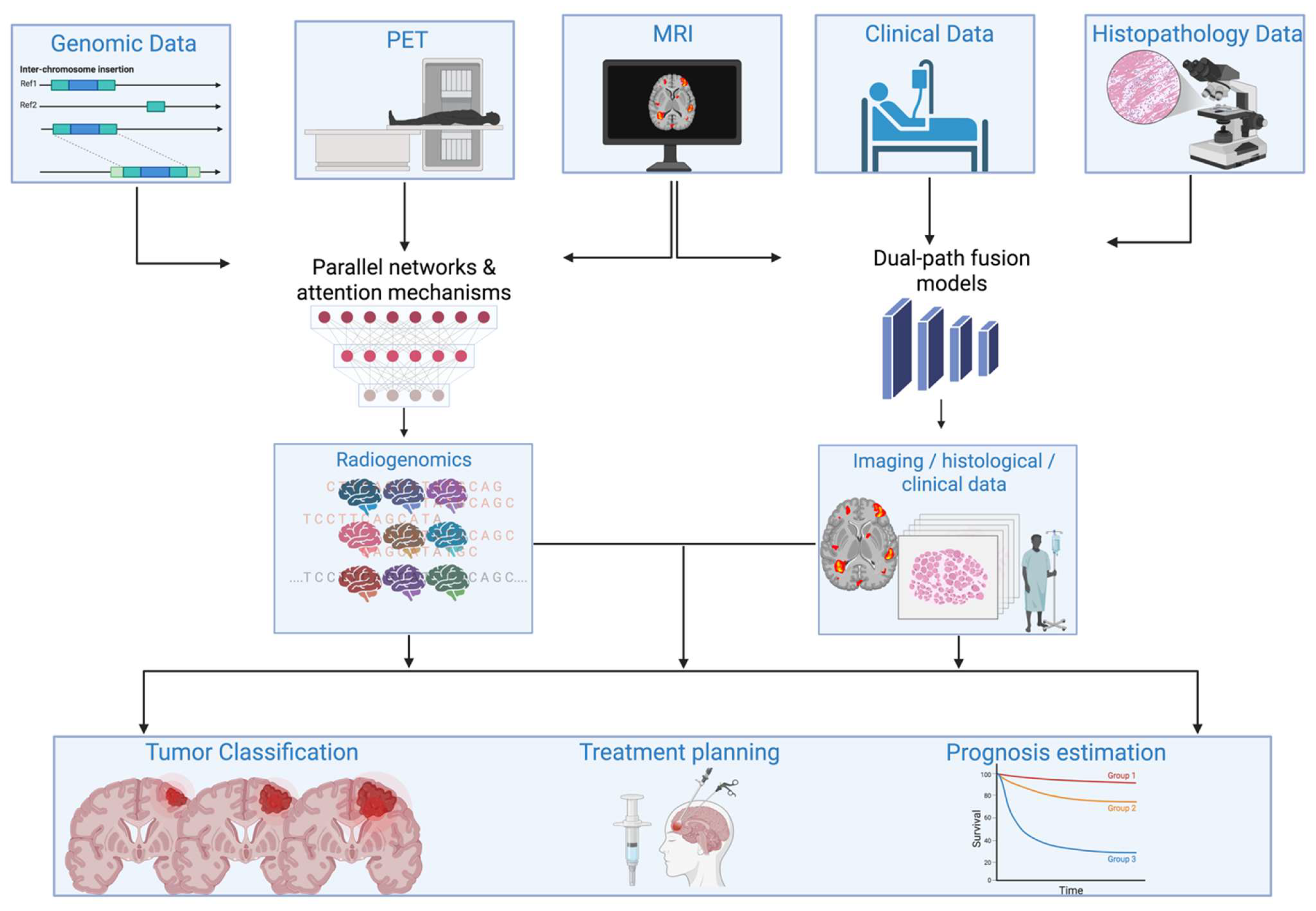
| Domain | Radiomics | Deep Learning |
|---|---|---|
| Feature Extraction | Relies on handcrafted features (e.g., shape, texture, intensity) extracted from imaging data | Learns features automatically from raw imaging data, without prior handcrafted design |
| Interpretability | More transparent; features can be linked to biological or pathological processes | Often considered a “black box” with limited interpretability unless explainable AI is applied |
| Data Requirements | Can be applied to smaller datasets with careful feature selection and robust modeling | Requires large, annotated datasets for effective training and generalization |
| Flexibility | Well-suited for combining imaging with clinical or genomic data | Highly adaptable to multimodal inputs and end-to-end tasks (e.g., segmentation + classification) |
| Performance | Good predictive performance but may plateau with highly complex tasks | Demonstrates superior accuracy in segmentation, classification, and molecular prediction |
| Reproducibility | Affected by differences in feature extraction protocols across centers | Model performance may vary with architecture, training data and preprocessing pipelines |
| Aspect | Traditional Methods (Histopathology and Imaging) | AI-Based Approaches (Radiomics and Deep Learning) |
|---|---|---|
| Invasiveness | Biopsy is required for definitive diagnosis; invasive and carries procedural risks [10,11] | Non-invasive, based on MRI and PET imaging features [3] |
| Time Efficiency | Histopathology and molecular profiling are time-consuming and can delay treatment decisions [16,17] | Rapid predictions generated directly from imaging data [3] |
| Sampling Bias | Biopsies may not capture tumor heterogeneity, leading to under- or misdiagnosis [14,15] | Analyzes the entire tumor volume, accounting for spatial heterogeneity [46] |
| Specificity | Imaging features alone lack sufficient specificity; overlap with treatment-related changes; biopsy considered a highly specific method [10,11] | Captures subtle, multidimensional patterns invisible to the human eye [3,24] |
| Reproducibility | Imaging interpretation varies across readers and institutions, limiting reproducibility [18,19,20] | Algorithms provide consistent and scalable outputs when trained on heterogeneous datasets and undergo external validation [63] |
| Molecular Insight | Requires histopathology and genetic testing for markers like IDH, MGMT, 1p/19q [2,12,13] | Can non-invasively predict molecular features such as IDH mutation, MGMT promoter methylation, and 1p/19q codeletion [26,27] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Christodoulou, R.C.; Pitsillos, R.; Papageorgiou, P.S.; Petrou, V.; Vamvouras, G.; Rivera, L.; Papageorgiou, S.G.; Solomou, E.E.; Georgiou, M.F. Artificial Intelligence in Glioma Diagnosis: A Narrative Review of Radiomics and Deep Learning for Tumor Classification and Molecular Profiling Across Positron Emission Tomography and Magnetic Resonance Imaging. Eng 2025, 6, 262. https://doi.org/10.3390/eng6100262
Christodoulou RC, Pitsillos R, Papageorgiou PS, Petrou V, Vamvouras G, Rivera L, Papageorgiou SG, Solomou EE, Georgiou MF. Artificial Intelligence in Glioma Diagnosis: A Narrative Review of Radiomics and Deep Learning for Tumor Classification and Molecular Profiling Across Positron Emission Tomography and Magnetic Resonance Imaging. Eng. 2025; 6(10):262. https://doi.org/10.3390/eng6100262
Chicago/Turabian StyleChristodoulou, Rafail C., Rafael Pitsillos, Platon S. Papageorgiou, Vasileia Petrou, Georgios Vamvouras, Ludwing Rivera, Sokratis G. Papageorgiou, Elena E. Solomou, and Michalis F. Georgiou. 2025. "Artificial Intelligence in Glioma Diagnosis: A Narrative Review of Radiomics and Deep Learning for Tumor Classification and Molecular Profiling Across Positron Emission Tomography and Magnetic Resonance Imaging" Eng 6, no. 10: 262. https://doi.org/10.3390/eng6100262
APA StyleChristodoulou, R. C., Pitsillos, R., Papageorgiou, P. S., Petrou, V., Vamvouras, G., Rivera, L., Papageorgiou, S. G., Solomou, E. E., & Georgiou, M. F. (2025). Artificial Intelligence in Glioma Diagnosis: A Narrative Review of Radiomics and Deep Learning for Tumor Classification and Molecular Profiling Across Positron Emission Tomography and Magnetic Resonance Imaging. Eng, 6(10), 262. https://doi.org/10.3390/eng6100262






