Comparative Inhibition Study by Nanomaterial, Plant Extract and Chemical Microcide on the Screaming Mummy in Egyptian Museum Store
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
- Enzyme Production
- Enzymes Assay
2.2.1. Determine Minimal Inhibitory Concentration (MIC) of Antimicrobial Agents against the Isolated Microorganisms
2.2.2. Evaluate Treatment for Isolated Microorganisms from Mummy
3. Results
3.1. Identification of the Microbial Isolates
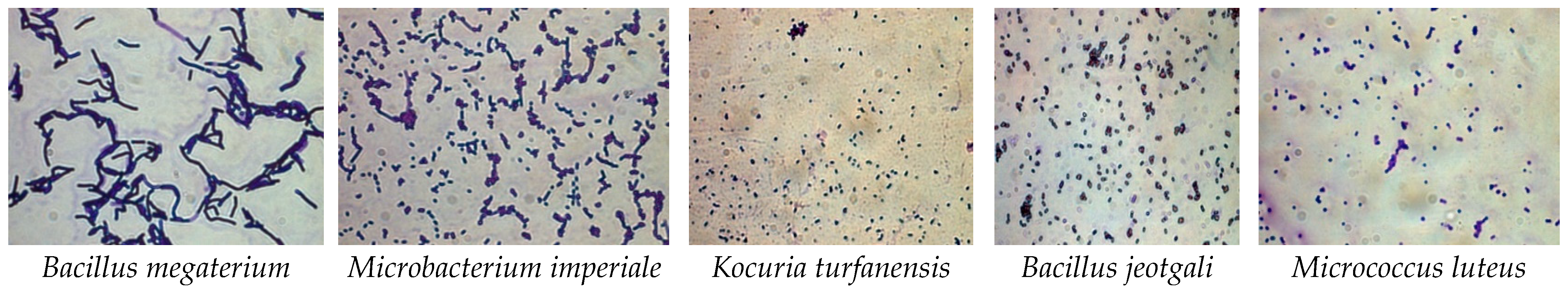
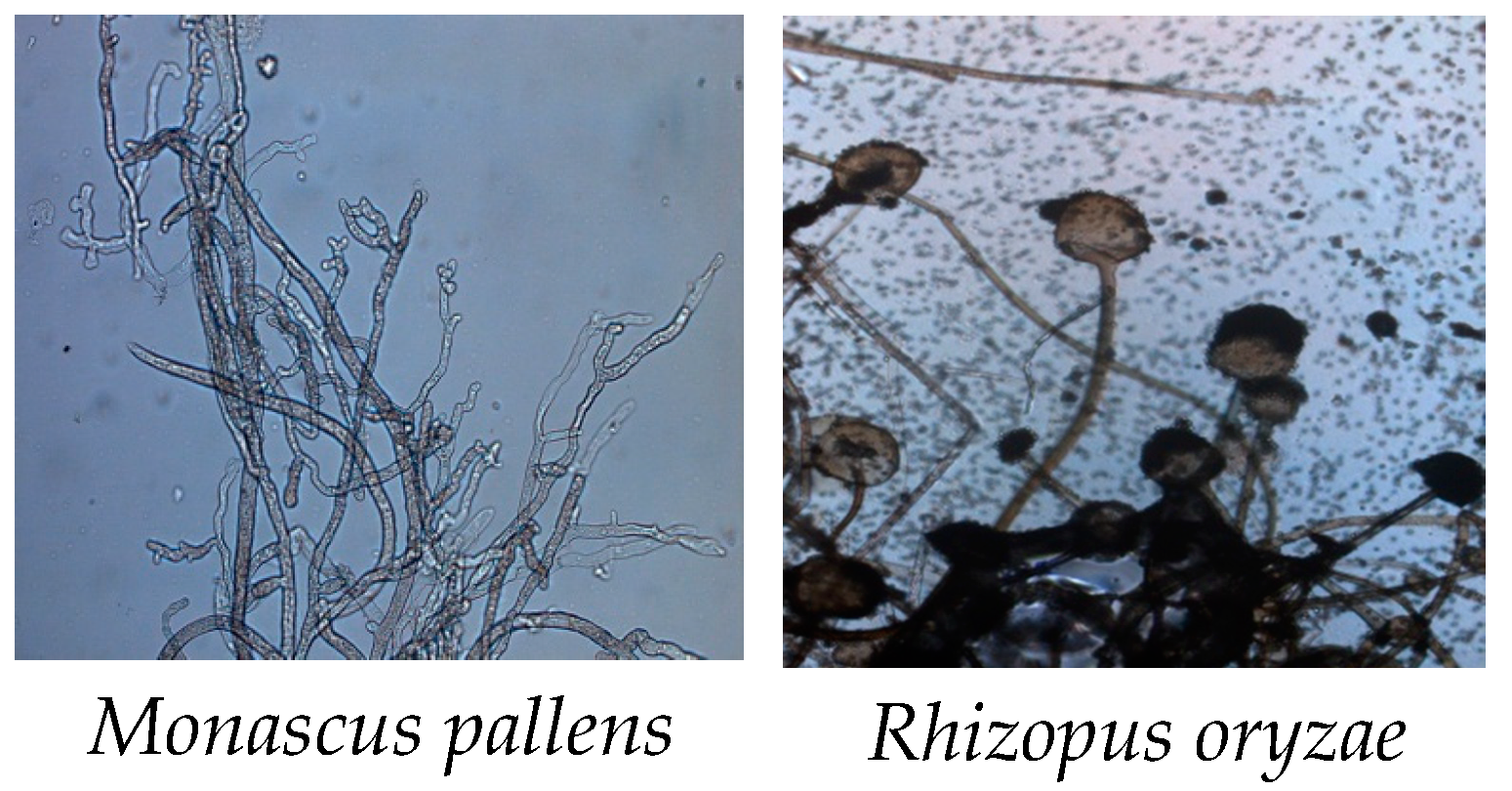
3.1.1. Optical Microscope Identification
3.1.2. rRNA Sequencing Identification
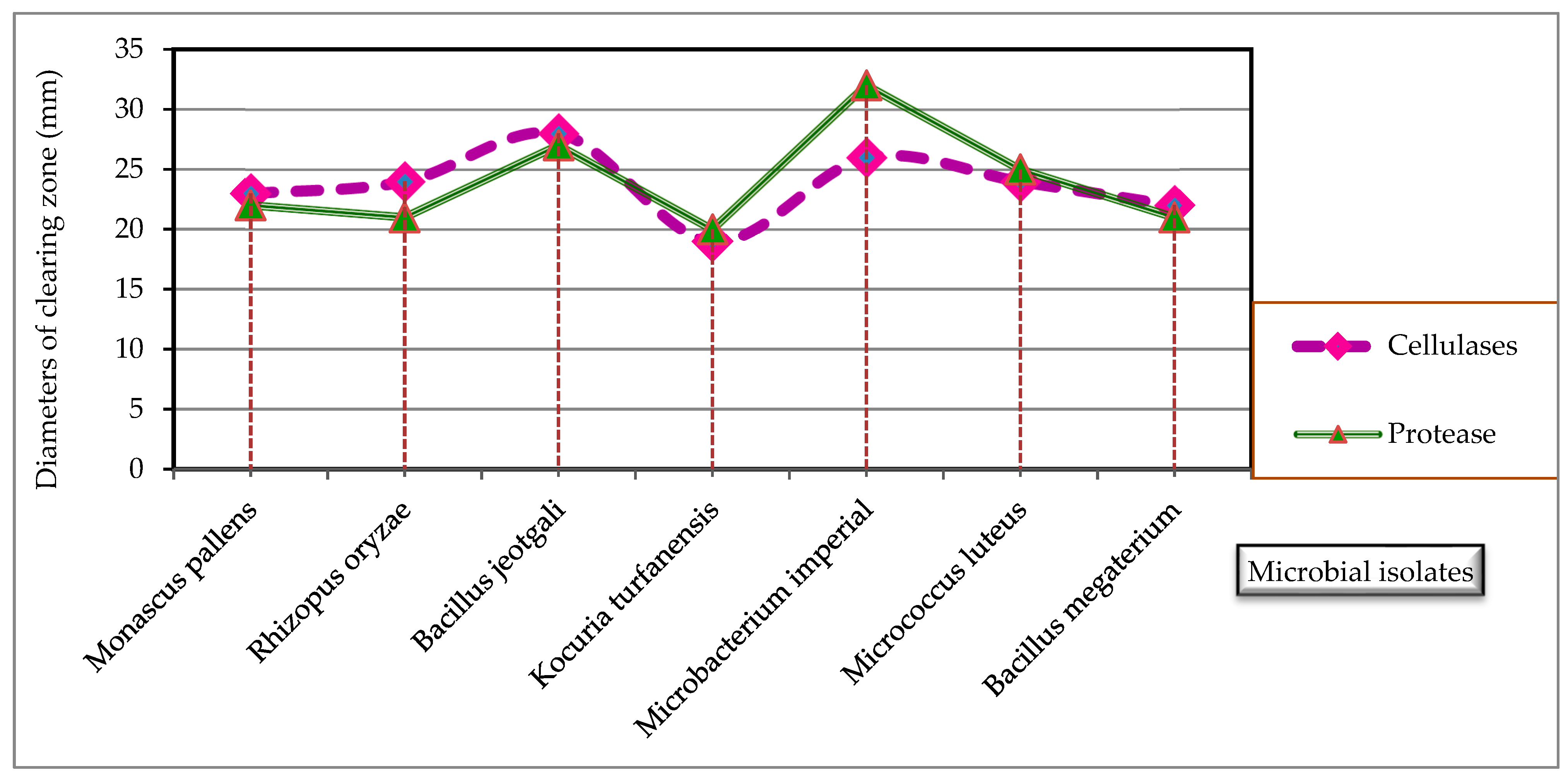
3.2. Determination of Cellulases and Protease Produced by the Isolated Microorganisms by Cup Plate Technique
3.3. The MIC of Isolated Micro-Organisms
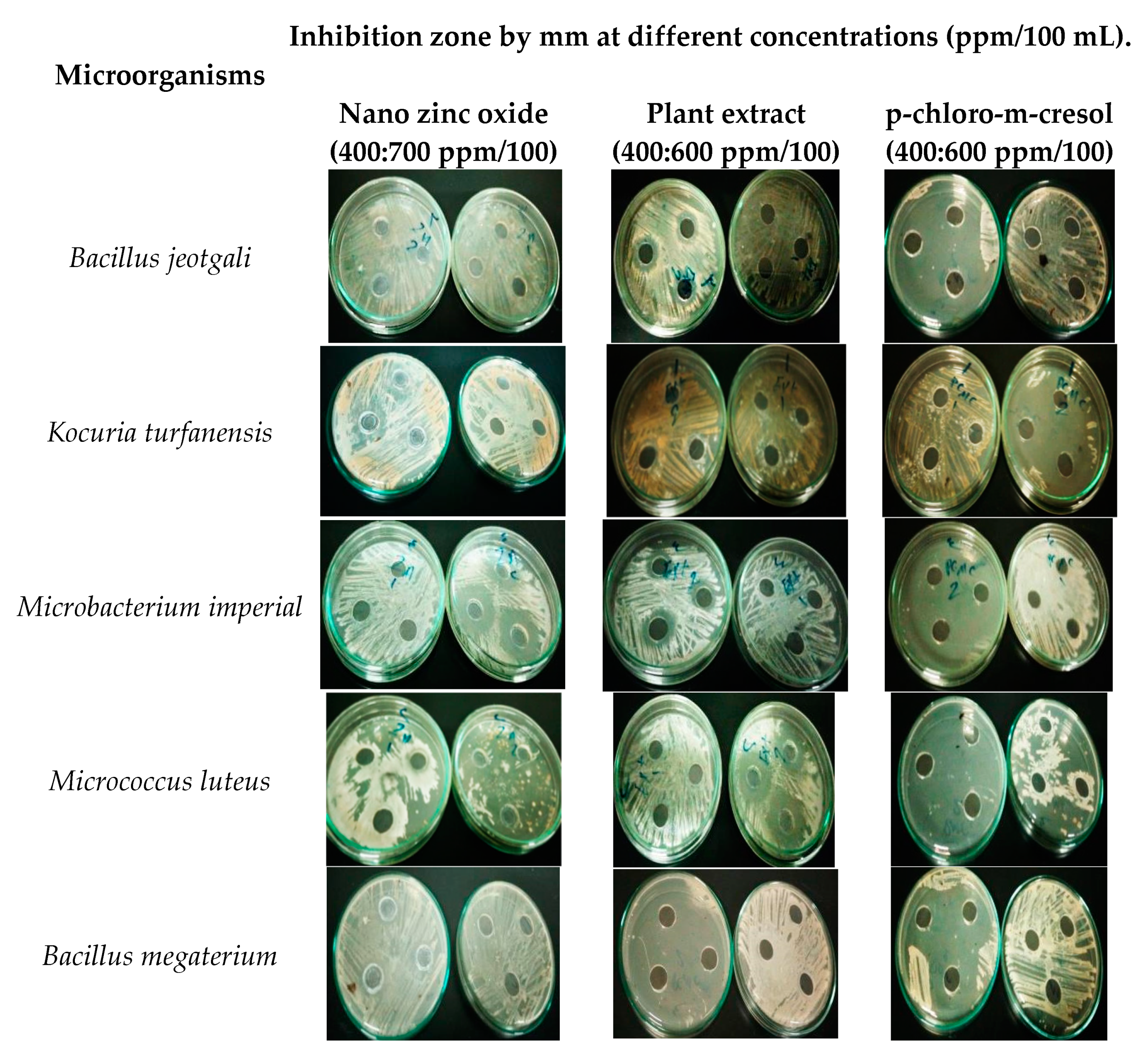
3.4. Evaluation of Different Methods for Treatment of Deteriorated Mummy
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Museum of Natural History Smithsonian Institution. Egyptian Mummies. 2012. Available online: https://www.si.edu/spotlight/ancient-egypt/mummies (accessed on 15 May 2021).
- Abdel-Maksoud, G.; El-Amin, A. A review on the materials used during the mummification processes in ancient Egypt. Mediterr. Archaeol. Archaeom. 2011, 11, 129–150. [Google Scholar]
- Magdy, M.; Ismail, M.; Issa, Y.; Abdel-Maksoudc, G.; Ibrahim, M. An analytical study for understanding the degradation process of a late period mummy. Adv. Res. Conserv. Sci. 2020, 1, 13–30. [Google Scholar]
- Hawass, Z.; Saleem, S. Computed tomography examination of the screaming mummy “Unknown-Woman-A”. Egypt. J. Radiol. Nucl. Med. 2020, 51, 139. [Google Scholar] [CrossRef]
- Arya, A.; Shah, A.R.; Sadasivan, S. Indoor aeromycoflora of Baroda museum and deterioration of Egyptian mummy. Curr. Sci. 2001, 81, 793–799. [Google Scholar]
- Naji, M.K.; Abdullah, Q.Y.; AL-Zaqri, A.Q. Evaluating the biodeterioration enzymatic activities of fungal contamination isolated from some ancient Yemeni mummies preserved in the national museum. Biochem. Res. Int. 2014, 2014, 481508. [Google Scholar] [CrossRef]
- Omar, A.M.; Taha, A.S.; Mohamed, A.A. Microbial deterioration of some archaeological artifacts: Manipulation and treatment. Eur. J. Exp. Biol. 2018, 8, 21. [Google Scholar] [CrossRef][Green Version]
- Elamin, A.; Takatori, K.; Matsuda, Y.; Tsukada, M.; Kirino, F. Microbialogical, morphological and spectroscopic study on the effect of resinous materials in the preservation of wrapping textiles of mummies. Mediterr. Archaeol. Archaeom. 2018, 18, 1–10. [Google Scholar] [CrossRef]
- David, M.E.; Ion, R.M.; Grigorescu, R.M.; Iancu, L.; Andrei, E.R. Nanomaterials used in conservation and restoration of cultural heritage: An up-to-date overview. Mater. J. 2020, 13, 2064. [Google Scholar] [CrossRef]
- Bonini, M.; Lenz, S.; Giorgi, R.; Baglioni, P. Nanomagnetic sponges for the cleaning of works of art. Langmuir J. 2007, 23, 8681–8685. [Google Scholar] [CrossRef]
- Giorgi, R.; Chelazzi, D.; Fratini, E.; Langer, S.; Niklasson, A.; Rådemar, M.; Svensson, J.E.; Baglioni, P. Nanoparticles of calcium hydroxide for wood deacidification: Decreasing the emissions of organic acid vapors in church organ environments. J. Cult. Herit. 2009, 10, 206–213. [Google Scholar] [CrossRef]
- Helmi, F.M.; Ali, N.M.; Ismael, S.M. Nanomaterials for the inhibition of microbial growth on Ancient Egyptian Funeral masks. Mediterr. Archaeol. Archaeom. 2015, 15, 87–95. [Google Scholar] [CrossRef]
- Sricharussin, W.; Threepopnatkul, P.; Neamjan, N. Effect of various shapes of zinc oxide on cotton fabric for UV-Blocking and anti-bacterial properties. Fibers Polym. J. 2011, 12, 1037–1041. [Google Scholar] [CrossRef]
- Taghizadeh, S.M.; Lal, N.; Ebrahiminezhad, A.; Moeini, F.; Seifan, M.; Ghasemi, Y.; Berenjian, A. Green and economic fabrication of Zinc Oxide (ZnO) nanorods as a broadband UV blocker and antimicrobial agent. Nanomater. J. 2020, 10, 530. [Google Scholar] [CrossRef]
- Khamseh, H. Conservation of leather historical object by Nano technology in archaeology found (Sample study cover leather book). J. Fundam. Appl. Sci. 2016, 8, 276–283. [Google Scholar] [CrossRef]
- Mansour, M.M.; EL-Hefny, M.; Salem, M.; Ali, H.M. The Biofungicide activity of some plant essential oils for the cleaner production of model linen fibers similar to those used in ancient Egyptian mummification. Process. J. 2020, 8, 79. [Google Scholar] [CrossRef]
- Othman, M.; Saadat, H.; Matsuda, Y. Antifungal activity of some plant extracts and essential oils against Fungi-Infested organic archaeological artefacts. Archaeometry 2020, 62, 187–199. [Google Scholar] [CrossRef]
- Elsayed, Y.; Shabana, Y. The effect of some essential oils on aspergillus Niger and Alternaria Alternata infestation in archaeological oil paintings. Mediterr. Archaeol. Archaeom. 2018, 18, 71–87. [Google Scholar] [CrossRef]
- Geweely, N.S.; Afifi, H.A.; Ibrahim, D.M.; Soliman, M.M. Efficacy of essential oils on Fungi isolated from archaeological objects in Saqqara excavation, Egypt. Geomicrobiol. J. 2019, 36, 148–168. [Google Scholar] [CrossRef]
- Khalaphallah, R.; El-Derby, A. The effect of nano TiO2 and plant extracts on microbial strains isolated from Theban ancient Egyptian royal tomb painting. Afr. J. Microbiol. Res. 2015, 9, 1424–1430. [Google Scholar] [CrossRef]
- Metwally, F.E.; Mohamed, A.A.; Mahalel, U.A.; Sheded, M.G. Evaluation of certain cosmopolitan hydrophytes in the Nile River, Aswan district for their ecological and bioactivity potentials: A review. Int. J. Sci. Technol. Res. 2020, 9, 1595–1606, ISSN 2277–8616. [Google Scholar]
- Omar, A.M.; Salah, T.A.; Mohamed, A.A.; Sheded, M.G. Attenuation of microbial induced deterioration of cellulose fibers by hornwort (Ceratophyllum demersum L.) methanolic extract. Int. J. Biol. Res. 2017, 5, 48–58. [Google Scholar] [CrossRef]
- Abdel-wahab, H.M.; Abdel-kader, A.E.; Yousef, R.S. Population dynamics of terrestrial insects in Saluga and Ghazal protected area, Aswan, Egypt. Egypt. Soc. Environ. Sci. 2019, 18, 117–123. [Google Scholar]
- Omar, A.; Taha, A.; El-Wekeel, F. Microbial degradation of ancient textiles in the Egyptian textile museum and methods of its control. Egypt. J. Archaeol. Restor. Stud. 2019, 9, 27–37. [Google Scholar]
- US EPA (United States Environmental Protection Agency). Reregistration Eligibility Decision (RED) p-Chloro-m-cresol, Prevention, Pesticides and Toxic Substances. EPA-738-R-96-008. 1997. Available online: https://archive.epa.gov/pesticides/reregistration/web/pdf/3046red.pdf (accessed on 15 May 2021).
- Andersen, F.A. Final report on the safety assessment of p-chloro-m-cresol. Int. J. Toxicol. 1997, 16, 268. [Google Scholar]
- Zimbro, M.J.; Power, D.A.; Miller, S.M.; Wilson, G.E. and Johnson, J.A. Difco & BBL Manual of Microbiological Culture Media, 2nd ed.; Becton, Dickinson and Company: New Jersey, MD, USA, 2009. [Google Scholar]
- Lane, D.J. ; 16S/23S rRNA Sequencing. Nucleic Acid Techniques in Bacterial Systematics; John Wiley and Sons: Chichester, UK, 1991; pp. 115–175. Available online: http://lycofs01.lycoming.edu/~newman/NovelMicrobe/16SrRNAprimers.pdf (accessed on 15 June 2021).
- Pitt, J.I.; Hocking, A.D. Fungi and Food Spoilage, 3rd ed.; Springer Science & Business Media LLC: Berlin/Heidelberg, Germany, 2009. [Google Scholar]
- White, T.J.; Bruns, T.; Lee, S. and Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Pangallo, D.; Simonovičová, A.; Chovanova, K.; Ferianc, P. Wooden art objects and the museum environment: Identification and biodegradative characteristics of isolated microflora. Soc. Appl. Microbiol. Lett. Appl. Microbiol. 2007, 45, 87–94. [Google Scholar] [CrossRef]
- Omar, A.M.; Taha, A.S.; Mohamed, A.A.; El-Wekeel, F. Fumigation is the ideal method in treating damaged archaeological paper using Ceratophyllum demersum L extract: A case study. J. Basic Environ. Sci. 2019, 6, 17–22. [Google Scholar]


| Swab Cite | Media Used | ||
|---|---|---|---|
| Cellulose Agar | Protein Agar | Nutrient Agar | |
| Head | Fungus 1 Rhizopus oryzae | G+ve Shortbacilli spore former1 | G+ve Shortbacilli spore former2 Microbacillus spp. |
| Wig | Microbacillus spp. Fungus 1 | Microbacillus spp. G+ve Shortbacilli spore former3 | G+ve Shortbacilli spore former3 |
| Rib cage | Rhizopus oryzae | G+ve Shortbacilli spore former2 | G+ve Shortbacilli spore former4 Microbacillus spp. |
| Abdominal and pelvic cavity | Fungus 1 | Microbacillus spp. | G+ve Shortbacilli spore former3 Micrococus spp. |
| Right leg | Rhizopus oryzae | Microbacillus spp. G+ve Shortbacilli spore former1 | G+ve Shortbacilli spore former3 |
| Left leg | Rhizopus oryzae Microbacillus spp. | G+ve Shortbacilli spore former3 G+ve Shortbacilli spore former4 | G+ve Shortbacilli spore former3 Microbacillus spp. |
| Mouth | Rhizopus oryzae Fungus 1 | Micrococus spp. | G+ve Shortbacilli spore former2 Micrococus spp. |
| Swab Cite | Identification | Closely Related Microbial Strains Accessed from Gen Bank | ||
|---|---|---|---|---|
| Strain No. | Accession No. | Similarity | ||
| Head | Bacillus jeotgali | YKJ-10 | NR025060T | 99.93% |
| Kocuria turfanensis | HO-9042 | NR043899T | 88.22% | |
| Microbacterium imperiale | DSM 20530 | NR026161T | 99.86% | |
| Wig | Microbacterium imperiale | DSM 20530 | NR026161T | 99.86% |
| Bacillus jeotgali | YKJ-10 | NR025060T | 99.93% | |
| Rib cage | Microbacterium imperiale | DSM 20530 | NR026161T | 99.86% |
| Bacillus jeotgali | YKJ-10 | NR025060T | 99.93% | |
| Abdominal and pelvic cavity | Microbacterium imperial | DSM 20530 | NR026161T | 99.86% |
| Micrococcus luteu | MB5 | MH450098 | 99.71% | |
| Bacillus jeotgali | YKJ-10 | NR025060T | 99.93% | |
| Right leg | Bacillus megaterium | NBRC 15308 = ATCC 14581 | NR112636T | 100% |
| Microbacterium imperial | DSM 20530 | NR026161T | 99.86% | |
| Kocuria turfanensis | HO-9042 | NR043899T | 88.22% | |
| Left leg | Microbacterium imperial | DSM 20530 | NR026161T | 99.86% |
| Bacillus jeotgali | YKJ-10 | NR025060T | 99.93% | |
| Bacillus megaterium | NBRC 15308 = ATCC 14581 | NR112636T | 100% | |
| Mouth | Bacillus jeotgali | YKJ-10 | NR025060T | 99.93% |
| Micrococcus luteus | MB5 | MH450098 | 99.71% | |
| Scheme | Identification | Closely Related Microbial Strains Accessed from Gen Bank | ||
|---|---|---|---|---|
| Strain No. | Accession No. | Similarity | ||
| Head | Monascus pallens | IMI 356820 | GU733328 | 100% |
| Rhizopus oryzae | IDI 354840 | GU732332 | 99.88% | |
| Wig | Microbacterium imperial | DSM 20530 | NR026161T | 99.86% |
| Monascus pallens | IMI 356820 | GU733328 | 100% | |
| Rib cage | Rhizopus oryzae | IDI 354840 | 2GU732332 | 99.88% |
| Abdominal and pelvic cavity | Monascus pallens | IMI 356820 | GU733328 | 100% |
| Right leg | Rhizopus oryzae | IDI 354840 | GU732332 | 99.88% |
| Left leg | Microbacterium imperial | DSM 20530 | NR026161T | 99.86% |
| Rhizopus oryzae | IDI 354840 | GU732332 | 99.88% | |
| Mouth | Monascus pallens | IMI 356820 | GU733328 | 100% |
| Rhizopus oryzae | IDI 354840 | GU732332 | 99.88% | |
| Microbial Isolates | Diameter of Clearing Zone (mm) | |
|---|---|---|
| Cellulases | Protease | |
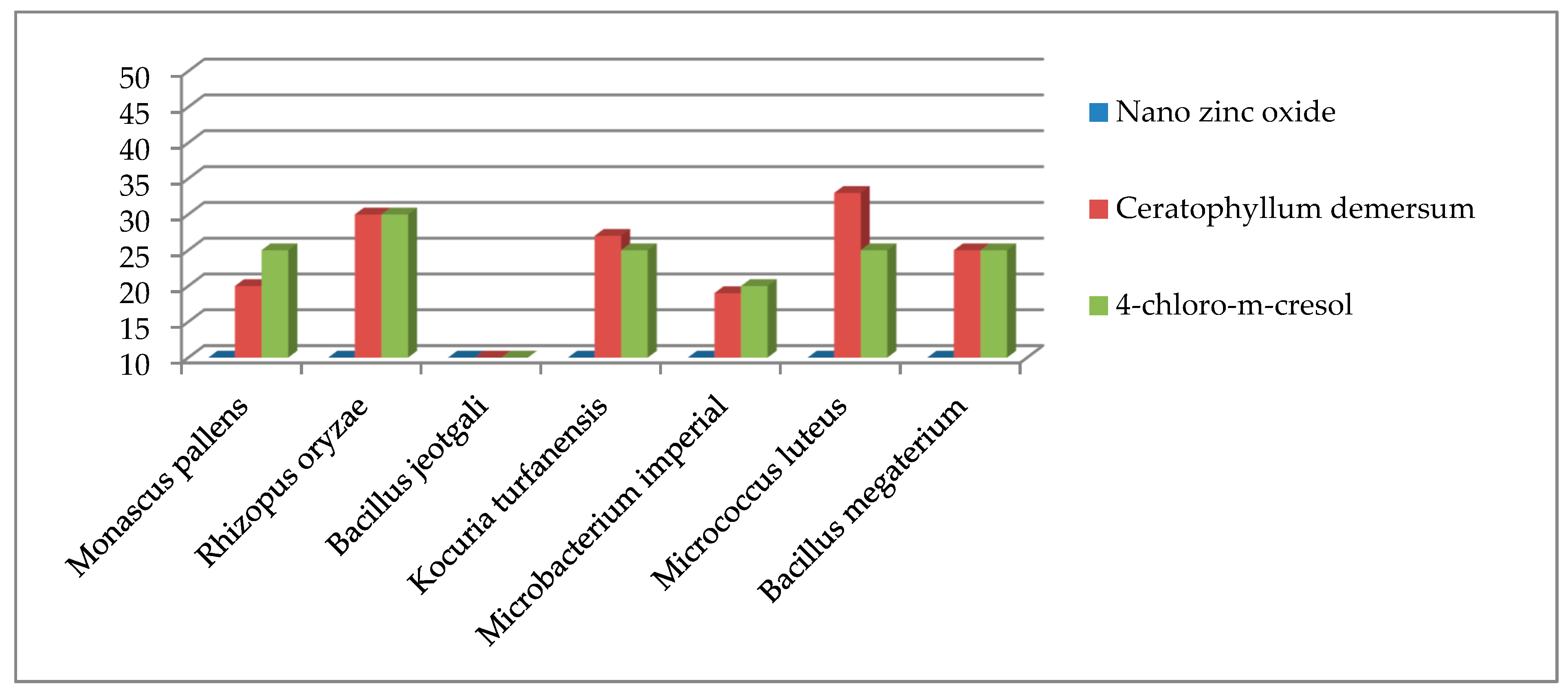
| Monascus pallens | 23 | 22 |
| Rhizopus oryzae | 24 | 21 |
| Bacillus jeotgali | 28 | 27 |
| Kocuria turfanensis | 19 | 20 |
| Microbacterium imperial | 26 | 32 |
| Micrococcus luteus | 24 | 25 |
| Bacillus megaterium | 22 | 21 |
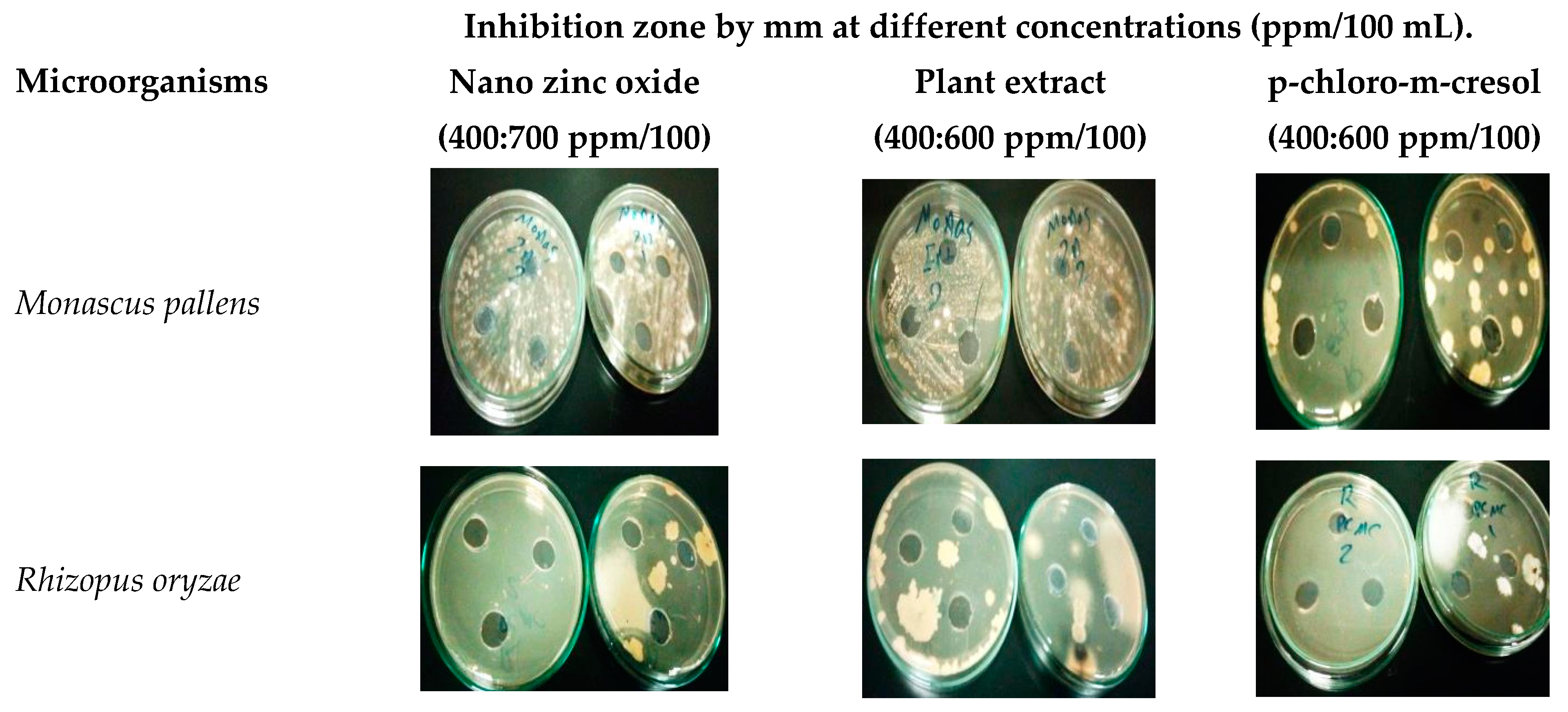
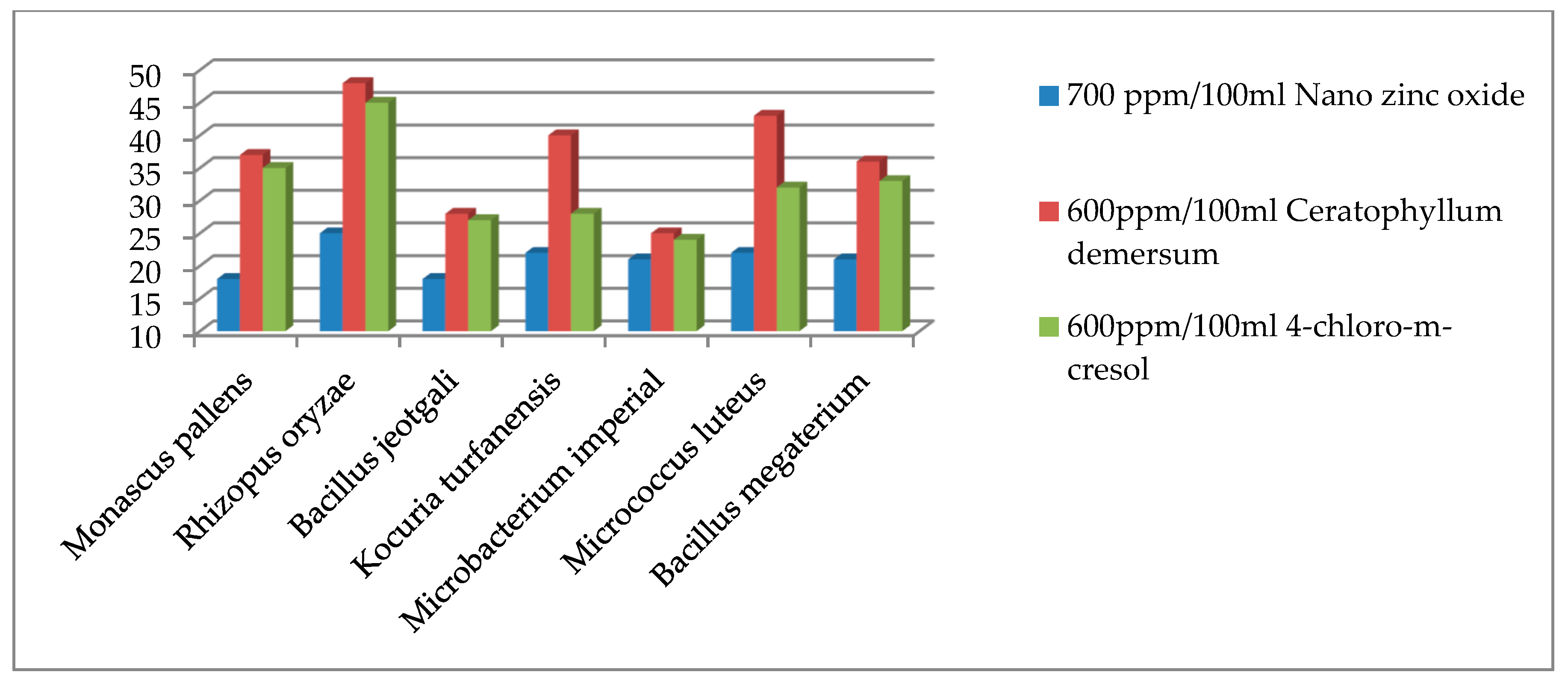
| Microorganisms | Inhibition Zone by mm at Different Concentrations (ppm/100 mL). | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Nano Zinc Oxide | Plant Extract | 4-chloro-m-cresol | ||||||||
| 400 | 500 | 600 | 700 | 400 | 500 | 600 | 400 | 500 | 600 | |
| Monascus pallens | 0 | 0 | 0 | 18 | 0 | 20 | 37 | 0 | 25 | 35 |
| Rhizopus oryzae | 0 | 0 | 0 | 25 | 0 | 30 | 48 | 0 | 30 | 45 |
| Bacillus jeotgali | 0 | 0 | 0 | 18 | 0 | 0 | 28 | 0 | 0 | 27 |
| Kocuria turfanensis | 0 | 0 | 0 | 22 | 0 | 27 | 40 | 0 | 25 | 28 |
| Microbacterium imperial | 0 | 0 | 0 | 21 | 0 | 19 | 25 | 0 | 20 | 24 |
| Micrococcus luteus | 0 | 0 | 0 | 22 | 0 | 33 | 43 | 0 | 25 | 32 |
| Bacillus megaterium | 0 | 0 | 0 | 21 | 0 | 25 | 36 | 0 | 25 | 33 |
| Growth Detection Time | Growth of the Infectious Isolates after Treatment with (Ceratophyllum demersum L.) at 600 ppm/100 mL |
|---|---|
| 48 h | No Growth |
| 3 Month | No Growth |
| 6 Month | No Growth |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ismael, S.; Omar, A.; Maher, M. Comparative Inhibition Study by Nanomaterial, Plant Extract and Chemical Microcide on the Screaming Mummy in Egyptian Museum Store. Heritage 2021, 4, 2481-2493. https://doi.org/10.3390/heritage4030140
Ismael S, Omar A, Maher M. Comparative Inhibition Study by Nanomaterial, Plant Extract and Chemical Microcide on the Screaming Mummy in Egyptian Museum Store. Heritage. 2021; 4(3):2481-2493. https://doi.org/10.3390/heritage4030140
Chicago/Turabian StyleIsmael, Sahar, Ali Omar, and Manal Maher. 2021. "Comparative Inhibition Study by Nanomaterial, Plant Extract and Chemical Microcide on the Screaming Mummy in Egyptian Museum Store" Heritage 4, no. 3: 2481-2493. https://doi.org/10.3390/heritage4030140
APA StyleIsmael, S., Omar, A., & Maher, M. (2021). Comparative Inhibition Study by Nanomaterial, Plant Extract and Chemical Microcide on the Screaming Mummy in Egyptian Museum Store. Heritage, 4(3), 2481-2493. https://doi.org/10.3390/heritage4030140





