Enhanced Performance Electrochemical Biosensor for Detection of Prostate Cancer Biomarker PCA3 Using Specific Aptamer
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents and Apparatus
2.2. Nucleic-Acid-Based Sensor Development
2.2.1. Fabrication of the Sensor
2.2.2. Electrochemical Detection of PCA3
3. Results and Discussion
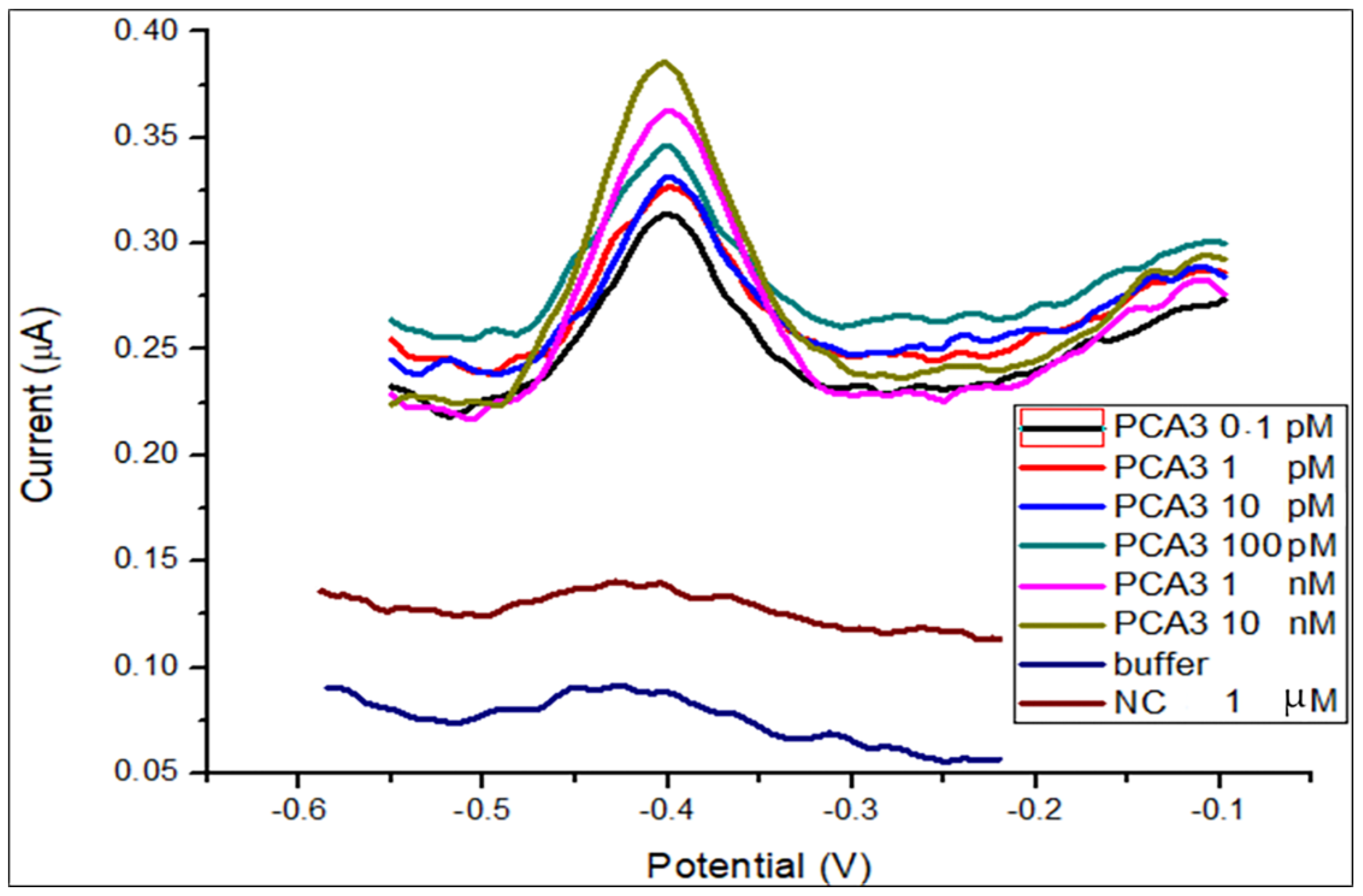
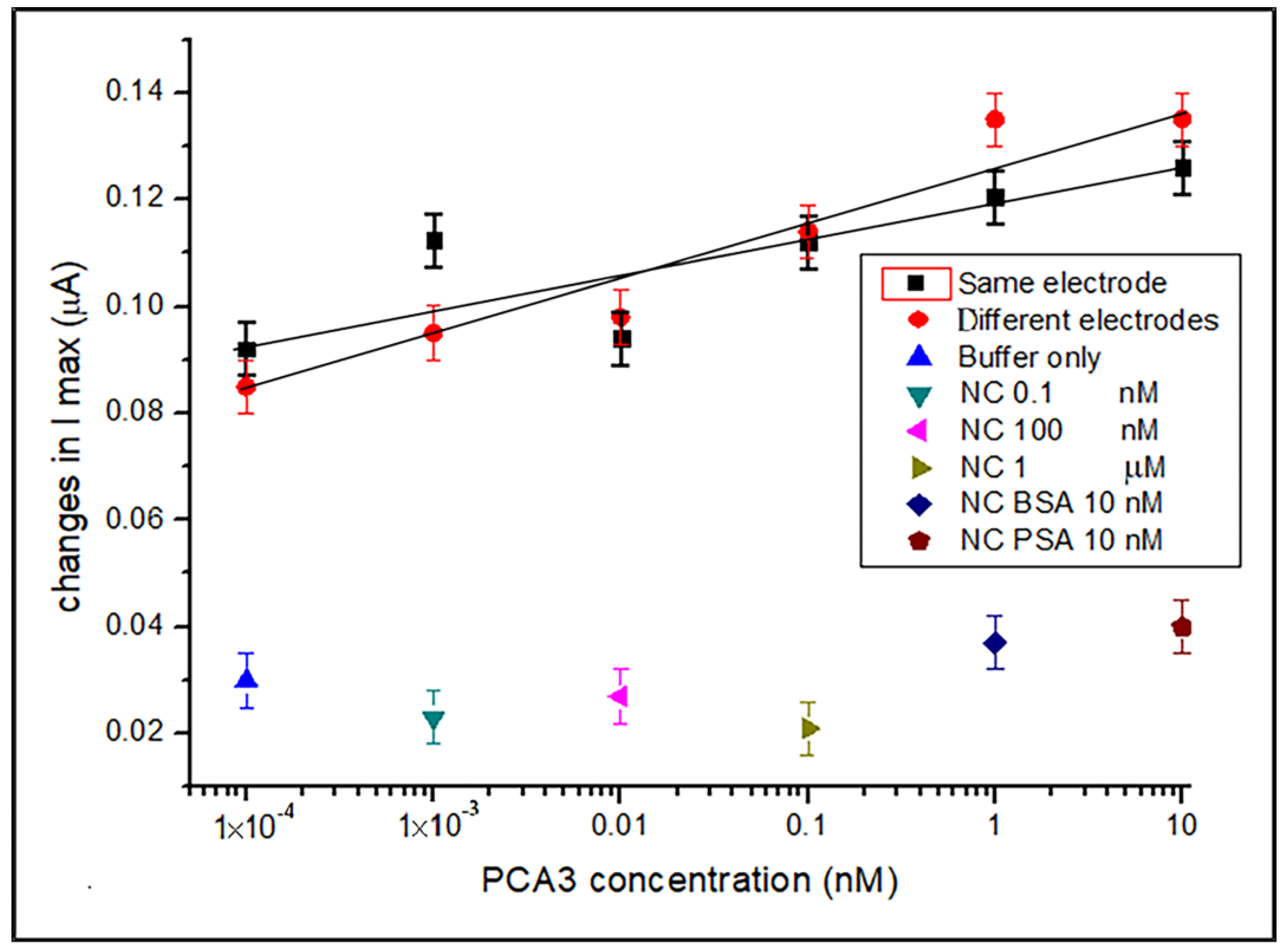
3.1. DPV Detection of PCA3 in Buffer
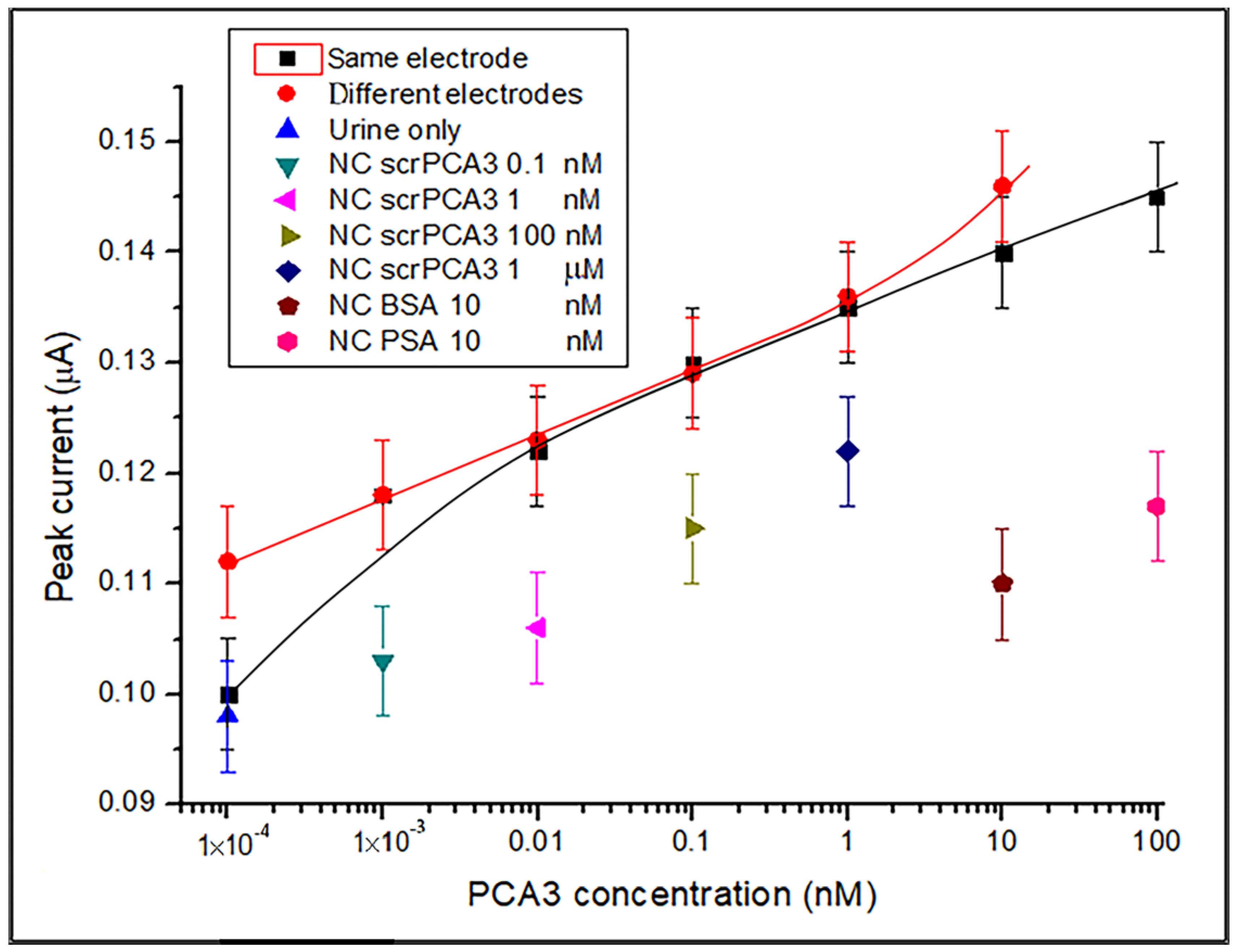
3.2. DPV Detection of PCA3 in Synthetic Urine
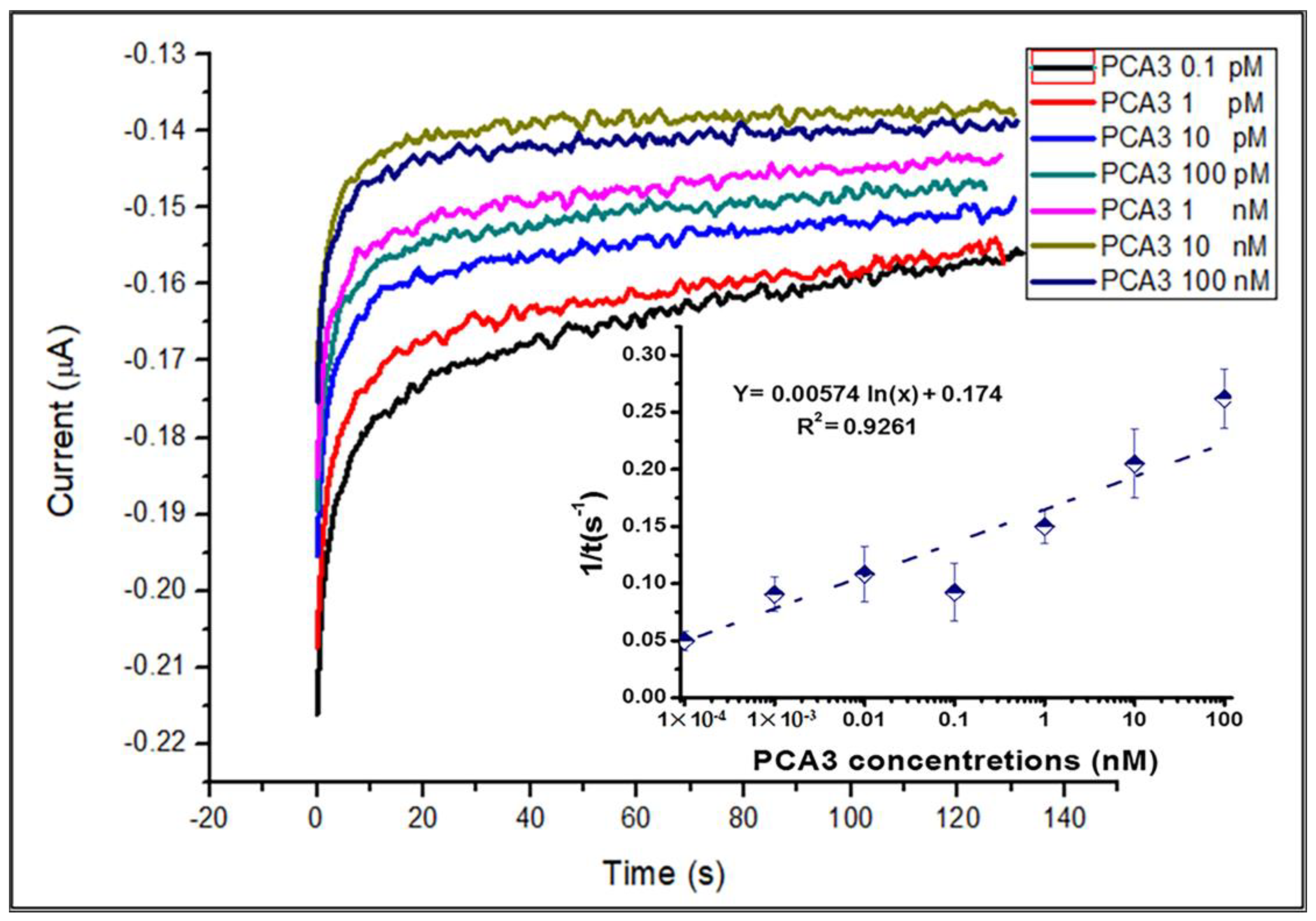
3.3. Aptamer/PCA3 Binding Kinetics Study
3.4. AFM Study of PCA3 t Aptamer Binding
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zadra, G.; Priolo, C.; Patnaik, A.; Loda, M. New Strategies in Prostate Cancer: Targeting Lipogenic Pathways and the Energy Sensor AMPK. Clin. Cancer Res. 2010, 16, 3322–3328. [Google Scholar] [CrossRef] [PubMed]
- Haga, Y.; Uemura, M.; Baba, S.; Inamura, K.; Takeuchi, K.; Nonomura, N.; Ueda, K. Identification of Multisialylated LacdiNAc Structures as Highly Prostate Cancer Specific Glycan Signatures on PSA. Anal. Chem. 2019, 91, 2247–2254. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Zhang, K.; He, Y.; Xu, X.; Li, D.; Cheng, S.; Zheng, X. Synergistic effects and mechanisms of impressic acid or acankoreanogein in combination with docetaxel on prostate cancer. RSC Adv. 2018, 8, 2768–2776. [Google Scholar] [CrossRef]
- Mangadlao, J.D.; Wang, X.; McCleese, C.; Escamilla, M.; Ramamurthy, G.; Wang, Z.; Govande, M.; Basilion, J.P.; Burda, C. Prostate-Specific Membrane Antigen Targeted Gold Nanoparticles for Theranostics of Prostate Cancer. ACS Nano 2018, 12, 3714–3725. [Google Scholar] [CrossRef]
- Catalona, W.J.; Richie, J.P.; Ahmann, F.R.; Hudson, M.A.; Scardino, P.T.; Flanigan, R.C.; Dekernion, J.B.; Ratliff, T.L.; Kavoussi, L.R.; Dalkin, B.L.; et al. Comparison of Digital Rectal Examination and Serum Prostate Specific Antigen in the Early Detection of Prostate Cancer: Results of a Multicenter Clinical Trial of 6630 Men. J. Urol. 1994, 151, 1283–1290. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef]
- Mottet, N.; van den Bergh, R.C.N.; Briers, E.; Van den Broeck, T.; Cumberbatch, M.G.; De Santis, M.; Fanti, S.; Fossati, N.; Gandaglia, G.; Gillessen, S.; et al. EAU-EANM-ESTRO-ESUR-SIOG Guidelines on Prostate Cancer—2020 Update. Eur. Urol. 2021, 79, 243–262. [Google Scholar] [CrossRef]
- Dall’Era, M.A.; Albertsen, P.C.; Bangma, C.; Carroll, P.R.; Carter, H.B.; Cooperberg, M.R.; Freedland, S.J.; Klotz, L.H.; Parker, C.; Soloway, M.S. Active Surveillance for Prostate Cancer: A Systematic Review of the Literature. Eur. Urol. 2012, 62, 976–983. [Google Scholar] [CrossRef]
- Tokudome, S.; Ando, R.; Koda, Y. Discoveries and application of prostate-specific antigen, and some proposals to optimize prostate cancer screening. Cancer Manag. Res. 2016, 8, 45–47. [Google Scholar] [CrossRef]
- Stephan, C.; Cammann, H.; Meyer, H.-A.; Lein, M.; Jung, K. PSA and new biomarkers within multivariate models to improve early detection of prostate cancer. Cancer Lett. 2007, 249, 18–29. [Google Scholar] [CrossRef]
- Stamey, T.A.; Caldwell, M.; McNeal, J.E.; Nolley, R.; Hemenez, M.; Downs, J. The prostate specific antigen era in the United States is over for prostate cancer: What happened in the last 20 years? J. Urol. 2004, 172, 1297–1301. [Google Scholar] [CrossRef] [PubMed]
- Adamaki, M.; Zoumpourlis, V. Prostate Cancer Biomarkers: From diagnosis to prognosis and precision-guided therapeutics. Pharmacol. Ther. 2021, 228, 107932. [Google Scholar] [CrossRef] [PubMed]
- Landers, K.A.; Burger, M.J.; Tebay, M.A.; Purdie, D.M.; Scells, B.; Samaratunga, H.; Lavin, M.F.; Gardiner, R.A. Use of multiple biomarkers for a molecular diagnosis of prostate cancer. Int. J. Cancer 2005, 114, 950–956. [Google Scholar] [CrossRef]
- Cary, K.C.; Cooperberg, M.R. Biomarkers in prostate cancer surveillance and screening: Past, present, and future. Ther. Adv. Urol. 2013, 5, 318–329. [Google Scholar] [CrossRef]
- Truong, M.; Yang, B.; Jarrard, D.F. Toward the Detection of Prostate Cancer in Urine: A Critical Analysis. J. Urol. 2013, 189, 422–429. [Google Scholar] [CrossRef]
- Salagierski, M.; Schalken, J.A. Molecular Diagnosis of Prostate Cancer: PCA3 and TMPRSS2:ERG Gene Fusion. J. Urol. 2012, 187, 795–801. [Google Scholar] [CrossRef] [PubMed]
- Auprich, M.; Bjartell, A.; Chun, F.K.-H.; de la Taille, A.; Freedland, S.J.; Haese, A.; Schalken, J.; Stenzl, A.; Tombal, B.; van der Poel, H. Contemporary Role of Prostate Cancer Antigen 3 in the Management of Prostate Cancer. Eur. Urol. 2011, 60, 1045–1054. [Google Scholar] [CrossRef]
- Pettersson, A.; Graff, R.E.; Bauer, S.R.; Pitt, M.J.; Lis, R.T.; Stack, E.C.; Martin, N.E.; Kunz, L.; Penney, K.L.; Ligon, A.H.; et al. The TMPRSS2:ERG Rearrangement, ERG Expression, and Prostate Cancer Outcomes: A Cohort Study and Meta-analysis. Cancer Epidemiol. Biomark. Prev. 2012, 21, 1497–1509. [Google Scholar] [CrossRef]
- Warrick, J.I.; Tomlins, S.A.; Carskadon, S.L.; Young, A.M.; Siddiqui, J.; Wei, J.T.; Chinnaiyan, A.M.; Kunju, L.P.; Palanisamy, N. Evaluation of tissue PCA3 expression in prostate cancer by RNA in situ hybridization—A correlative study with urine PCA3 and TMPRSS2-ERG. Mod. Pathol. 2014, 27, 609–620. [Google Scholar] [CrossRef]
- Groskopf, J.; Aubin, S.M.; Deras, I.L.; Blase, A.; Bodrug, S.; Clark, C.; Brentano, S.; Mathis, J.; Pham, J.; Meyer, T.; et al. APTIMA PCA3 Molecular Urine Test: Development of a Method to Aid in the Diagnosis of Prostate Cancer. Clin. Chem. 2006, 52, 1089–1095. [Google Scholar] [CrossRef]
- Gittelman, M.C.; Hertzman, B.; Bailen, J.; Williams, T.; Koziol, I.; Henderson, R.J.; Efros, M.; Bidair, M.; Ward, J.F. PCA3 Molecular Urine Test as a Predictor of Repeat Prostate Biopsy Outcome in Men with Previous Negative Biopsies: A Prospective Multicenter Clinical Study. J. Urol. 2013, 190, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Htoo, K.P.P.; Yamkamon, V.; Yainoy, S.; Suksrichavalit, T.; Viseshsindh, W.; Eiamphungporn, W. Colorimetric detection of PCA3 in urine for prostate cancer diagnosis using thiol-labeled PCR primer and unmodified gold nanoparticles. Clin. Chim. Acta 2019, 488, 40–49. [Google Scholar] [CrossRef] [PubMed]
- De Kok, J.B.; Verhaegh, G.W.; Roelofs, R.W.; Hessels, D.; Kiemeney, L.A.; Aalders, T.W.; Swinkels, D.W.; Schalken, J.A. DD3(PCA3), a very sensitive and specific marker to detect prostate tumors. Cancer Res. 2002, 62, 2695–2698. [Google Scholar] [PubMed]
- Hessels, D.; Gunnewiek, J.M.T.K.; van Oort, I.; Karthaus, H.F.M.; van Leenders, G.J.L.; van Balken, B.; Kiemeney, L.A.; Witjes, J.A.; Schalken, J.A. DD3PCA3-based Molecular Urine Analysis for the Diagnosis of Prostate Cancer. Eur. Urol. 2003, 44, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Kanyong, P.; Rawlinson, S.; Davis, J. Immunochemical Assays and Nucleic-Acid Detection Techniques for Clinical Diagnosis of Prostate Cancer. J. Cancer 2016, 7, 523–531. [Google Scholar] [CrossRef][Green Version]
- Takita, S.; Nabok, A.; Smith, D.; Lishchuk, A. Spectroscopic Ellipsometry Detection of Prostate Cancer Bio-Marker PCA3 Using Specific Non-Labeled Aptamer: Comparison with Electrochemical Detection. Chem. Proc. 2021, 5, 65. [Google Scholar] [CrossRef]
- Chen, A.; Chatterjee, S. Nanomaterials based electrochemical sensors for biomedical applications. Chem. Soc. Rev. 2013, 42, 5425–5438. [Google Scholar] [CrossRef]
- Maduraiveeran, G.; Sasidharan, M.; Ganesan, V. Electrochemical sensor and biosensor platforms based on advanced nanomaterials for biological and biomedical applications. Biosens. Bioelectron. 2018, 103, 113–129. [Google Scholar] [CrossRef]
- Labib, M.; Sargent, E.H.; Kelley, S.O. Electrochemical Methods for the Analysis of Clinically Relevant Biomolecules. Chem. Rev. 2016, 116, 9001–9090. [Google Scholar] [CrossRef]
- Syedmoradi, L.; Daneshpour, M.; Alvandipour, M.; Gomez, F.A.; Hajghassem, H.; Omidfar, K. Point of care testing: The impact of nanotechnology. Biosens. Bioelectron. 2017, 87, 373–387. [Google Scholar] [CrossRef]
- Beitollahi, H.; Mohammadi, S.Z.; Safaei, M.; Tajik, S. Applications of electrochemical sensors and biosensors based on modified screen-printed electrodes: A review. Anal. Methods 2020, 12, 1547–1560. [Google Scholar] [CrossRef]
- Antuña-Jiménez, D.; González-García, M.B.; Hernández-Santos, D.; Fanjul-Bolado, P. Screen-Printed Electrodes Modified with Metal Nanoparticles for Small Molecule Sensing. Biosensors 2020, 10, 9. [Google Scholar] [CrossRef] [PubMed]
- Jolly, P.; Formisano, N.; Tkáč, J.; Kasák, P.; Frost, C.G.; Estrela, P. Label-free impedimetric aptasensor with antifouling surface chemistry: A prostate specific antigen case study. Sens. Actuators B Chem. 2015, 209, 306–312. [Google Scholar] [CrossRef]
- Rodrigues, V.C.; Soares, J.C.; Soares, A.C.; Braz, D.C.; Melendez, M.E.; Ribas, L.C.; Scabini, L.F.S.; Bruno, O.M.; Carvalho, A.L.; Reis, R.M.; et al. Electrochemical and optical detection and machine learning applied to images of genosensors for diagnosis of prostate cancer with the biomarker PCA3. Talanta 2021, 222, 121444. [Google Scholar] [CrossRef]
- Moranova, L.; Stanik, M.; Hrstka, R.; Campuzano, S.; Bartosik, M. Electrochemical LAMP-based assay for detection of RNA biomarkers in prostate cancer. Talanta 2022, 238, 123064. [Google Scholar] [CrossRef]
- Sánchez-Salcedo, R.; Miranda-Castro, R.; De-los-Santos-Álvarez, N.; Lobo-Castañón, M.J. Dual electrochemical genosensor for early diagnosis of prostate cancer through lncRNAs detection. Biosens. Bioelectron. 2021, 192, 113520. [Google Scholar] [CrossRef]
- Nabok, A.; Abu-Ali, H.; Takita, S.; Smith, D.P. Electrochemical detection of prostate cancer biomarker pca3 using specific rna-based aptamer labelled with ferrocene. Chemosensors 2021, 9, 59. [Google Scholar] [CrossRef]
- Takita, S.; Nabok, A.; Lishchuk, A.; Smith, D. Optimization of Apta-Sensing Platform for Detection of Prostate Cancer Marker PCA3. Int. J. Mol. Sci. 2021, 22, 12701. [Google Scholar] [CrossRef]
- Takita, S.; Nabok, A.; Lishchuk, A.; Mussa, M.H.; Smith, D. Detection of Prostate Cancer Biomarker PCA3 with Electrochemical Aptasensor. Eng. Proc. 2022, 16, 8. [Google Scholar] [CrossRef]
- Deshmukh, M.A.; Patil, H.K.; Bodkhe, G.A.; Yasuzawa, M.; Koinkar, P.; Ramanaviciene, A.; Shirsat, M.D.; Ramanavicius, A. EDTA-modified PANI/SWNTs nanocomposite for differential pulse voltammetry based determination of Cu(II) ions. Sens. Actuators B Chem. 2018, 260, 331–338. [Google Scholar] [CrossRef]
- Adeloju, S.B.; Bond, A.M.; Briggs, M.H. Multielement determination in biological materials by differential pulse voltammetry. Anal. Chem. 1985, 57, 1386–1390. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Xu, N.; Men, H.; Li, S.; Lu, Y.; Low, S.S.; Li, X.; Zhu, L.; Cheng, C.; Xu, G.; et al. Salivary Cortisol Determination on Smartphone-Based Differential Pulse Voltammetry System. Sensors 2020, 20, 1422. [Google Scholar] [CrossRef] [PubMed]
- Marangoni, K.; Neves, A.F.; Rocha, R.M.; Faria, P.R.; Alves, P.T.; Souza, A.G.; Fujimura, P.T.; Santos, F.A.A.; Araújo, T.G.; Ward, L.S.; et al. Prostate-specific RNA aptamer: Promising nucleic acid antibody-like cancer detection. Sci. Rep. 2015, 5, 12090. [Google Scholar] [CrossRef] [PubMed]
- Sharafeldin, M.; Fleschhut, F.; James, T.; Davis, J.J. A Quantification of Target Protein Biomarkers in Complex Media by Faradaic Shotgun Tagging. Anal. Chem. 2022, 94, 2375–2382. [Google Scholar] [CrossRef]
- Reddy, N.R.; Rhodes, S.; Fang, J. Colorimetric Detection of Dopamine with J-Aggregate Nanotube-Integrated Hydrogel Thin Films. ACS Omega 2020, 5, 18198–18204. [Google Scholar] [CrossRef]
- Zheng, H.; Han, X.; Wei, Q.; Liu, X.; Li, Y.; Zhou, J. A green flexible and wearable biosensor based on carbon nanofibers for sensitive detection of uric acid in artificial urine. J. Mater. Chem. B 2022, 10, 8450–8461. [Google Scholar] [CrossRef]
- Attar, A.M.; Richardson, M.B.; Speciale, G.; Majumdar, S.; Dyer, R.P.; Sanders, E.C.; Penner, R.M.; Weiss, G.A. Electrochemical Quantification of Glycated and Non-glycated Human Serum Albumin in Synthetic Urine. ACS Appl. Mater. Interfaces 2019, 11, 4757–4765. [Google Scholar] [CrossRef]
- Mehennaoui, S.; Poorahong, S.; Jimenez, G.C.; Siaj, M. Selection of high affinity aptamer-ligand for dexamethasone and its electrochemical biosensor. Sci. Rep. 2019, 9, 6600. [Google Scholar] [CrossRef]
- Du, Y.; Chen, C.; Yin, J.; Li, B.; Zhou, M.; Dong, S.; Wang, E. Solid-State Probe Based Electrochemical Aptasensor for Cocaine: A Potentially Convenient, Sensitive, Repeatable, and Integrated Sensing Platform for Drugs. Anal. Chem. 2010, 82, 1556–1563. [Google Scholar] [CrossRef]
- Su, W.; Kim, S.E.; Cho, M.; Nam, J.D.; Choe, W.S.; Lee, Y. Selective detection of endotoxin using an impedance aptasensor with electrochemically deposited gold nanoparticles. Innate Immun. 2013, 19, 388–397. [Google Scholar] [CrossRef]



| Samples | Roughness Parameters | ||
|---|---|---|---|
| Ra (nm) | Rz (nm) | Rmax (nm) | |
| (a) | 0.85 | 2.52 | 3.10 |
| (b) | 0.58 | 1.53 | 2.85 |
| (c) | 2.12 | 5.20 | 5.60 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Takita, S.; Nabok, A.; Lishchuk, A.; Mussa, M.H.; Smith, D. Enhanced Performance Electrochemical Biosensor for Detection of Prostate Cancer Biomarker PCA3 Using Specific Aptamer. Eng 2023, 4, 367-379. https://doi.org/10.3390/eng4010022
Takita S, Nabok A, Lishchuk A, Mussa MH, Smith D. Enhanced Performance Electrochemical Biosensor for Detection of Prostate Cancer Biomarker PCA3 Using Specific Aptamer. Eng. 2023; 4(1):367-379. https://doi.org/10.3390/eng4010022
Chicago/Turabian StyleTakita, Sarra, Alexei Nabok, Anna Lishchuk, Magdi H. Mussa, and David Smith. 2023. "Enhanced Performance Electrochemical Biosensor for Detection of Prostate Cancer Biomarker PCA3 Using Specific Aptamer" Eng 4, no. 1: 367-379. https://doi.org/10.3390/eng4010022
APA StyleTakita, S., Nabok, A., Lishchuk, A., Mussa, M. H., & Smith, D. (2023). Enhanced Performance Electrochemical Biosensor for Detection of Prostate Cancer Biomarker PCA3 Using Specific Aptamer. Eng, 4(1), 367-379. https://doi.org/10.3390/eng4010022








