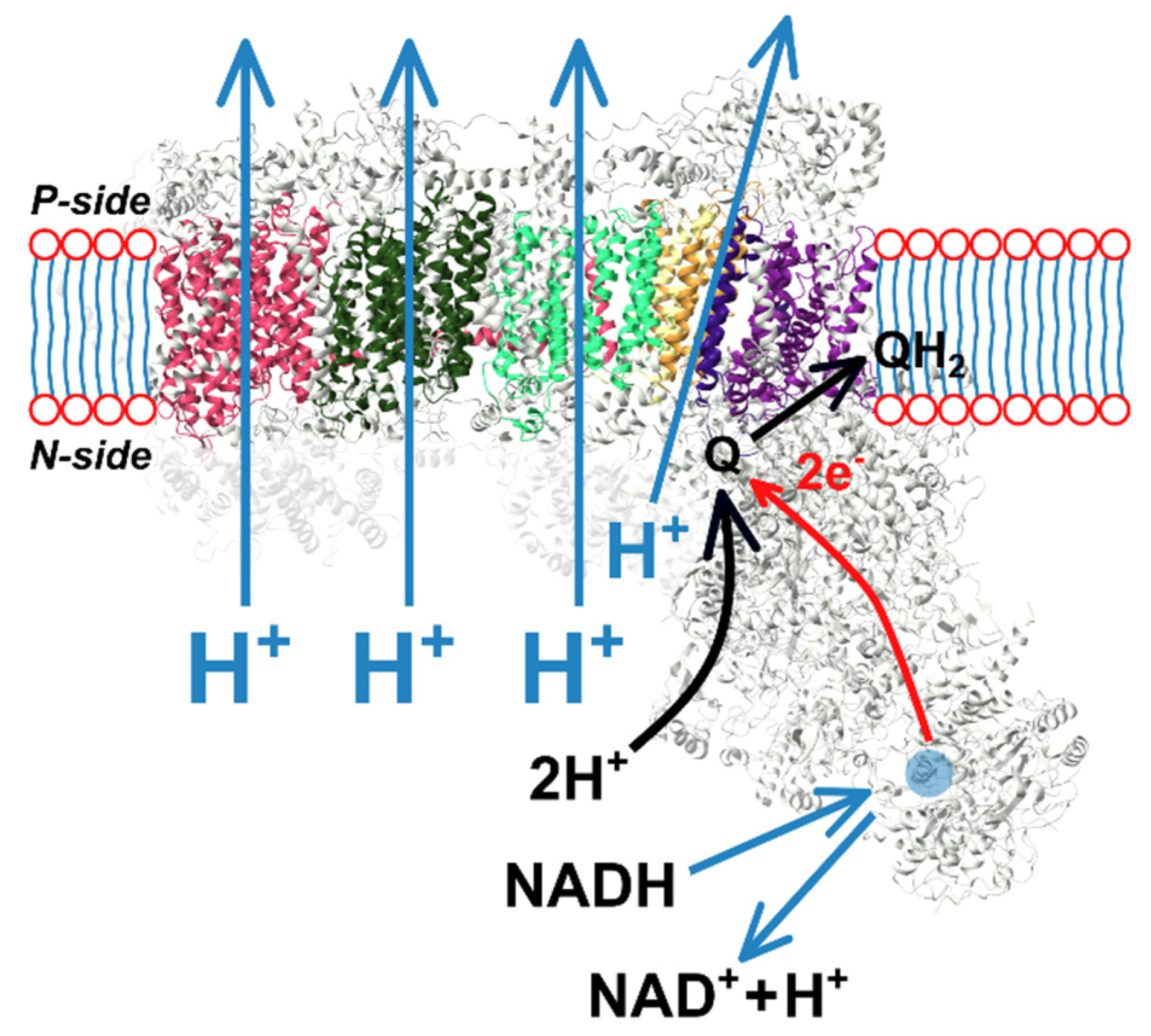
1. Introduction
The respiratory electron-transport chain of the inner mitochondrial membrane enables eukaryotic cells to store chemical energy from nutrients in the form of adenosine triphosphate (ATP), which serves as the “energy currency” of the cell [
1]. The conversion of food energy requires the excitation of highly energetic electrons above 1 eV. This excess energy will dissipate if not converted to a more stable form. In the first step of ATP synthesis, proton-pumping complexes stockpile this electron energy by generating and maintaining a proton gradient across the membrane, which manifests itself as a proton-motive force (PMF). In the second step, the PMF facilitates a proton current which drives the rotation of a mechanical nanomotor, the ATP synthase enzyme. In the third step, mechanical energy provides the means for ATP synthesis.
Complex I (NADH-quinone oxidoreductase) is the largest and most elaborate enzyme complex of the respiratory chain [
2,
3,
4,
5]. It serves as the first electron-acceptor for the incoming reducing equivalents in the respiratory chain. The structure of Complex I was resolved recently in a series of X-ray and cryo-electron microscopy experiments [
6,
7,
8,
9,
10,
11,
12]. Complex I consists of an L-shaped assembly of a hydrophobic arm embedded in the lipid membrane and a hydrophilic peripheral arm, which protrudes into the mitochondrial matrix (
Figure 1). Electron transfer from nicotinamide adenine dinucleotide (NADH) to quinone occurs in the hydrophilic domain via a set of FeS complexes, while the four proton pumps are located within the membrane region [
13]. The electron and proton pathways are spatially separated (up to 30 nm from the tip of the electron entry to the terminal proton-pumping subunit). The physical mechanism of the electron–proton energy exchange remains elusive. It is commonly accepted that electron transfer events facilitate conformational changes along the complex (manifesting as an electrostatic wave) that lead to proton transfer against the established PMF (about 200 mV across the 3 nm thin membrane) [
2,
12,
14,
15,
16]. In our previous work [
17], we revealed the physical mechanism of energy transfer from such an electrostatic wave to a pumped proton. For simplicity, we replaced the electron system in our model with an external periodic force. In the present paper, we complete our model by introducing electron transfer as the cause of conformational changes.
Our analysis is based on a single-particle approach exploring resemblances between processes in semiconductor structures and living organisms at the nanoscale. Indeed, within the mitochondrial complexes, electron transport occurs as hopping between metal atoms (or FeS clusters) embedded into the proteins, similar to the hopping between semiconductor quantum dots. Such 0D nanostructures are frequently called “artificial atoms”, and we extend the similarity by calling atoms “natural quantum dots”. Non-radiative energy transfer process (Förster transfer) in quantum dots is caused by a two-particle electron–hole Coulomb interaction. In proton-pumping complexes, electrostatics is the only possible way to transfer energy from electrons to protons, and this process can also be treated within a two-particle picture. Finally, the temperature for nanostructure operations (4 K) scales to physiological temperatures of 300 K, similarly to the scale of typical quantum dot energies (few meV) to the redox drop from NADH to O
2 (1.1 eV, in several steps). In other words, in biological systems, energy levels are so well separated that they remain discrete even at elevated temperatures. We successfully applied this approach in several papers [
17,
18,
19,
20,
21,
22] explaining the operating principles of mitochondrial complexes. For many purposes, in physiological conditions, our quantum Heisenberg equations can be rewritten in the form of rate equations with possible quantum effects ignored. When needed, manifestations of quantum effects can be revealed, as in the case of exciton transfer in photosynthetic complexes [
23].
2. Methods
We examined the model exhibited in
Figure 2. It consists of three electron sites placed between two electron reservoirs (source and drain, representing NADH and quinone shuttles, respectively), and three proton sites placed between two proton reservoirs representing positive and negative sides of the membrane. The respective positions of the energy levels demonstrate that electron transport occurs in a usual way, from higher potential to lower, whereas protons are pumped to higher potential. As the energies of electron site
L and proton site
A are below their corresponding chemical potentials, these sites are initially populated from source reservoir and the negative side of the membrane, respectively (
Figure 2a). The electron can proceed to the next site
C where it is temporarily stuck because of the large energy mismatch between this site and the next site
R. At the central site, the electron facilitates conformational changes, which we model as a piston with positive charges at the edges. The positive charge at the right edge is attracted by the electron at the central site, and the piston moves to the right (
Figure 2b). When the piston is shifted to the right, the energy of the middle proton site
M decreases, facilitating proton transfer to this site. Simultaneously, the energy of electron site
C drops, decreasing the energy mismatch, and the electron can proceed to site
R (
Figure 2c) and to the drain reservoir. When the electron escapes, elastic forces return the piston to the left, increasing the energy of populated proton site
M, allowing the proton to move to site
B (
Figure 2d) and to the positive side of the membrane. Consequently, the mechanical motion of the piston mediates energy transfer from the electronic system to the protonic one. It should be noted that our model is quite simplified in comparison to the real structure. Electron transport occurs via eight FeS complexes and protons are transferred via various residues and water molecules. We included just three sites in both cases, using two of them for the prevention of back current and allowing for the energy modulation of the middle ones. However, in our simple model, we succeeded in revealing the physical principles both for the creation of the electrostatic wave and for its action into facilitating proton pumping.
To describe this model, we introduce the Hamiltonian:
The electron part of the Hamiltonian,
HE, is given by
where
are the electron creation/annihilation operators for the
σ-site (
σ =
L,
C,
R) with
Eσ being the electron energies at these sites;
and
are the electron creation/annihilation operators for the source and drain, respectively, with
ESk and
EDk being the energies for these electrons; ∆
σσ’ are the transfer amplitudes between the sites, where
TSk and
TDk are the amplitudes of the transfers from the source and drain, respectively, to the corresponding electron sites. The proton Hamiltonian has a similar form given by
where
are the proton creation/annihilation operators for the
τ-site (
τ =
A,
M,
B), with
Eτ being the proton energies at these sites;
and
are the proton creation/annihilation operators for the negative and positive sides of the membrane, respectively, with
ENq and
EPq being the energies for these protons; and ∆
ττ’ are the transfer amplitudes between the sites, where
TNq and
TPq are the amplitudes of the transfers from the negative and positive sides of the membrane, respectively, to the corresponding proton sites.
Henv describes the coupling of electrons and protons to the protein environment, represented by the set of independent harmonic oscillators, as
Here,
pj and
xj are the momentum and coordinate of the
j-th harmonic oscillator with mass
mj and frequency
ωj, and
cσj and
Cτj are the electron- and proton-environment coupling strengths, respectively.
The energies of the middle electron (
C) and proton (
M) sites depend on the position of the piston,
x, as
and
where
le,p and
re,p are the horizontal separations of the piston from the corresponding sites (along the direction of the motion) at the equilibrium position and the vertical shifts, respectively.
We assume that the piston is in the overdamped regime, and its dynamics can be described by the phenomenological Langevin equation given by
where
ζ is the drag coefficient; the first term on the right side is responsible for the elastic forces returning the piston back to the equilibrium position; the second and third terms are the electrostatic forces from the electrons and protons of the middle sites, respectively, with
nC and
NM being the populations of these sites; and the last term represents white noise with zero mean and correlation function
The equations of motion for the electron and proton operators can be obtained from the corresponding Hamiltonians. It was shown previously [
16,
19,
22] that in physiological conditions they can be rewritten in terms of the rate equations for the site populations averaged over the environment. In our model, these equations have forms
for electrons, and
for protons. Here, the angular brackets mean both thermal and quantum-mechanical averaging. The reservoir coupling constants are calculated as
and assumed to be frequency-independent. The reservoir distribution Fermi functions are given by
where
μS,D.N,P are the chemical potentials of the corresponding reservoirs. Kinetic coefficients Φ
σ (
σ =
L,
R) and Φ
τ (
τ =
A,
B) have the forms
where
are the Marcus rates, and
are the reorganization energies of the environment due to electron and proton transfer events. The Marcus rates are not postulated here but derived microscopically [
16,
19,
22].
When electron and proton populations are determined, the corresponding currents can be calculated. As expected, the incoming and outgoing currents are equal in magnitude and opposite in sign for protons and electrons and given by
Correspondingly, the quantum yield of the system is defined as the ratio of the proton and electron current magnitudes,
3. Results
We solved the set of coupled equations, Equations (7), (9), and (10), numerically using the MATLAB software package [
24]. To determine the system parameters for numerical procedure, we held constant known values from the literature. In particular, in order for the voltage across the membrane to be 160 mV, we fixed the proton chemical potentials at
μN = −80 meV and
μ,P = 80 meV, while the energies of the proton sites were
EA = −60 meV,
EB = 100 meV, and
EM0 = −150 meV. The drag coefficient,
ζ = 4.14 nN m
−1s, corresponds to the diffusion coefficient
D =
T/
ζ = 10
−12 m
2s
−1, known for lipid membranes [
25]. The reorganization energy associated with the electron transfer between the FeS complexes is
λσ = 700 meV [
26]. As the proton coupling to the environment is weaker, Λ
τ = 50 meV. For the other parameters, we generated a cost-function and used fmincon to find the optimal parameters for maximal proton-pumping current. In this, the inter-site transfer amplitudes and the coupling to reservoirs were initially assumed to be about 1 meV and 0.01 meV, respectively. The resulting optimal values were ∆
AM = 0.42 meV, Δ
BM = 0.31 meV, ∆
LC = 0.7 meV, ∆
RC = 0.45 meV, Γ
N = 0.0052 meV, Γ
P = 0.0047 meV,
γS = 0.059 meV, and
γD = 0.026 meV
The obtained time dependencies of piston position as well as electron and proton site populations are shown in
Figure 3 and
Figure 4, respectively. One can see that the piston motion exhibits noisy dynamics superimposed on quasiperiodic motion caused by electrostatic forces from populated middle sites. The same periodicity can be seen in the proton population as well. Larger electron reorganization energy leads to smoother dynamics. Both electrostatic forces on the piston from electrons and protons are exerted to the right (see
Figure 2). Correspondingly, a new equilibrium position is set near 4 nm and the piston oscillates around this point.
To obtain the proton-pumping current, we performed both time averaging and averaging over various realizations of white noise. The temperature dependence of this current and its dependence on the energy of the middle proton site are shown in
Figure 5.
It is evident from
Figure 5a that for the present set of parameters, proton pumping is most effective at physiological temperatures. Site
M dependence (
Figure 5b) exhibits clear resonant character with maximal value at −100 meV and drops to zero when the energy level is outside the working regime for the model of
Figure 2.
We examined the geometrical conditions for the functionality of our model by plotting the dependencies of the currents on separations of the piston equilibrium position from the electron and proton sites in
Figure 6. When these separations are large enough, the equations for particle operators and for piston position become decoupled; the electron and proton parts of the system are separated, and there is no more energy transfer between them. Accordingly, the proton-pumping current should vanish. Indeed, when the separation between the piston and proton site
M increases, this current decreases monotonically (see
Figure 6a). However, the dependence of the proton current on the separation between the piston and electron site
C reaches a maximum when control of the piston dynamics by the site population is the most efficient.