Characterization of Fungal Melanins from Black Stains on Paper Artefacts
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal Cultures
2.2. Extraction and Purification of Melanins
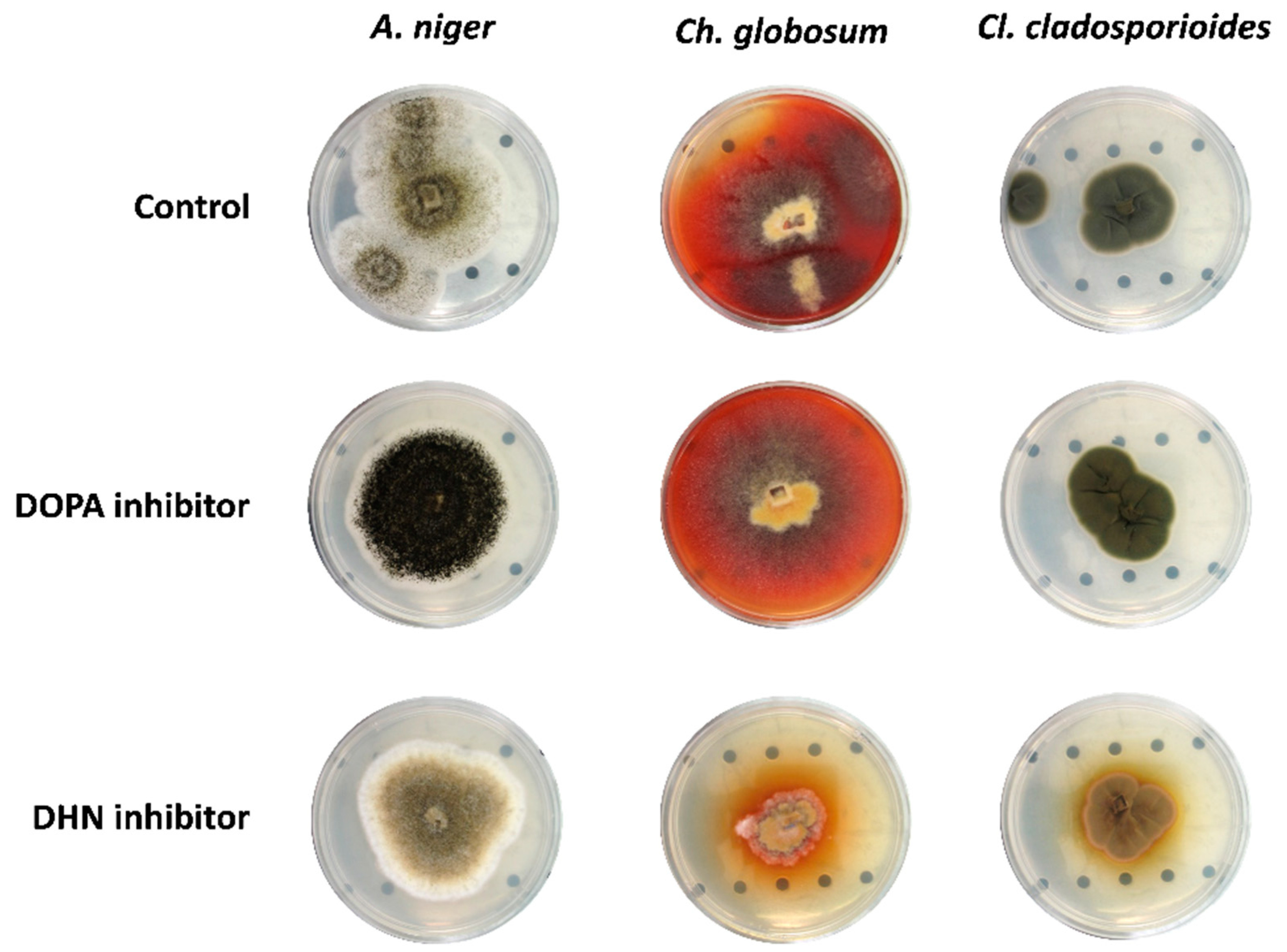
2.3. Inhibition of Melanin Formation by Pathway-Specific Inhibitors
2.4. μ-FTIR Analysis
2.5. UV-Vis Analysis
2.6. μ-Raman Analysis
2.7. Solid-State (SS) NMR Analysis
2.8. Mass Spectral Analysis
3. Results
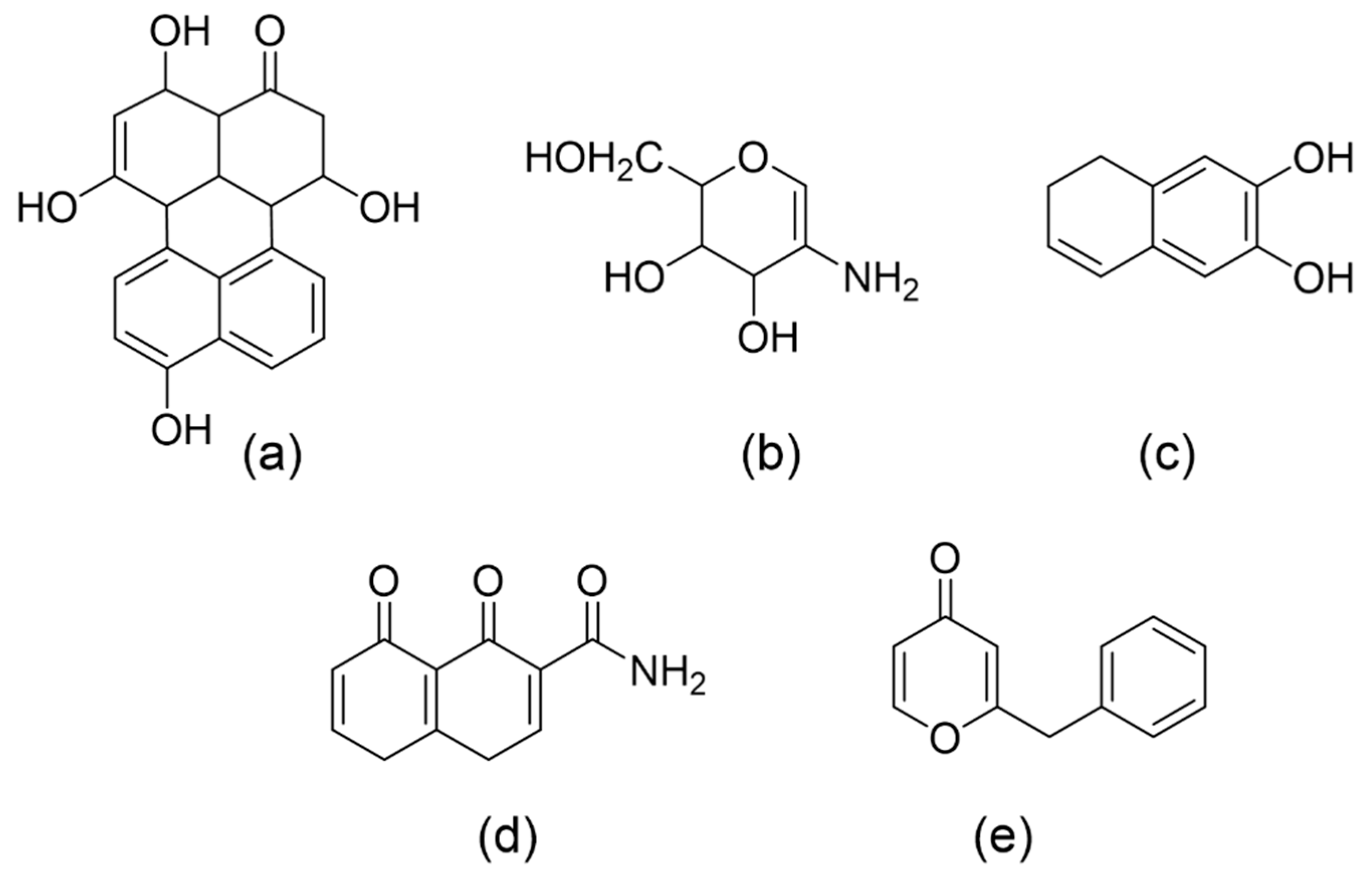
3.1. Melanin Synthesis Pathway
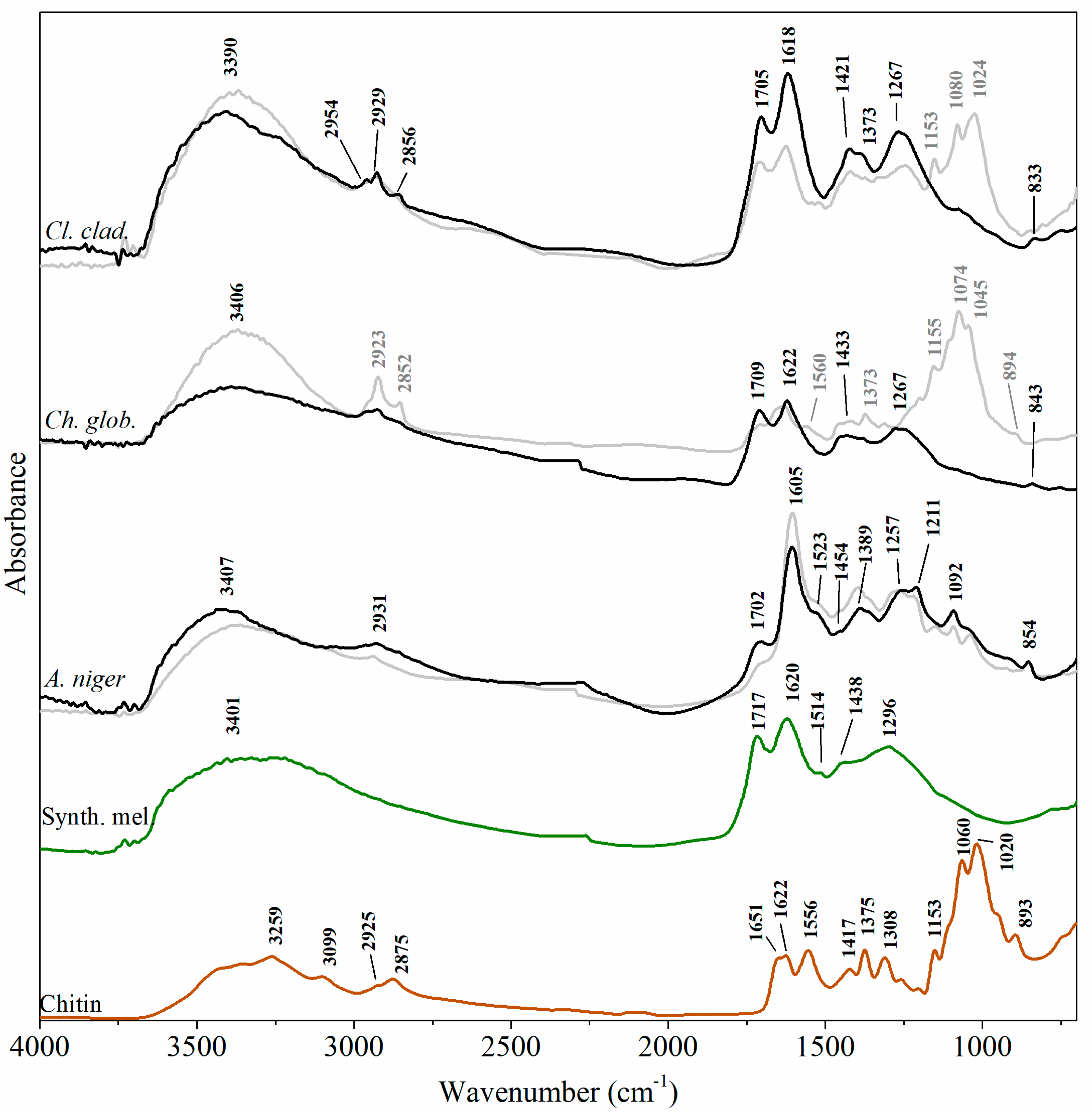
3.2. µ-FTIR Analysis
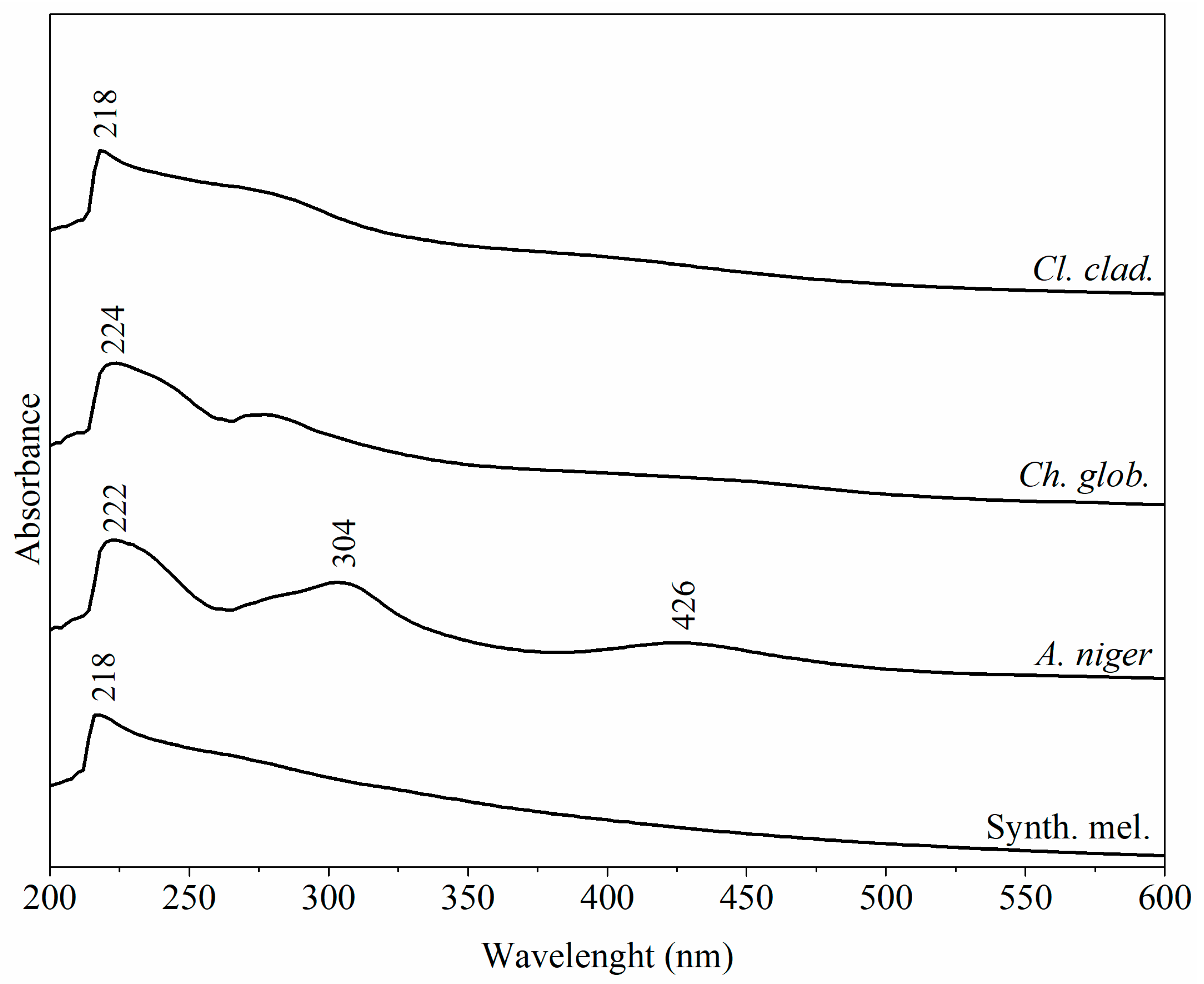
3.3. UV-Vis Analysis
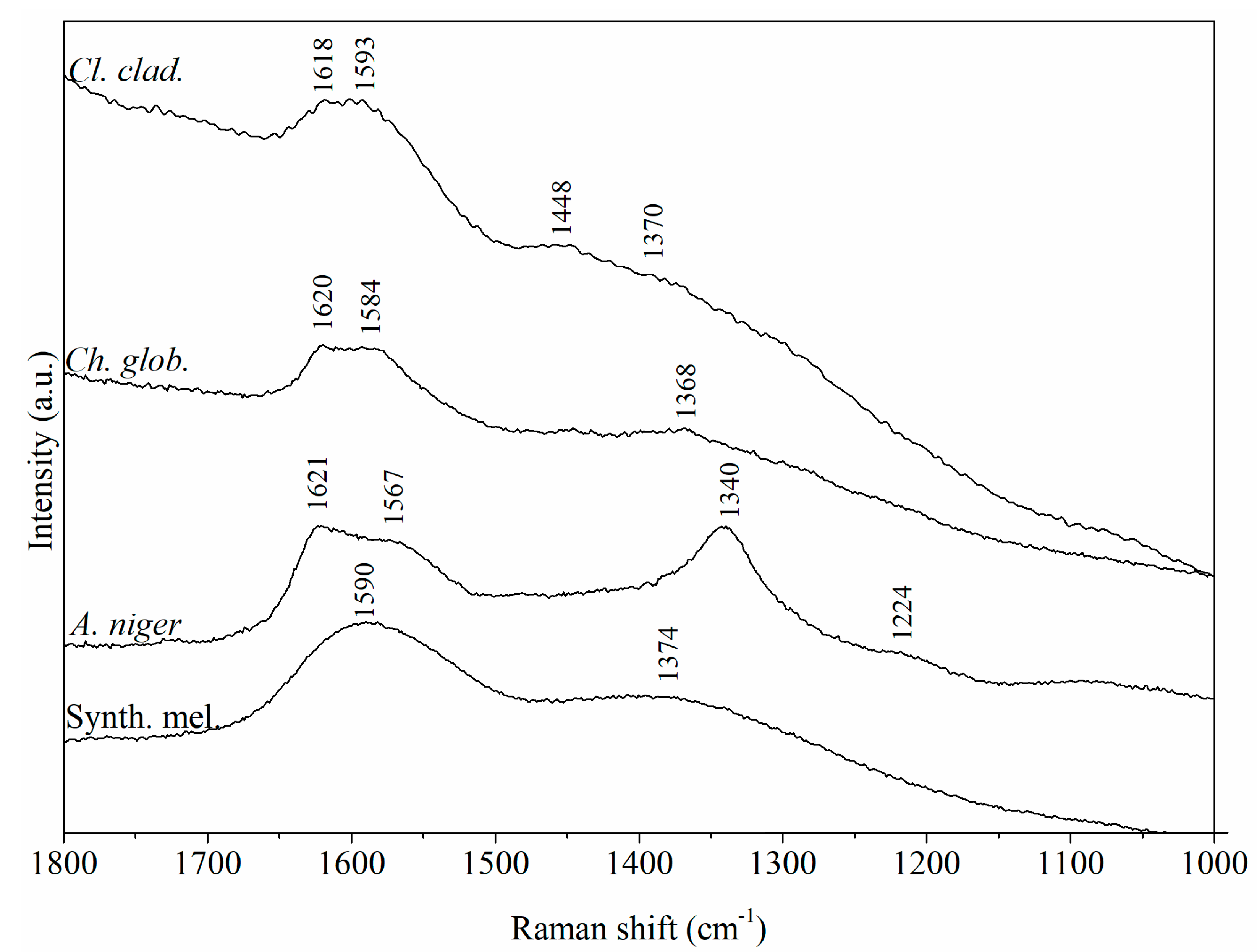
3.4. μ-Raman Analysis
3.5. Solid-State (SS) NMR Analysis
3.6. Mass Spectral Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Area, M.C.; Cheradame, H. Paper Aging and Degradation: Recent Findings and Research Methods. Bioresources 2011, 6, 5307–5337. [Google Scholar] [CrossRef]
- Caneva, G.; Maggi, O.; Nugari, M.P.; Pietrini, A.M.; Piervittori, R.; Ricci, S.; Roc-cardi, A. The Biological Aerosol as a Factor of Biodeterioration. In Cultural Heritage and Aerobiology—Methods and Measurement Techniques for Biodeterioration Monitoring; Mandrioli, P., Caneva, G., Sabbioni, C., Eds.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 2003; pp. 3–29. [Google Scholar]
- Zotti, M.; Ferroni, A.; Calvini, P. Inhibition Properties of Simple Fungistatic Compounds on Fungi Isolated from Foxing Spots. Restaurator 2007, 28, 201–217. [Google Scholar] [CrossRef]
- Sequeira, S.O.; Cabrita, E.J.; Macedo, M.F. Fungal Biodeterioration of Paper: How Are Paper and Book Conservators Dealing with It ? An International Survey. Restaurator 2014, 35, 181–199. [Google Scholar] [CrossRef]
- Melo, D.; Sequeira, S.O.; Lopes, J.A.; Macedo, M.F. Stains versus Colourants Produced by Fungi Colonising Paper Cultural Heritage: A Review. J. Cult. Herit. 2019, 35, 161–182. [Google Scholar] [CrossRef]
- Nitiu, D.S.; Mallo, A.C.; Saparrat, M.C.N. Fungal Melanins That Deteriorate Paper Cultural Heritage: An Overview. Mycologia 2020, 112, 859–870. [Google Scholar] [CrossRef]
- Barrulas, R.V.; Nunes, A.D.; Sequeira, S.O.; Casimiro, M.H.; Corvo, M.C. Cleaning Fungal Stains on Paper with Hydrogels: The Effect of PH Control. Int. Biodeterior Biodegrad. 2020, 152, 104996. [Google Scholar] [CrossRef]
- Gao, J.; Wenderoth, M.; Doppler, M.; Schuhmacher, R.; Marko, D.; Fischer, R. Fungal Melanin Biosynthesis Pathway as Source for Fungal Toxins. mBio 2022, 13, e0021922. [Google Scholar] [CrossRef]
- Xu, X.; Cao, R.; Li, K.; Wan, Q.; Wu, G.; Lin, Y.; Huang, T.; Wen, G. The Protective Role and Mechanism of Melanin for Aspergillus Niger and Aspergillus Flavus against Chlorine-Based Disinfectants. Water Res. 2022, 223, 119039. [Google Scholar] [CrossRef]
- Cordero, R.J.B.; Casadevall, A. Functions of Fungal Melanin beyond Virulence. Fungal Biol. Rev. 2017, 31, 99–112. [Google Scholar] [CrossRef]
- Chang, P.K.; Scharfenstein, L.L.; Mack, B.; Wei, Q.; Gilbert, M.; Lebar, M.; Cary, J.W. Identification of a Copper-Transporting ATPase Involved in Biosynthesis of A. Flavus Conidial Pigment. Appl. Microbiol. Biotechnol. 2019, 103, 4889–4897. [Google Scholar] [CrossRef]
- Eisenman, H.C.; Casadevall, A. Synthesis and Assembly of Fungal Melanin. Appl. Microbiol. Biotechnol. 2012, 93, 931–940. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.; Garcia-Rivera, J.; Yan, B.; Casadevall, A.; Stark, R.E. Unlocking the Molecular Structure of Fungal Melanin Using 13C Biosynthetic Labeling and Solid-State NMR. Society 2003, 42, 8105–8109. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, A.C.; Sequeira, S.O.; Macedo, M.F. Fungi in Archives, Libraries and Museums: A Review on Paper Conservation and Human Health. Crit. Rev. Microbiol. 2019, 45, 686–700. [Google Scholar] [CrossRef] [PubMed]
- Trovão, J.; Mesquita, N.; Paiva, D.S.; Paiva de Carvalho, H.; Avelar, L.; Portugal, A. Can Arthropods Act as Vectors of Fungal Dispersion in Heritage Collections? A Case Study on the Archive of the University of Coimbra, Portugal. Int. Biodeterior Biodegrad. 2013, 79, 49–55. [Google Scholar] [CrossRef]
- Sequeira, S.O.; De Carvalho, H.P.; Mesquita, N.; Portugal, A.; Macedo, M.F. Fungal Stains on Paper: Is What You See What You Get? Conserv. Patrim. 2019, 32, 18–27. [Google Scholar] [CrossRef]
- Pal, A.K.; Gajjar, D.U.; Vasavada, A.R. DOPA and DHN Pathway Orchestrate Melanin Synthesis in Aspergillus Species. Med. Mycol. 2014, 52, 10–18. [Google Scholar]
- Sequeira, S.O.; Laia, C.A.T.; Phillips, A.J.L.; Cabrita, E.J.; Macedo, M.F. Clotrimazole and Calcium Hydroxide Nanoparticles: A Low Toxicity Antifungal Alternative for Paper Conservation. J. Cult. Herit. 2017, 24, 45–52. [Google Scholar] [CrossRef]
- Beltrán-García, M.J.; Prado, F.M.; Oliveira, M.S.; Ortiz-Mendoza, D.; Scalfo, A.C.; Pessoa, A.; Medeiros, M.H.G.; White, J.F.; Di Mascio, P. Singlet Molecular Oxygen Generation by Light-Activated DHN-Melanin of the Fungal Pathogen Mycosphaerella Fijiensis in Black Sigatoka Disease of Bananas. PLoS ONE 2014, 9, e91616. [Google Scholar] [CrossRef]
- Zhang, J.; Gonzalez, E.; Hestilow, T.; Haskins, W.; Huang, Y. Review of Peak Detection Algorithms in Liquid-Chromatography-Mass Spectrometry. Curr. Genomics. 2009, 10, 388–401. [Google Scholar] [CrossRef]
- Scigelova, M.; Hornshaw, M.; Giannakopulos, A.; Makarov, A. Fourier Transform Mass Spectrometry. Mol. Cell. Proteom. 2011, 10, M111.009431. [Google Scholar] [CrossRef]
- Llorente, C.; Bárcena, A.; Vera Bahima, J.; Saparrat, M.C.N.; Arambarri, A.M.; Rozas, M.F.; Mirífico, M.V.; Balatti, P.A. Cladosporium Cladosporioides LPSC 1088 Produces the 1,8-Dihydroxynaphthalene-Melanin-Like Compound and Carries a Putative Pks Gene. Mycopathologia 2012, 174, 397–408. [Google Scholar] [CrossRef] [PubMed]
- Cárdenas, G.; Cabrera, G.; Taboada, E.; Miranda, S.P. Chitin Characterization by SEM, FTIR, XRD, and 13C Cross Polarization/Mass Angle Spinning NMR. J. Appl. Polym. Sci. 2004, 93, 1876–1885. [Google Scholar] [CrossRef]
- Kumirska, J.; Czerwicka, M.; Kaczyński, Z.; Bychowska, A.; Brzozowski, K.; Thöming, J.; Stepnowski, P. Application of Spectroscopic Methods for Structural Analysis of Chitin and Chitosan. Mar. Drugs 2010, 8, 1567–1636. [Google Scholar] [CrossRef] [PubMed]
- Šandula, J.; Kogan, G.; Kačuráková, M.; MacHová, E. Microbial (1→3)-β-D-Glucans, Their Preparation, Physico-Chemical Characterization and Immunomodulatory Activity. Carbohydr. Polym. 1999, 38, 247–253. [Google Scholar] [CrossRef]
- Pralea, I.E.; Moldovan, R.C.; Petrache, A.M.; Ilieș, M.; Hegheș, S.C.; Ielciu, I.; Nicoară, R.; Moldovan, M.; Ene, M.; Radu, M.; et al. From Extraction to Advanced Analytical Methods: The Challenges of Melanin Analysis. Int. J. Mol. Sci. 2019, 20, 3943. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.F.; Rhim, J.W. Isolation and Characterization of Melanin from Black Garlic and Sepia Ink. LWT 2019, 99, 17–23. [Google Scholar] [CrossRef]
- Tavzes, C.; Palcic, J.; Fackler, K.; Pohleven, F.; Koestler, R.J. Biomimetic System for Removal of Fungal Melanin Staining on Paper. Int. Biodeterior Biodegrad. 2013, 84, 307–313. [Google Scholar] [CrossRef]
- El-Naggar, N.E.-A.; El-Ewasy, S.M. Bioproduction, Characterization, Anticancer and Antioxidant Activities of Extracellular Melanin Pigment Produced by Newly Isolated Microbial Cell Factories Streptomyces Glaucescens NEAE-H. Sci. Rep. 2017, 7, 42129. [Google Scholar] [CrossRef]
- Gonçalves, R.C.R.; Lisboa, H.C.F.; Pombeiro-Sponchiado, S.R. Characterization of Melanin Pigment Produced by Aspergillus Nidulans. World J. Microbiol. Biotechnol. 2012, 28, 1467–1474. [Google Scholar] [CrossRef]
- Seelam, S.D.; Agsar, D.; Halmuthur, M.S.K.; Reddy Shetty, P.; Vemireddy, S.; Reddy, K.M.; Umesh, M.K.; Rajitha, C.H. Characterization and Photoprotective Potentiality of Lime Dwelling Pseudomonas Mediated Melanin as Sunscreen Agent against UV-B Radiations. J. Photochem. Photobiol. B 2021, 216, 112126. [Google Scholar] [CrossRef]
- Choque, E.; El Rayess, Y.; Raynal, J.; Mathieu, F. Fungal Naphtho-γ-Pyrones—Secondary Metabolites of Industrial Interest. Appl. Microbiol. Biotechnol. 2015, 99, 1081–1096. [Google Scholar] [CrossRef] [PubMed]
- Jørgensen, T.R.; Park, J.; Arentshorst, M.; van Welzen, A.M.; Lamers, G.; vanKuyk, P.A.; Damveld, R.A.; van den Hondel, C.A.M.; Nielsen, K.F.; Frisvad, J.C.; et al. The Molecular and Genetic Basis of Conidial Pigmentation in Aspergillus Niger. Fungal Genet. Biol. 2011, 48, 544–553. [Google Scholar] [CrossRef] [PubMed]
- Ray, A.C.; Eakin, R.E. Studies on the Biosynthesis of Aspergillin by Aspergillus Niger. Appl. Microbiol. 1975, 30, 909–915. [Google Scholar] [CrossRef] [PubMed]
- Centeno, S.A.; Shamir, J. Surface Enhanced Raman Scattering (SERS) and FTIR Characterization of the Sepia Melanin Pigment Used in Works of Art. J. Mol. Struct. 2008, 873, 149–159. [Google Scholar] [CrossRef]
- Culka, A.; Jehlička, J.; Ascaso, C.; Artieda, O.; Casero, C.M.; Wierzchos, J. Raman Microspectrometric Study of Pigments in Melanized Fungi from the Hyperarid Atacama Desert Gypsum Crust. J. Raman Spectrosc. 2017, 48, 1487–1493. [Google Scholar] [CrossRef]
- Huang, Z.; Lui, H.; Chen, X.K.; Alajlan, A.; McLean, D.I.; Zeng, H. Raman Spectroscopy of in Vivo Cutaneous Melanin. J. Biomed. Opt. 2004, 9, 1198–1205. [Google Scholar] [CrossRef]
- Martin-Sanchez, P.M.; Sanchez-Cortes, S.; Lopez-Tobar, E.; Jurado, V.; Bastian, F.; Alabouvette, C.; Saiz-Jimenez, C. The Nature of Black Stains in Lascaux Cave, France, as Revealed by Surface-Enhanced Raman Spectroscopy. J. Raman Spectrosc. 2012, 43, 464–467. [Google Scholar] [CrossRef]
- Agarwal, U.P.; Reiner, R.S. Near-IR Surface-Enhanced Raman Spectrum of Lignin. J. Raman Spectrosc. 2009, 40, 1527–1534. [Google Scholar] [CrossRef]
- MERCK. Melanin. Available online: https://www.sigmaaldrich.com/PT/en/product/sigma/m8631 (accessed on 11 January 2022).
- Adhyaru, B.; Akhmedov, N.; Katritzky, A.; Bowers, C. Solid-State Cross-Polarization Magic Angle Spinning 13C and 15N NMR Characterization of Sepia Melanin, Sepia Melanin Free Acid and Human Hair Melanin in Comparison with Several Model Compounds. Magn. Reson. Chem. 2003, 41, 466–474. [Google Scholar] [CrossRef]
- Dreyer, D.R.; Miller, D.J.; Freeman, B.D.; Paul, D.R.; Bielawski, C.W. Elucidating the Structure of Poly(Dopamine). Langmuir 2012, 28, 6428–6435. [Google Scholar] [CrossRef]
- Hu, Y.; Hao, X.R.; Lou, J.; Zhang, P.; Pan, J.; Zhu, X.D. A PKS Gene, Pks-1, Is Involved in Chaetoglobosin Biosynthesis, Pigmentation and Sporulation in Chaetomium Globosum. Sci. China Life Sci. 2012, 55, 1100–1108. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sankawa, U.; Shimada, H.; Sato, T.; Kinoshita, T.; Yamasaki, K. Biosynthesis of Scytalone. Tetrahedron Lett. 1977, 18, 483–486. [Google Scholar] [CrossRef]
- Yang, Y.; Lowry, M.; Xu, X.; Escobedo, J.O.; Sibrian-Vazquez, M.; Wong, L.; Schowalter, C.M.; Jensen, T.J.; Fronczek, F.R.; Warner, I.M.; et al. Seminaphthofluorones Are a Family of Water-Soluble, Low Molecular Weight, NIR-Emitting Fluorophores. Proc. Natl. Acad. Sci. USA 2008, 105, 8829–8834. [Google Scholar] [CrossRef]
- Snyder, S.A.; Tang, Z.-Y.; Gupta, R. Enantioselective Total Synthesis of (?)-Napyradiomycin A1 via Asymmetric Chlorination of an Isolated Olefin. J. Am. Chem. Soc. 2009, 131, 5744–5745. [Google Scholar] [CrossRef] [PubMed]
- Conradt, D.; Schätzle, M.A.; Husain, S.M.; Müller, M. Diversity in Reduction with Short-Chain Dehydrogenases: Tetrahydroxynaphthalene Reductase, Trihydroxynaphthalene Reductase, and Glucose Dehydrogenase. ChemCatChem 2015, 7, 3116–3120. [Google Scholar] [CrossRef]
- de Oliveira, K.T.; Miller, L.Z.; McQuade, D.T. Exploiting Photooxygenations Mediated by Porphyrinoid Photocatalysts under Continuous Flow Conditions. RSC Adv. 2016, 6, 12717–12725. [Google Scholar] [CrossRef]
- Korytowski, W.; Sarna, T. Bleaching of Melanin Pigments. Role of Copper Ions and Hydrogen Peroxide in Autooxidation and Photooxidation of Synthetic Dopa-Melanin. J. Biol. Chem. 1990, 265, 12410–12416. [Google Scholar] [CrossRef]
- Watt, A.A.R.; Bothma, J.P.; Meredith, P. The Supramolecular Structure of Melanin. Soft Matter 2009, 5, 3754. [Google Scholar] [CrossRef]
- Olofsson, M.A.; Bylund, D. Liquid Chromatography with Electrospray Ionization and Tandem Mass Spectrometry Applied in the Quantitative Analysis of Chitin-Derived Glucosamine for a Rapid Estimation of Fungal Biomass in Soil. Int. J. Anal. Chem. 2016, 2016, 9269357. [Google Scholar] [CrossRef]
- Xin, C.; Ma, J.; Tan, C.; Yang, Z.; Ye, F.; Long, C.; Ye, S.; Hou, D. Preparation of Melanin from Catharsius Molossus L. and Preliminary Study on Its Chemical Structure. J. Biosci. Bioeng. 2015, 119, 446–454. [Google Scholar] [CrossRef]
- Belozerskaya, T.A.; Gessler, N.N.; Aver’yanov, A.A. Melanin Pigments of Fungi. In Fungal Metabolites; Mérillo, J.-M., Ramawat, K.G., Eds.; Springer International Publishing: Berlin/Heidelberg, Germany, 2017; pp. 263–291. [Google Scholar]
- Araújo, M.F.C. Melaninas de Origem Marinha: Caracterização Quimica e Termo-Física. Master’s Thesis, Universidade de Lisboa, Lisboa, Portugal, December 2010. [Google Scholar]
- Chang, P.K.; Cary, J.W.; Lebar, M.D. Biosynthesis of Conidial and Sclerotial Pigments in Aspergillus Species. Appl. Microbiol. Biotechnol. 2020, 104, 2277–2286. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, M.H. Comparisons of Fungal Melanin Biosynthesis in Ascomycetous, Imperfect and Basidiomycetous Fungi. Trans. Br. Mycol. Soc. 1983, 81, 29–36. [Google Scholar] [CrossRef]
- Geib, E.; Brock, M. Comment on: “Melanisation of Aspergillus Terreus—Is Butyrolactone I Involved in the Regulation of Both DOPA and DHN Types of Pigments in Submerged Culture? Microorganisms 2017, 5, 22”. Microorganisms 2017, 5, 34. [Google Scholar] [CrossRef] [PubMed]
- Yu, R.; Liu, J.; Wang, Y.; Wang, H.; Zhang, H. Aspergillus Niger as a Secondary Metabolite Factory. Front. Chem. 2021, 9, 701022. [Google Scholar] [CrossRef] [PubMed]
- Chiang, Y.M.; Meyer, K.M.; Praseuth, M.; Baker, S.E.; Bruno, K.S.; Wang, C.C.C. Characterization of a Polyketide Synthase in Aspergillus Niger Whose Product Is a Precursor for Both Dihydroxynaphthalene (DHN) Melanin and Naphtho-γ-Pyrone. Fungal Genet. Biol. 2011, 48, 430–437. [Google Scholar] [CrossRef]
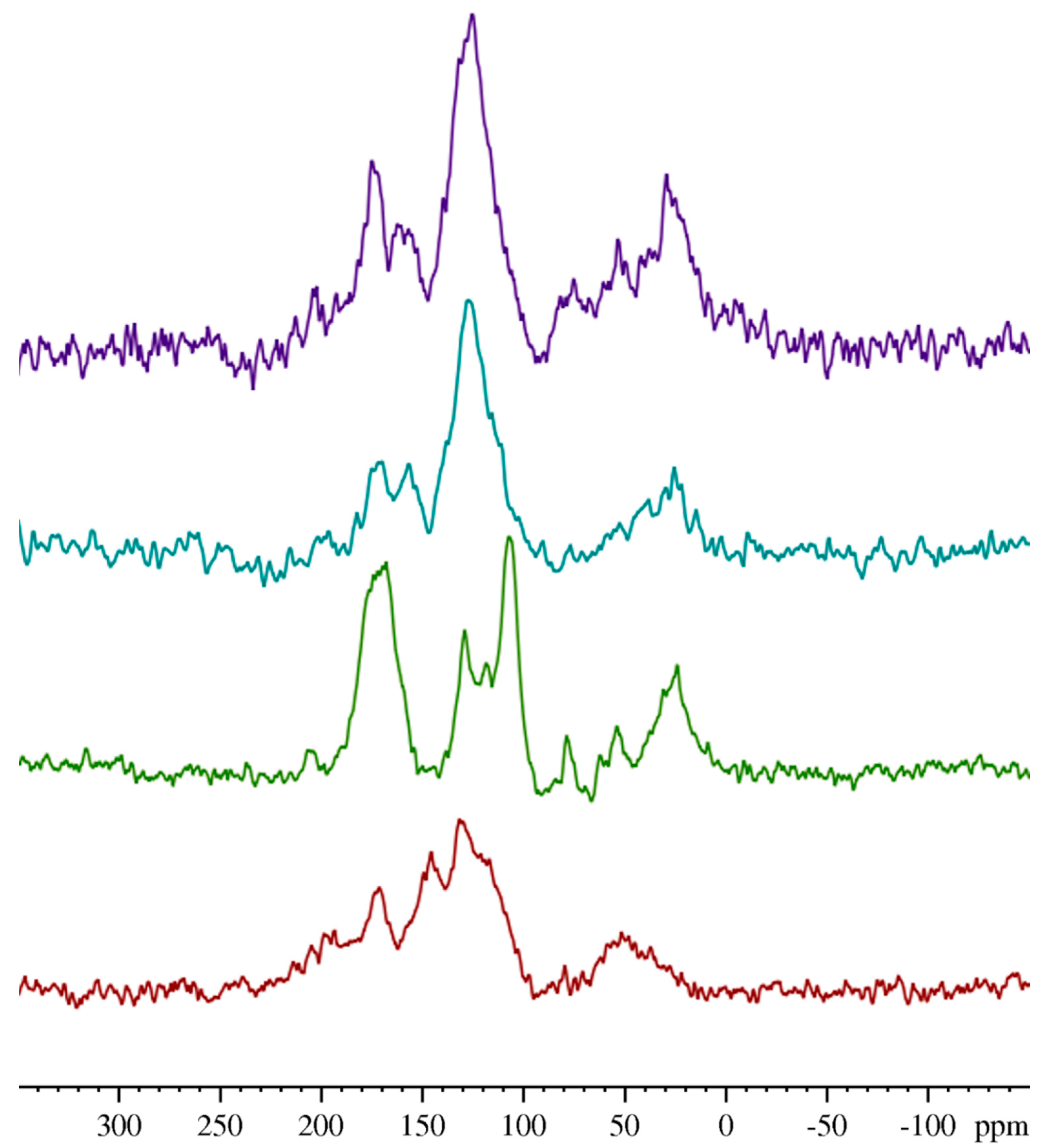
| Wavenumber (cm−1) | Assignment | References | |||
|---|---|---|---|---|---|
| Synth. | A. niger | Ch. globosum | Cl. cladosporioides | ||
| 3401 | 3407 | 3406 | 3390 | νO-H; νN-H | [27,28,29] |
| 2931 | 2923 | 2954; 2929; 2856 | Aliphatic νC-H2 and νC-H3; νN-H | [27,29,30] | |
| 1717 | 1702 | 1709 | 1705 | ν C=O unconjugated with benzene ring | [28] |
| 1620 | 1605 | 1622 | 1618 | νC=O; νC=C aromatic (from conjugated quinone structures) | [22,28,30] |
| 1514 | 1523 | δN-H; νC-N | [22,28,29,30] | ||
| 1438 | 1454; 1389 | 1421 | 1432 | νC-N; δC-H3 and δC-H2) | [22,27,28,29,30] |
| 1296 | 1257 | 1267 | 1267 | νC-OH (alcoholic, carboxylic, phenolic groups); | [22,27,28,30] |
| 1092 | 1080 | Alcoholic C-O; C-H in-plane of aliphatic structure | [22,27,28,29] | ||
| 854 | 843 | 833 | δC-H out-of-plane in aromatic structure | [27,29] | |
| Melanin | Carbonyl | Aromatic | Aliphatic | |
|---|---|---|---|---|
| Cq | CH | |||
| A. niger | 188–153 (Carbonyl + Cq) | 135–100 | 80–10 | |
| Cl. cladosporioides | 180–165 | 165–150 | 145–95 | 66–10 |
| Ch. globosum | 180–165 | 160–147 | 145–95 | 50–10 |
| Synthetic | 180–160 | 157–142 | 140–100 | 67–30 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Melo, D.; Paiva, T.G.; Lopes, J.A.; Corvo, M.C.; Sequeira, S.O. Characterization of Fungal Melanins from Black Stains on Paper Artefacts. Heritage 2022, 5, 3049-3065. https://doi.org/10.3390/heritage5040158
Melo D, Paiva TG, Lopes JA, Corvo MC, Sequeira SO. Characterization of Fungal Melanins from Black Stains on Paper Artefacts. Heritage. 2022; 5(4):3049-3065. https://doi.org/10.3390/heritage5040158
Chicago/Turabian StyleMelo, Daniela, Tiago G. Paiva, João A. Lopes, Marta C. Corvo, and Sílvia O. Sequeira. 2022. "Characterization of Fungal Melanins from Black Stains on Paper Artefacts" Heritage 5, no. 4: 3049-3065. https://doi.org/10.3390/heritage5040158
APA StyleMelo, D., Paiva, T. G., Lopes, J. A., Corvo, M. C., & Sequeira, S. O. (2022). Characterization of Fungal Melanins from Black Stains on Paper Artefacts. Heritage, 5(4), 3049-3065. https://doi.org/10.3390/heritage5040158








