Towards Early Maternal Morbidity Risk Identification by Concept Extraction from Clinical Notes in Spanish Using Fine-Tuned Transformer-Based Models
Abstract
1. Introduction
2. Materials and Methods
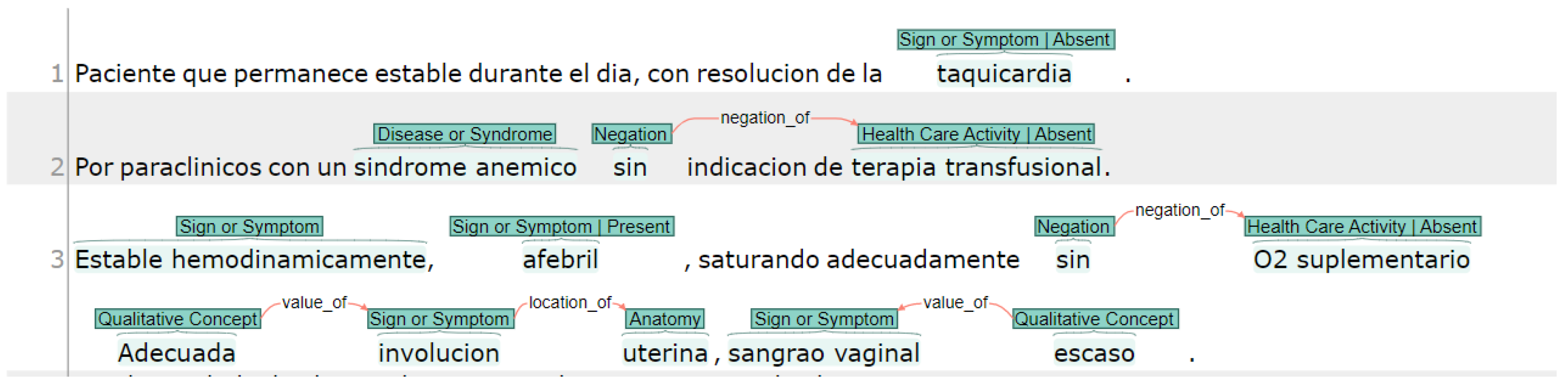
2.1. Dataset and Annotations
- Anatomy (T023): Body part, organ, or organ component.
- Disease or Syndrome (T047): Observations made by patients or clinicians about the body or mind that are considered abnormal or caused by disease.
- Healthcare activity (T058): Descriptions of diagnostics and laboratory procedures.
- Idea or Concept:
- (a)
- Negation (C1518422): Those concepts that indicated something does not exist or that it lacks veracity.
- (b)
- Quantitative concepts (T081): Those concepts that refer to the numerical nature of data, such as measurements obtained in a laboratory test.
- (c)
- Qualitative concepts (T080): Concepts that are used to name what is linked to the quality, such as the way of being or the properties of something.
- (d)
- Temporal concepts (T079): These refer to a point in time that can be a time, but they exclude the concepts that refer to duration (duration is an amount).
- Sign or Symptom (T184): An observable manifestation of a disease or condition based on clinical judgment, even if a disease or condition experienced by the patient and is reported as a subjective observation.
2.2. Preprocessing
2.3. Deep Learning Models
- bert: Bidirectional Encoder Representations from Transformers [27] is a pre-trained deep learning model developed by Google for natural language processing tasks such as question answering and language inference. It is based on the transformer architecture and is trained on a large corpus of text data using unsupervised learning. BERT can be fine-tuned for a wide range of natural language understanding tasks and has achieved state-of-the-art results on a number of benchmarks, including the GLUE benchmark and SQuAD.
- roberta: It is an optimized bert model [28], which has been pre-trained by Text Mining Unit (TeMU) at the Barcelona Supercomputing Center, using the most extensive Spanish corpus known to date, with a total of 201,080,084 documents, compiled from the web crawlings performed by the National Library of Spain (Biblioteca Nacional de España) from 2009 to 2019.
- gpt-2: Generative Pre-trained Transformer is a language generation model developed by OpenAI. GPT-2 is trained on a massive dataset of over 40 GB of text data and can generate human-like text in various languages and styles. It can be used for various natural language processing tasks such as language translation, text summarization, and question answering. We used a fine-tuned version of GPT-2 available in Hugging Face (https://huggingface.co/brad1141/gpt2-finetuned-comp2, accessed on 2 October 2024). This version was fine-tuned on the token classification task. This supervised fine-tuning allowed the model to adapt its generative capabilities toward the specific task of entity recognition and categorization. This aligns with recent trends in NLP in which generative models are used for information extraction [29].
- bio-es: A model produced by the Barcelona Supercomputing Center based on the roberta model using a large biomedical corpora from a variety of sources in Spanish with a total of 1.1 B tokens across 2.5 M documents, allowing us to acquire specialized knowledge and language patterns relevant to the field of biomedicine in Spanish. Additionally, this model was fine-tuned on three NER tasks: PharmaCoNER, CANTEMIST, and ICTUSnet [30].
- bio-ehr-es: Like the bio-es model, bio-ehr-es is based on the roberta model and was fine-tuned in the same NER tasks as bio-es. The key difference is that bio-ehr-es incorporates electronic health record (EHR) information drawn from a corpus with more than 514k clinical documents containing 95 million tokens [30].
2.4. Fine-Tuning
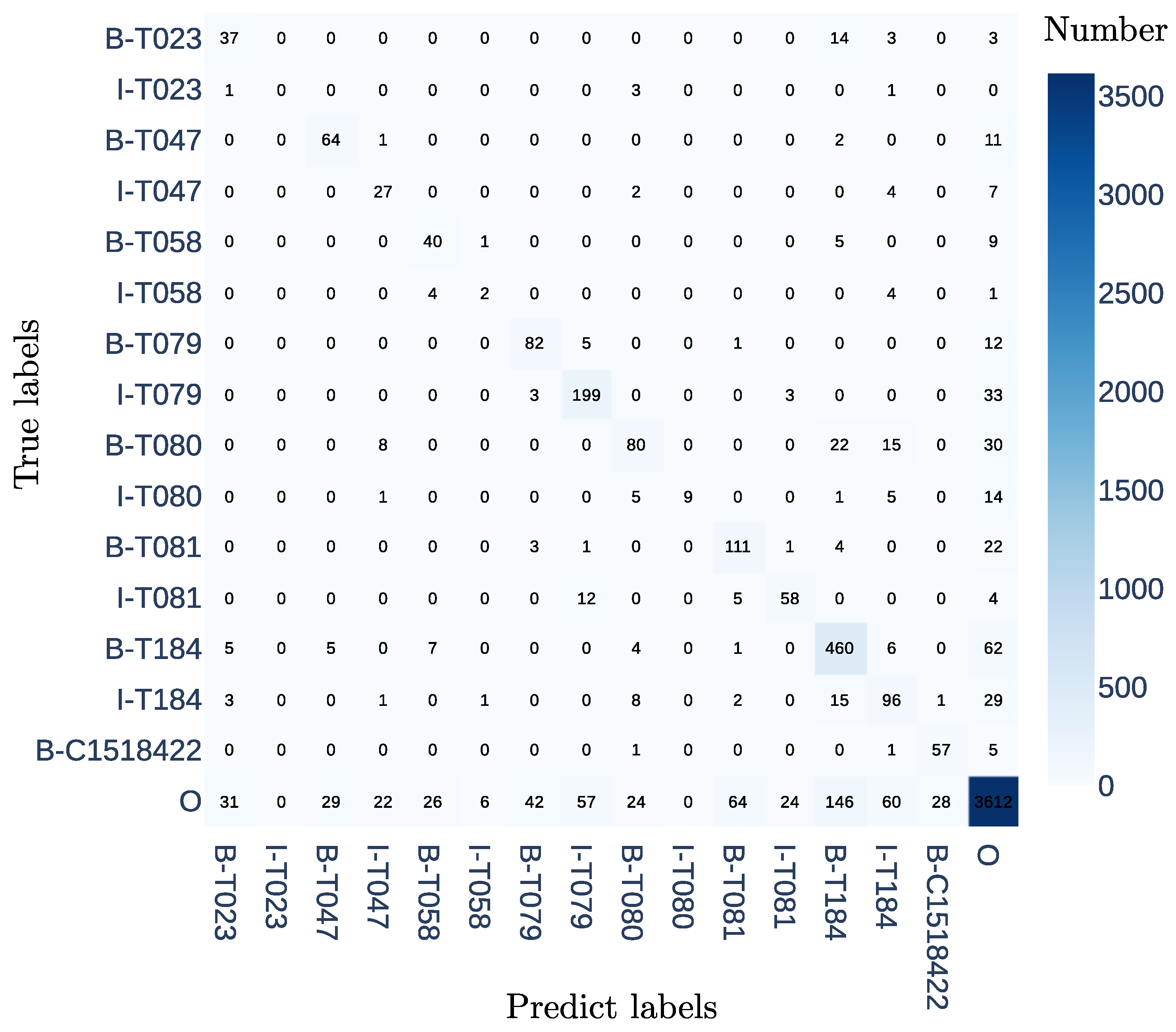
2.5. Evaluation Metrics
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Centers for Disease Control and Prevention: CDC. Pregnancy Mortality Surveillance System; CDC: Atlanta, GA, USA, 2022. [Google Scholar]
- World Health Organization: WHO. Maternal Mortality; WHO: Geneva, Switzerland, 2023. [Google Scholar]
- Vanderkruik, R.C.; Tunçalp, Ö.; Chou, D.; Say, L. Framing maternal morbidity: WHO scoping exercise. BMC Pregnancy Childbirth 2013, 13, 213. [Google Scholar] [CrossRef] [PubMed]
- Pan American Health Organization. Recommendations for Establishing a National Maternal Near-Miss Surveillance System in Latin America and the Caribbean; Pan American Health Organization: Washington, DC, USA, 2022. [Google Scholar]
- Abdollahpour, S.; Miri, H.H.; Khadivzadeh, T. The global prevalence of maternal near miss: A systematic review and meta-analysis. Health Promot. Perspect. 2019, 9, 255. [Google Scholar] [CrossRef]
- Aleman, A.; Colomar, M.; Colistro, V.; Tomaso, G.; Sosa, C.; Serruya, S.; de Francisco, L.A.; Ciganda, A.; De Mucio, B. Predicting severe maternal outcomes in a network of sentinel sites in Latin-American countries. Int. J. Gynecol. Obstet. 2023, 160, 939–946. [Google Scholar] [CrossRef]
- Kasthurirathne, S.N.; Mamlin, B.W.; Purkayastha, S.; Cullen, T. Overcoming the maternal care crisis: How can lessons learnt in global health informatics address US maternal health outcomes? In Proceedings of the AMIA Annual Symposium, Washington, DC, USA, 4–8 November 2017; American Medical Informatics Association: Washington, DC, USA, 2017; Volume 2017, p. 1034. [Google Scholar]
- Chen, P.; Zhang, M.; Yu, X.; Li, S. Named entity recognition of Chinese electronic medical records based on a hybrid neural network and medical MC-BERT. BMC Med. Inform. Decis. Mak. 2022, 22, 315. [Google Scholar] [CrossRef]
- López-García, G.; Jerez, J.M.; Ribelles, N.; Alba, E.; Veredas, F.J. Explainable clinical coding with in-domain adapted transformers. J. Biomed. Inform. 2023, 139, 104323. [Google Scholar] [CrossRef] [PubMed]
- Shickel, B.; Tighe, P.J.; Bihorac, A.; Rashidi, P. Deep EHR: A survey of recent advances in deep learning techniques for electronic health record (EHR) analysis. IEEE J. Biomed. Health Inform. 2017, 22, 1589–1604. [Google Scholar] [CrossRef]
- Torres-Silva, E.A.; Rúa, S.; Giraldo-Forero, A.F.; Durango, M.C.; Flórez-Arango, J.F.; Orozco-Duque, A. Classification of Severe Maternal Morbidity from Electronic Health Records Written in Spanish Using Natural Language Processing. Appl. Sci. 2023, 13, 10725. [Google Scholar] [CrossRef]
- Wu, Y.; Jiang, M.; Xu, J.; Zhi, D.; Xu, H. Clinical named entity recognition using deep learning models. In Proceedings of the AMIA Annual Symposium, Washington, DC, USA, 4–8 November 2017; American Medical Informatics Association: Washington, DC, USA, 2017; Volume 2017, p. 1812. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, L.; Polosukhin, I. Attention Is All You Need. arXiv 2017, arXiv:1706.03762. [Google Scholar]
- Harnoune, A.; Rhanoui, M.; Mikram, M.; Yousfi, S.; Elkaimbillah, Z.; El Asri, B. BERT based clinical knowledge extraction for biomedical knowledge graph construction and analysis. Comput. Methods Programs Biomed. Update 2021, 1, 100042. [Google Scholar] [CrossRef]
- Scaboro, S.; Portelli, B.; Chersoni, E.; Santus, E.; Serra, G. Extensive evaluation of transformer-based architectures for adverse drug events extraction. Knowl.-Based Syst. 2023, 275, 110675. [Google Scholar] [CrossRef]
- Wolf, T.; Debut, L.; Sanh, V.; Chaumond, J.; Delangue, C.; Moi, A.; Cistac, P.; Rault, T.; Louf, R.; Funtowicz, M.; et al. Huggingface’s transformers: State-of-the-art natural language processing. arXiv 2019, arXiv:1910.03771. [Google Scholar]
- Kalyan, K.S.; Rajasekharan, A.; Sangeetha, S. Ammus: A survey of transformer-based pretrained models in natural language processing. arXiv 2021, arXiv:2108.05542. [Google Scholar]
- Touvron, H.; Lavril, T.; Izacard, G.; Martinet, X.; Lachaux, M.A.; Lacroix, T.; Rozière, B.; Goyal, N.; Hambro, E.; Azhar, F.; et al. Llama: Open and efficient foundation language models. arXiv 2023, arXiv:2302.13971. [Google Scholar]
- Team, G.; Anil, R.; Borgeaud, S.; Alayrac, J.B.; Yu, J.; Soricut, R.; Schalkwyk, J.; Dai, A.M.; Hauth, A.; Millican, K.; et al. Gemini: A family of highly capable multimodal models. arXiv 2023, arXiv:2312.11805. [Google Scholar]
- Montoya-Lora, D.; Giraldo-Forero, A.F.; Torres-Silva, E.A.; Rua, S.; Arango-Valencia, S.; Barrientos-Gómez, J.G.; Florez-Arango, J.F.; Orozco-Duque, A. Prompt Engineering of GPT-3.5 for Extracting Signs and Symptoms and Their Assertions in Maternal Electronic Health Records: A Pilot Study. In Proceedings of the Latin American Conference on Biomedical Engineering, Panama City, Panama, 2–5 October 2024; Springer: Berlin/Heidelberg, Germany, 2024; pp. 53–62. [Google Scholar]
- Yuan, K.; Yoon, C.H.; Gu, Q.; Munby, H.; Walker, A.S.; Zhu, T.; Eyre, D.W. Transformers and large language models are efficient feature extractors for electronic health record studies. Commun. Med. 2025, 5, 83. [Google Scholar] [CrossRef] [PubMed]
- Jahan, I.; Laskar, M.T.R.; Peng, C.; Huang, J.X. A comprehensive evaluation of large language models on benchmark biomedical text processing tasks. Comput. Biol. Med. 2024, 171, 108189. [Google Scholar] [CrossRef]
- Rohanian, O.; Nouriborji, M.; Kouchaki, S.; Nooralahzadeh, F.; Clifton, L.; Clifton, D.A. Exploring the effectiveness of instruction tuning in biomedical language processing. Artif. Intell. Med. 2024, 158, 103007. [Google Scholar] [CrossRef]
- Klie, J.C.; Bugert, M.; Boullosa, B.; De Castilho, R.E.; Gurevych, I. The inception platform: Machine-assisted and knowledge-oriented interactive annotation. In Proceedings of the 27th International Conference on Computational Linguistics: System Demonstrations, Santa Fe, NM, USA, 21–25 August 2018; pp. 5–9. [Google Scholar]
- Landis, J.R.; Koch, G.G. The measurement of observer agreement for categorical data. Biometrics 1977, 33, 159–174. [Google Scholar] [CrossRef]
- Uronen, L.; Salanterä, S.; Hakala, K.; Hartiala, J.; Moen, H. Combining supervised and unsupervised named entity recognition to detect psychosocial risk factors in occupational health checks. Int. J. Med. Inform. 2022, 160, 104695. [Google Scholar] [CrossRef]
- Devlin, J.; Chang, M.W.; Lee, K.; Toutanova, K. Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv 2018, arXiv:1810.04805. [Google Scholar]
- Liu, Y.; Ott, M.; Goyal, N.; Du, J.; Joshi, M.; Chen, D.; Levy, O.; Lewis, M.; Zettlemoyer, L.; Stoyanov, V. Roberta: A robustly optimized bert pretraining approach. arXiv 2019, arXiv:1907.11692. [Google Scholar]
- Rezapour, M.; Ali, E. GPT-2 Based Named Entity Recognition, with back-translation data augmentation. ESS Open Arch. Eprints 2024, 172, 17284645. [Google Scholar]
- Carrino, C.P.; Llop, J.; Pàmies, M.; Gutiérrez-Fandiño, A.; Armengol-Estapé, J.; Silveira-Ocampo, J.; Valencia, A.; Gonzalez-Agirre, A.; Villegas, M. Pretrained Biomedical Language Models for Clinical NLP in Spanish. In Proceedings of the 21st Workshop on Biomedical Language Processing, Dublin, Ireland, 26 May 2022; pp. 193–199. [Google Scholar] [CrossRef]
- Miranda-Escalada, A.; Farré, E.; Krallinger, M. Named Entity Recognition, Concept Normalization and Clinical Coding: Overview of the Cantemist Track for Cancer Text Mining in Spanish, Corpus, Guidelines, Methods and Results. In Proceedings of the Iberian Languages Evaluation Forum (IberLEF 2020), Online, 23 September 2020; pp. 303–323. [Google Scholar]
- Gonzalez-Agirre, A.; Marimon, M.; Intxaurrondo, A.; Rabal, O.; Villegas, M.; Krallinger, M. Pharmaconer: Pharmacological substances, compounds and proteins named entity recognition track. In Proceedings of the 5th Workshop on BioNLP Open Shared Tasks, Hong Kong, China, 4 November 2019; pp. 1–10. [Google Scholar]
- Takahashi, K.; Yamamoto, K.; Kuchiba, A.; Koyama, T. Confidence interval for micro-averaged F 1 and macro-averaged F 1 scores. Appl. Intell. 2022, 52, 4961–4972. [Google Scholar] [CrossRef] [PubMed]
- Yadav, V.; Bethard, S. A survey on recent advances in named entity recognition from deep learning models. arXiv 2019, arXiv:1910.11470. [Google Scholar]
- Segura-Bedmar, I.; Martínez, P.; Herrero-Zazo, M. Semeval-2013 task 9: Extraction of drug-drug interactions from biomedical texts (ddiextraction 2013). In Proceedings of the Second Joint Conference on Lexical and Computational Semantics (* SEM), Volume 2: Proceedings of the Seventh International Workshop on Semantic Evaluation (SemEval 2013), Atlanta, Georgia, 14–15 June 2013; Association for Computational Linguistics: Stroudsburg, PA, USA, 2013; pp. 341–350. [Google Scholar]
- Tsai, R.T.H.; Wu, S.H.; Chou, W.C.; Lin, Y.C.; He, D.; Hsiang, J.; Sung, T.Y.; Hsu, W.L. Various criteria in the evaluation of biomedical named entity recognition. BMC Bioinform. 2006, 7, 92. [Google Scholar] [CrossRef]

| Mention Type | Label | Annotations | |
|---|---|---|---|
| Train | Test | ||
| Anatomy | B-T023 | 410 | 34 |
| I-T023 | 75 | 3 | |
| Disease or Syndrome | B-T047 | 272 | 40 |
| I-T047 | 194 | 37 | |
| Healthcare activity | B-T058 | 350 | 38 |
| I-T058 | 177 | 17 | |
| Temporal concept | B-T079 | 812 | 112 |
| I-T079 | 2097 | 341 | |
| Qualitative concept | B-T080 | 1224 | 158 |
| I-T080 | 325 | 37 | |
| Quantitative concept | B-T081 | 815 | 107 |
| I-T081 | 877 | 93 | |
| Sign or Symptom | B-T184 | 3056 | 369 |
| I-T184 | 1191 | 161 | |
| Negation | B-C1518422 | 502 | 64 |
| No entity | O | 30,473 | 3514 |
| Total | 42,850 | 5025 | |
| Model | Micro | Macro | Parameters | ||||
|---|---|---|---|---|---|---|---|
| F1 | P | R | F1 | LR | Epoch | Task | |
| gpt-2 | 0.776 ± 0.033 | 0.582 ± 0.031 | 0.426 ± 0.030 | 0.462 ± 0.033 | 8 × | 12 | TC |
| bert [27] | 0.768 ± 0.020 | 0.582 ± 0.044 | 0.488 ± 0.029 | 0.494 ± 0.028 | 5 × | 12 | MLM |
| roberta [28] | 0.810 ± 0.059 | 0.623 ± 0.051 | 0.543 ± 0.041 | 0.541 ± 0.059 | 5 × | 13 | MLM |
| bio-es [30] | 0.822 ± 0.062 | 0.588 ± 0.041 | 0.565 ± 0.070 | 0.552 ± 0.027 | 8 × | 13 | MLM |
| bio-ehr-es [30] | 0.823 ± 0.034 | 0.570 ± 0.035 | 0.531 ± 0.048 | 0.526 ± 0.034 | 5 × | 13 | MLM |
| Class | Partial | Exact | ||||
|---|---|---|---|---|---|---|
| P | R | F1 | P | R | F1 | |
| T023 | 0.616 ± 0.117 | 0.745 ± 0.143 | 0.663 ± 0.088 | 0.589 ± 0.122 | 0.713 ± 0.152 | 0.635 ± 0.101 |
| T047 | 0.636 ± 0.047 | 0.769 ± 0.036 | 0.695 ± 0.034 | 0.583 ± 0.06 | 0.704 ± 0.036 | 0.637 ± 0.048 |
| T058 | 0.605 ± 0.074 | 0.69 ± 0.088 | 0.64 ± 0.056 | 0.576 ± 0.08 | 0.654 ± 0.069 | 0.608 ± 0.053 |
| T079 | 0.564 ± 0.076 | 0.769 ± 0.08 | 0.649 ± 0.07 | 0.51 ± 0.087 | 0.693 ± 0.094 | 0.586 ± 0.086 |
| T080 | 0.582 ± 0.06 | 0.663 ± 0.092 | 0.618 ± 0.059 | 0.552 ± 0.062 | 0.628 ± 0.094 | 0.586 ± 0.064 |
| T081 | 0.634 ± 0.091 | 0.759 ± 0.164 | 0.684 ± 0.112 | 0.591 ± 0.091 | 0.709 ± 0.162 | 0.638 ± 0.113 |
| T184 | 0.687 ± 0.057 | 0.825 ± 0.035 | 0.749 ± 0.046 | 0.655 ± 0.059 | 0.785 ± 0.038 | 0.714 ± 0.049 |
| C1518422 | 0.635 ± 0.082 | 0.856 ± 0.051 | 0.726 ± 0.056 | 0.635 ± 0.082 | 0.856 ± 0.051 | 0.726 ± 0.056 |
| Class | Partial | Exact | ||||
|---|---|---|---|---|---|---|
| P | R | F1 | P | R | F1 | |
| T023 | 0.75 ± 0.113 | 0.631 ± 0.195 | 0.664 ± 0.119 | 0.716 ± 0.107 | 0.604 ± 0.192 | 0.635 ± 0.12 |
| T047 | 0.651 ± 0.065 | 0.748 ± 0.124 | 0.692 ± 0.071 | 0.58 ± 0.067 | 0.668 ± 0.128 | 0.617 ± 0.079 |
| T058 | 0.638 ± 0.064 | 0.588 ± 0.147 | 0.602 ± 0.068 | 0.604 ± 0.064 | 0.556 ± 0.135 | 0.569 ± 0.061 |
| T079 | 0.602 ± 0.045 | 0.786 ± 0.067 | 0.681 ± 0.049 | 0.553 ± 0.051 | 0.721 ± 0.072 | 0.625 ± 0.056 |
| T080 | 0.589 ± 0.088 | 0.583 ± 0.067 | 0.585 ± 0.073 | 0.548 ± 0.085 | 0.542 ± 0.063 | 0.544 ± 0.07 |
| T081 | 0.616 ± 0.086 | 0.721 ± 0.148 | 0.655 ± 0.084 | 0.572 ± 0.096 | 0.672 ± 0.165 | 0.61 ± 0.109 |
| T184 | 0.706 ± 0.024 | 0.755 ± 0.079 | 0.729 ± 0.045 | 0.667 ± 0.023 | 0.714 ± 0.079 | 0.688 ± 0.046 |
| C1518422 | 0.702 ± 0.059 | 0.747 ± 0.303 | 0.685 ± 0.194 | 0.702 ± 0.059 | 0.747 ± 0.303 | 0.685 ± 0.194 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Published by MDPI on behalf of the International Institute of Knowledge Innovation and Invention. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giraldo-Forero, A.F.; Durango, M.C.; Rúa, S.; Torres-Silva, E.A.; Arango-Valencia, S.; Florez-Arango, J.F.; Orozco-Duque, A. Towards Early Maternal Morbidity Risk Identification by Concept Extraction from Clinical Notes in Spanish Using Fine-Tuned Transformer-Based Models. Appl. Syst. Innov. 2025, 8, 78. https://doi.org/10.3390/asi8030078
Giraldo-Forero AF, Durango MC, Rúa S, Torres-Silva EA, Arango-Valencia S, Florez-Arango JF, Orozco-Duque A. Towards Early Maternal Morbidity Risk Identification by Concept Extraction from Clinical Notes in Spanish Using Fine-Tuned Transformer-Based Models. Applied System Innovation. 2025; 8(3):78. https://doi.org/10.3390/asi8030078
Chicago/Turabian StyleGiraldo-Forero, Andrés F., Maria C. Durango, Santiago Rúa, Ever A. Torres-Silva, Sara Arango-Valencia, José F. Florez-Arango, and Andrés Orozco-Duque. 2025. "Towards Early Maternal Morbidity Risk Identification by Concept Extraction from Clinical Notes in Spanish Using Fine-Tuned Transformer-Based Models" Applied System Innovation 8, no. 3: 78. https://doi.org/10.3390/asi8030078
APA StyleGiraldo-Forero, A. F., Durango, M. C., Rúa, S., Torres-Silva, E. A., Arango-Valencia, S., Florez-Arango, J. F., & Orozco-Duque, A. (2025). Towards Early Maternal Morbidity Risk Identification by Concept Extraction from Clinical Notes in Spanish Using Fine-Tuned Transformer-Based Models. Applied System Innovation, 8(3), 78. https://doi.org/10.3390/asi8030078







