VG-SAM: Visual In-Context Guided SAM for Universal Medical Image Segmentation
Abstract
1. Introduction
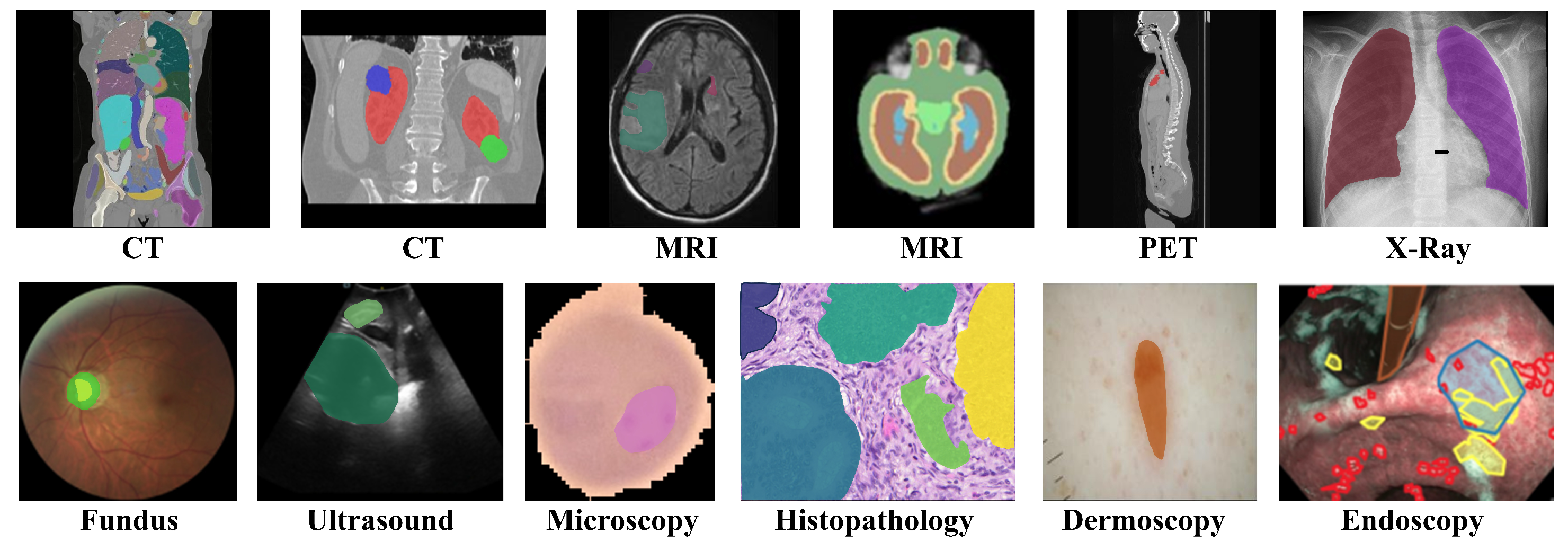
- We propose a novel VG-SAM method for universal medical image segmentation, which achieves deep integration of in-context learning with Segment Anything Model (SAM) and effectively addresses the performance degradation in unseen domains.
- We design an in-context retrieval strategy based on multi-level feature similarity to select the most relevant in-context examples for the query image and extract high-quality priors from them to construct semantic guidance signals.
- We propose a visual in-context generation strategy that leverages a visual prompt construction module to generate multi-granularity visual prompts tailored for SAM. Furthermore, an adaptive fusion module seamlessly integrates semantic guidance signals and visual prompts to effectively guide SAM’s segmentation process, ensuring the generation of high-quality segmentation masks.
- We evaluate our VG-SAM on multiple publicly available medical segmentation datasets encompassing diverse imaging modalities and anatomical structures. Extensive experiments demonstrate the effectiveness and superiority of our VG-SAM. Our source code is publicly available at: https://github.com/dailenson/VG-SAM (accessed on 13 October 2025).
2. Related Work
2.1. Task-Specific Medical Image Segmentation
2.2. Universal Medical Image Segmentation
3. Proposed Method
3.1. Overall Scheme
3.2. Multi-Scale In-Context Retrieval
3.3. Visual In-Context Guidance for SAM-Based Segmentation
4. Experimental Results and Discussions
4.1. Experimental Settings
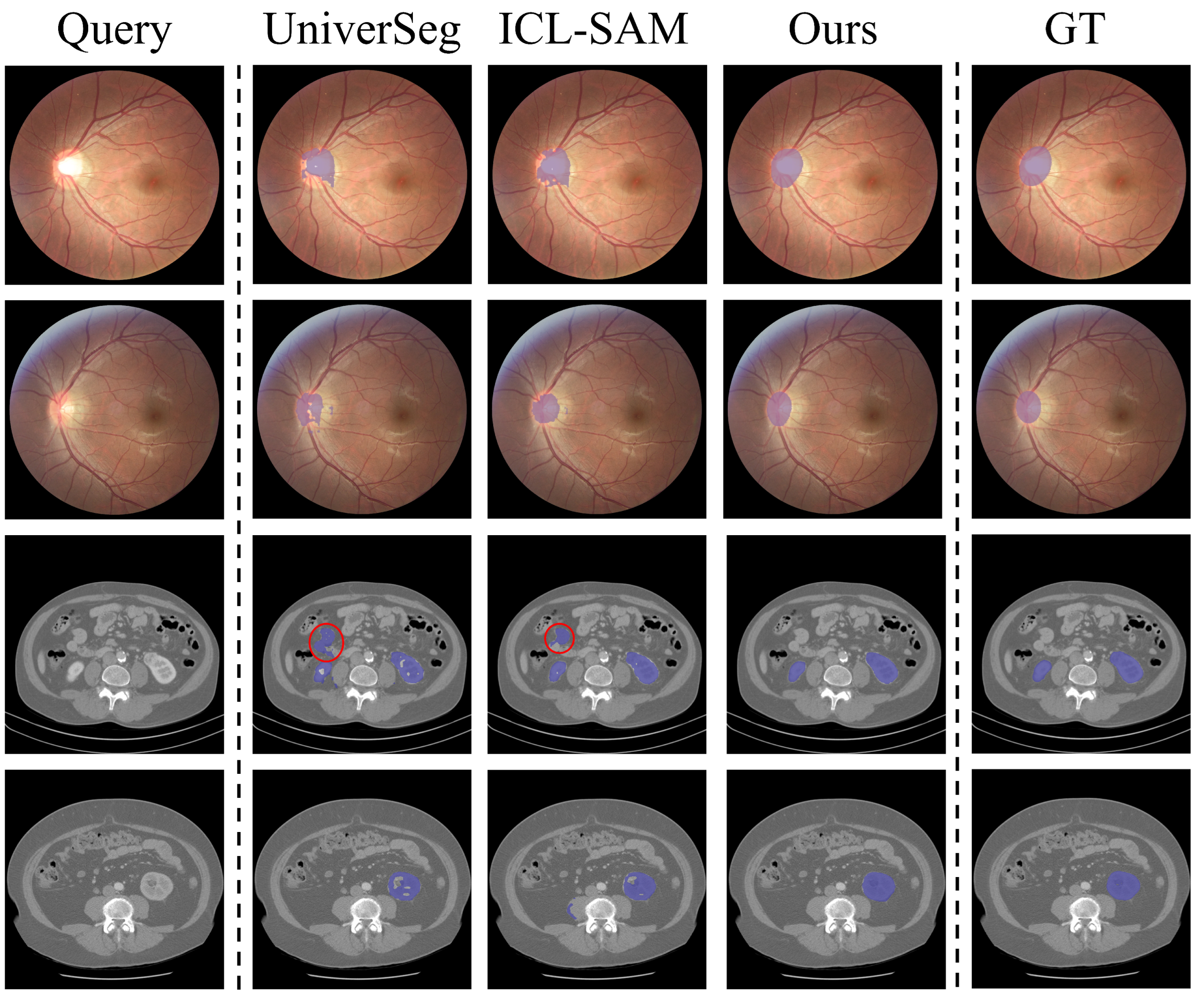
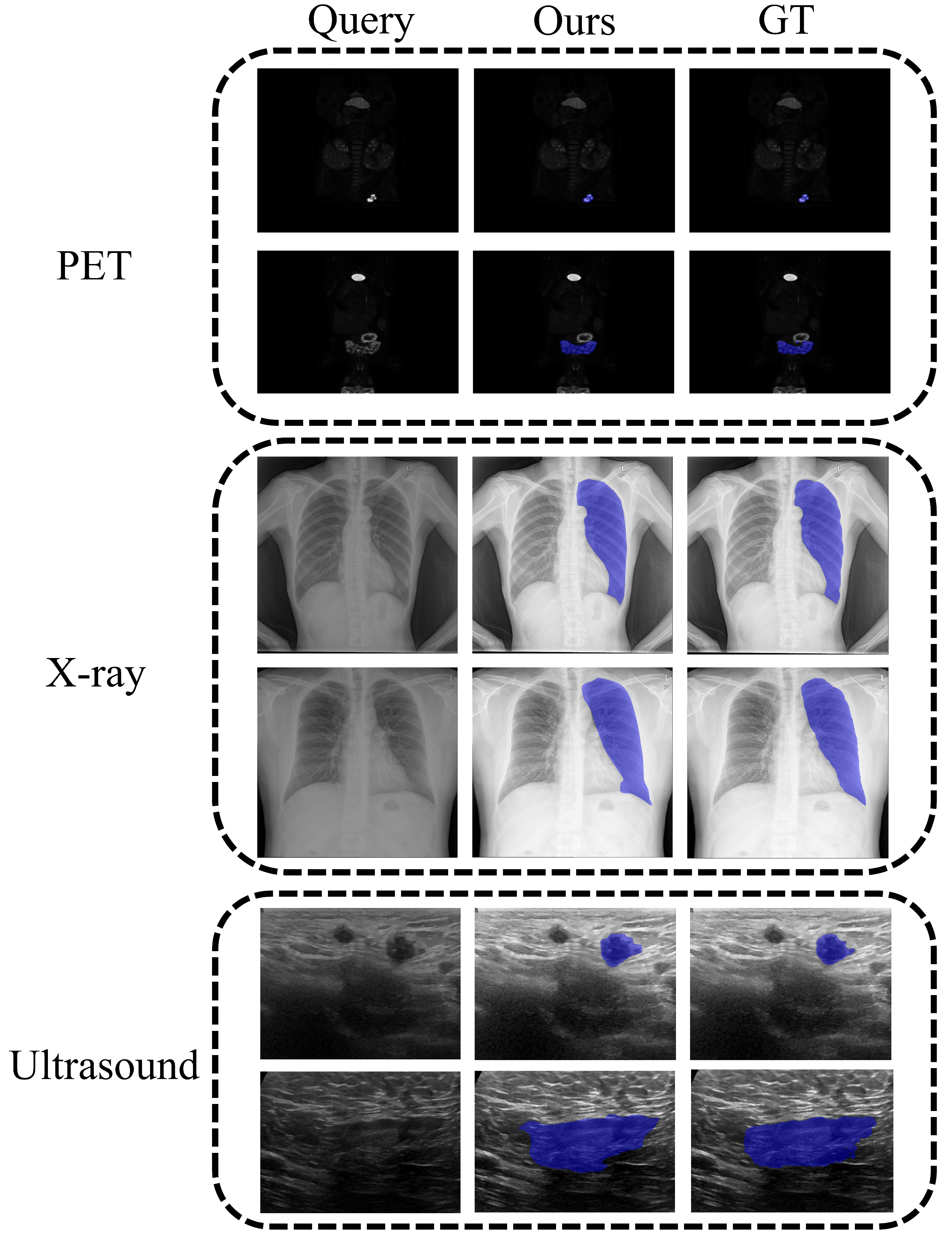
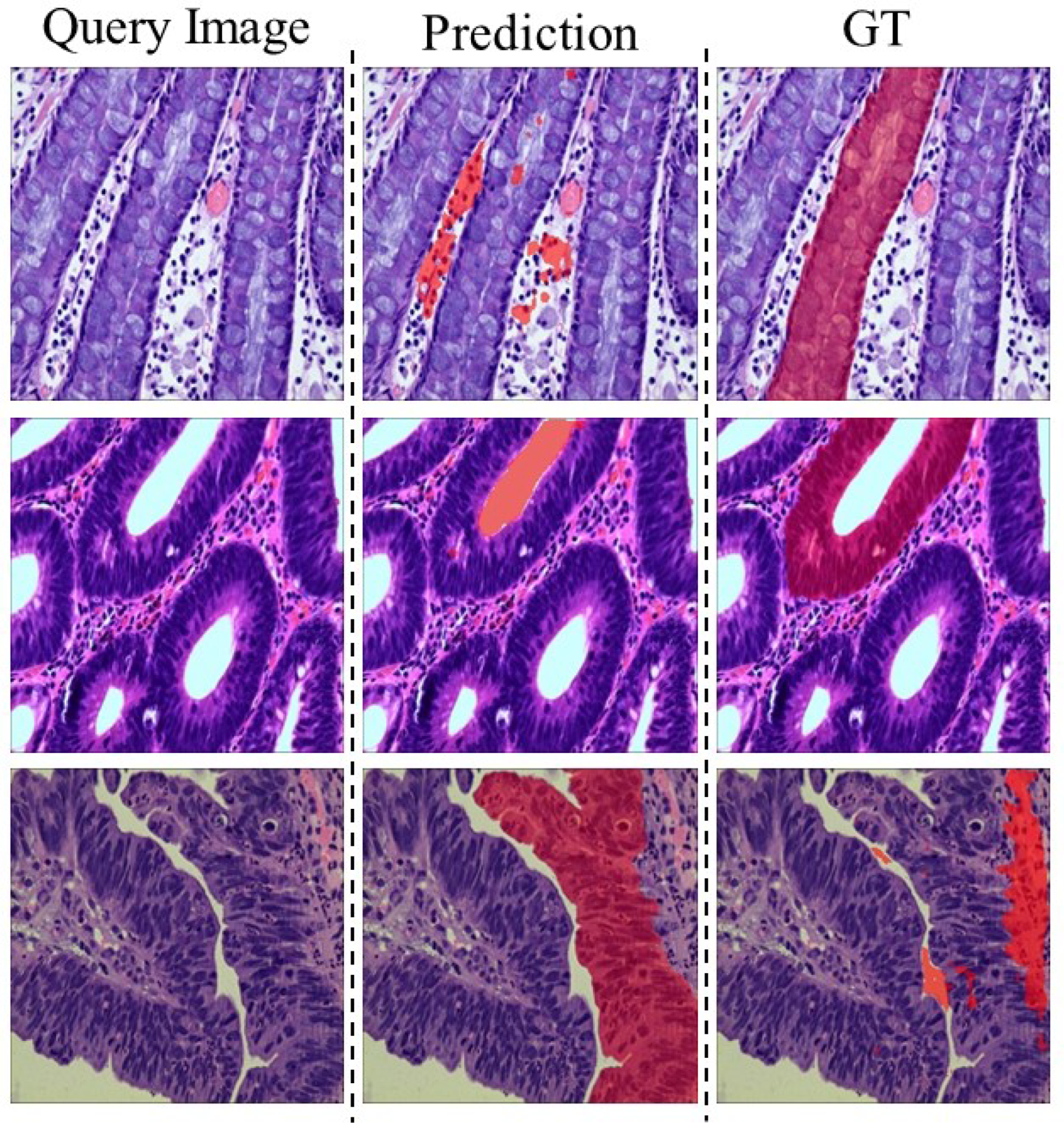
4.2. Main Results
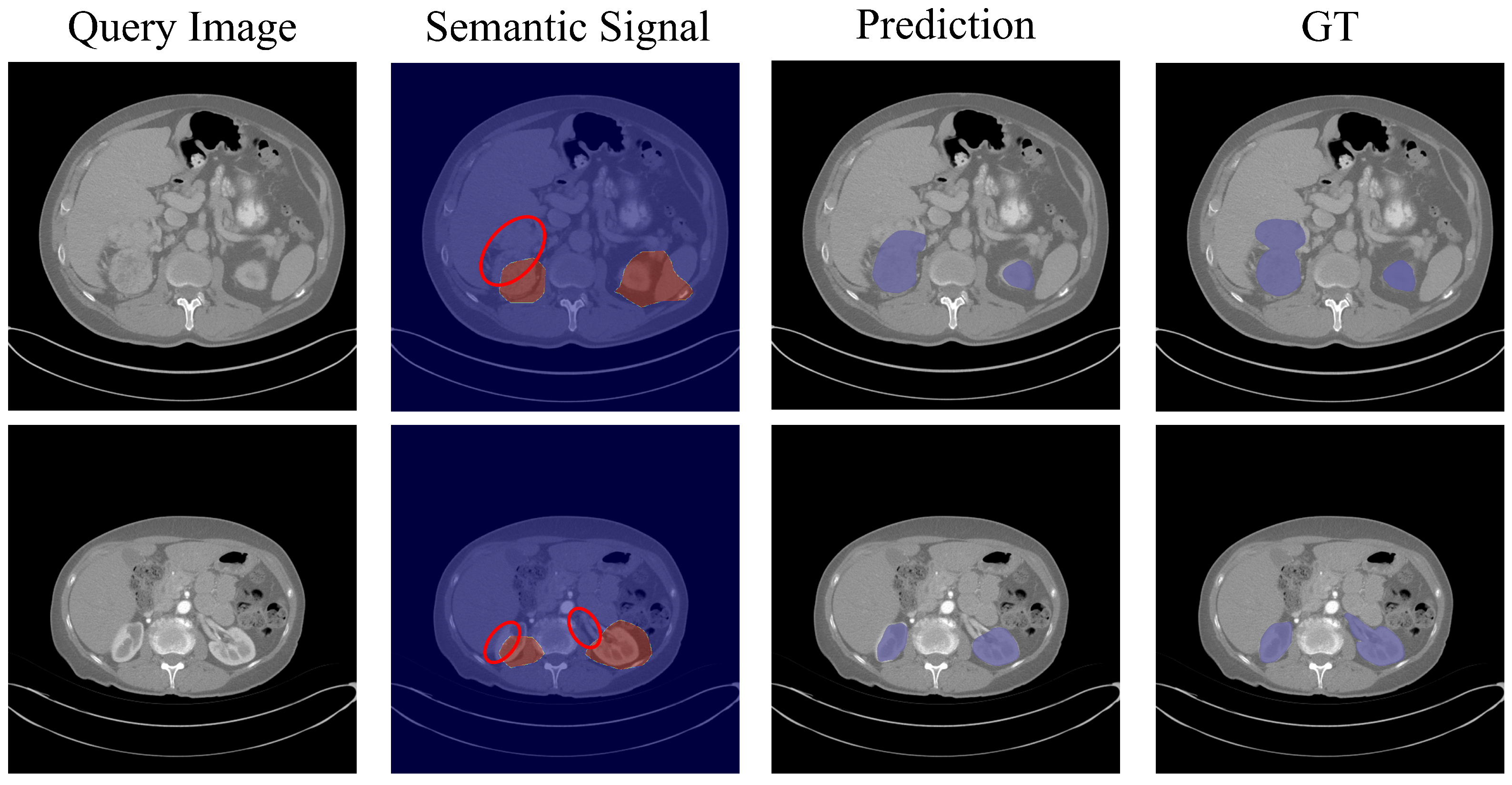
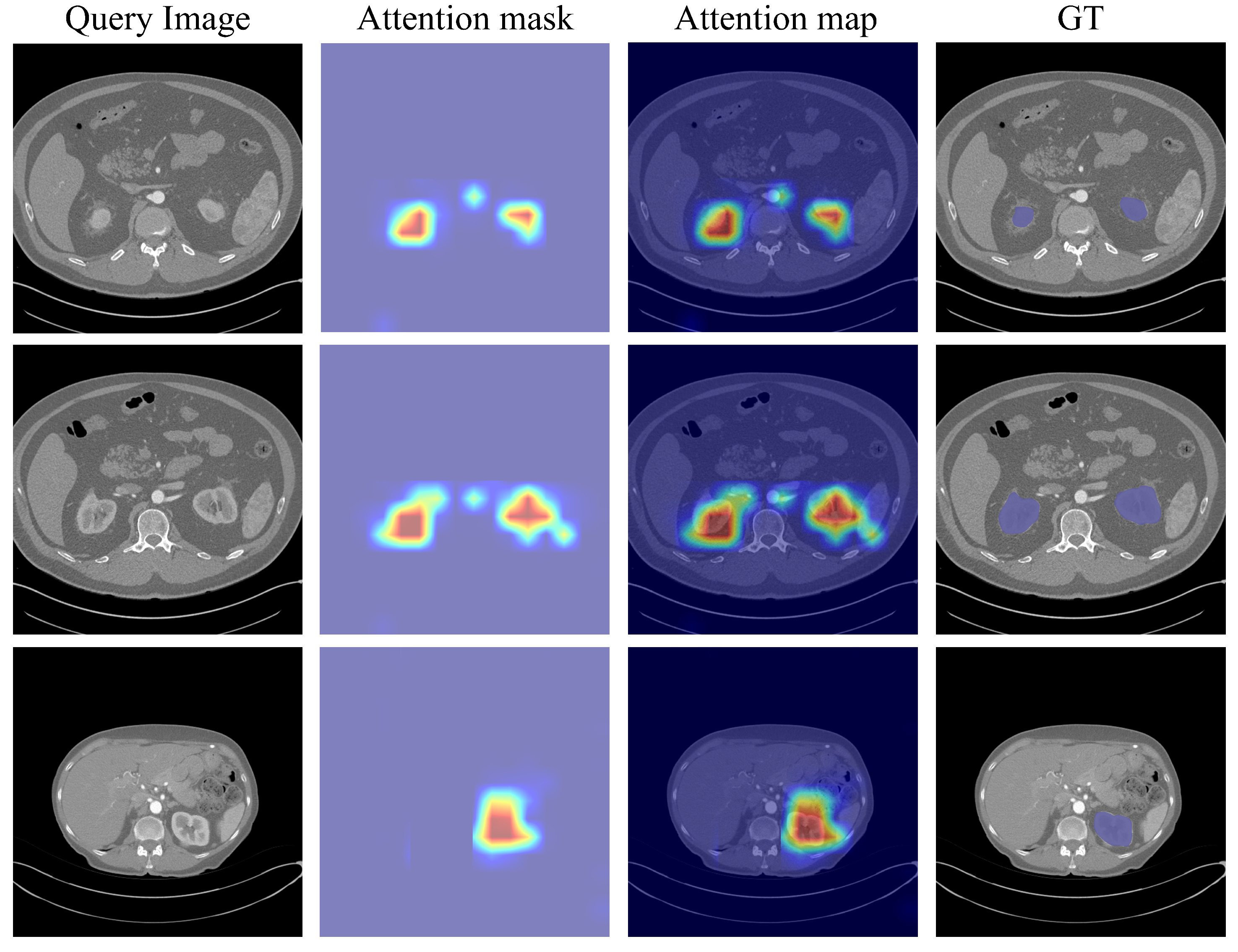
4.3. Analyses and Discussions
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Shao, Y.; Yang, J.; Zhou, W.; Sun, H.; Gao, Q. Fractal-Inspired Region-Weighted Optimization and Enhanced MobileNet for Medical Image Classification. Fractal Fract. 2025, 9, 511. [Google Scholar] [CrossRef]
- Isensee, F.; Jaeger, P.F.; Kohl, S.A.; Petersen, J.; Maier-Hein, K.H. nnU-Net: A self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 2021, 18, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Butoi, V.I.; Ortiz, J.J.G.; Ma, T.; Sabuncu, M.R.; Guttag, J.; Dalca, A.V. Universeg: Universal medical image segmentation. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Paris, France, 2–3 October 2023; pp. 21438–21451. [Google Scholar]
- Chen, H.; Cai, Y.; Wang, C.; Chen, L.; Zhang, B.; Han, H.; Guo, Y.; Ding, H.; Zhang, Q. Multi-organ foundation model for universal ultrasound image segmentation with task prompt and anatomical prior. IEEE Trans. Med. Imaging 2024, 44, 1005–1018. [Google Scholar] [CrossRef]
- Chen, Z.; Xu, Q.; Liu, X.; Yuan, Y. Un-sam: Universal prompt-free segmentation for generalized nuclei images. arXiv 2024, arXiv:2402.16663. [Google Scholar]
- Ye, J.; Cheng, J.; Chen, J.; Deng, Z.; Li, T.; Wang, H.; Su, Y.; Huang, Z.; Chen, J.; Jiang, L.; et al. Sa-med2d-20m dataset: Segment anything in 2d medical imaging with 20 million masks. arXiv 2023, arXiv:2311.11969. [Google Scholar]
- Guan, H.; Liu, M. Domain adaptation for medical image analysis: A survey. IEEE Trans. Biomed. Eng. 2021, 69, 1173–1185. [Google Scholar] [CrossRef] [PubMed]
- Ghamsarian, N.; Gamazo Tejero, J.; Márquez-Neila, P.; Wolf, S.; Zinkernagel, M.; Schoeffmann, K.; Sznitman, R. Domain adaptation for medical image segmentation using transformation-invariant self-training. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Vancouver, BC, Canada, 8–12 October 2023; pp. 331–341. [Google Scholar]
- Brown, T.; Mann, B.; Ryder, N.; Subbiah, M.; Kaplan, J.D.; Dhariwal, P.; Neelakantan, A.; Shyam, P.; Sastry, G.; Askell, A.; et al. Language models are few-shot learners. Adv. Neural Inf. Process. Syst. 2020, 33, 1877–1901. [Google Scholar]
- Czolbe, S.; Dalca, A.V. Neuralizer: General neuroimage analysis without re-training. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Vancouver, BC, Canada, 17–24 June 2023; pp. 6217–6230. [Google Scholar]
- Gao, J.; Lao, Q.; Kang, Q.; Liu, P.; Du, C.; Li, K.; Zhang, L. Boosting your context by dual similarity checkup for In-Context learning medical image segmentation. IEEE Trans. Med. Imaging 2024, 44, 310–319. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Kirillov, A.; Mintun, E.; Ravi, N.; Mao, H.; Rolland, C.; Gustafson, L.; Xiao, T.; Whitehead, S.; Berg, A.C.; Lo, W.Y.; et al. Segment anything. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Paris, France, 2–3 October 2023; pp. 4015–4026. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Las Vegas, NV, USA, 27–30 June2016; pp. 770–778. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. Adv. Neural Inf. Process. Syst. 2017, 30, 6000–6010. [Google Scholar]
- Zhou, Z.; Siddiquee, M.; Tajbakhsh, N.; Liang, J.U. A nested U-Net architecture for medical image segmentation. arXiv 2018, arXiv:1807.10165. [Google Scholar] [CrossRef]
- Oktay, O.; Schlemper, J.; Folgoc, L.L.; Lee, M.; Heinrich, M.; Misawa, K.; Mori, K.; McDonagh, S.; Hammerla, N.Y.; Kainz, B.; et al. Attention u-net: Learning where to look for the pancreas. arXiv 2018, arXiv:1804.03999. [Google Scholar] [CrossRef]
- Çiçek, Ö.; Abdulkadir, A.; Lienkamp, S.S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning dense volumetric segmentation from sparse annotation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Athens, Greece, 17–21 October 2016; pp. 424–432. [Google Scholar]
- Chen, J.; Lu, Y.; Yu, Q.; Luo, X.; Adeli, E.; Wang, Y.; Lu, L.; Yuille, A.L.; Zhou, Y. Transunet: Transformers make strong encoders for medical image segmentation. arXiv 2021, arXiv:2102.04306. [Google Scholar] [CrossRef]
- Cao, H.; Wang, Y.; Chen, J.; Jiang, D.; Zhang, X.; Tian, Q.; Wang, M. Swin-unet: Unet-like pure transformer for medical image segmentation. In Proceedings of the European Conference on Computer Vision, Tel Aviv, Israel, 23–27 October 2022; pp. 205–218. [Google Scholar]
- Hatamizadeh, A.; Yang, D.; Roth, H.; Xu, D.U. Transformers for 3d medical image segmentation. arXiv 2021, arXiv:2103.10504. [Google Scholar] [PubMed]
- Wasserthal, J.; Breit, H.C.; Meyer, M.T.; Pradella, M.; Hinck, D.; Sauter, A.W.; Heye, T.; Boll, D.T.; Cyriac, J.; Yang, S.; et al. TotalSegmentator: Robust segmentation of 104 anatomic structures in CT images. Radiol. Artif. Intell. 2023, 5, e230024. [Google Scholar] [CrossRef]
- Ouyang, C.; Biffi, C.; Chen, C.; Kart, T.; Qiu, H.; Rueckert, D. Self-supervised learning for few-shot medical image segmentation. IEEE Trans. Med. Imaging 2022, 41, 1837–1848. [Google Scholar] [CrossRef]
- Wu, J.; Ji, W.; Fu, H.; Xu, M.; Jin, Y.; Xu, Y. Medsegdiff-v2: Diffusion-based medical image segmentation with transformer. In Proceedings of the AAAI Conference on Artificial Intelligence, Vancouver, BC, Canada, 26–27 February 2024; pp. 6030–6038. [Google Scholar]
- Hu, X.; Xu, X.; Shi, Y. How to efficiently adapt large segmentation model (sam) to medical images. arXiv 2023, arXiv:2306.13731. [Google Scholar] [CrossRef]
- Cheng, Z.; Wei, Q.; Zhu, H.; Wang, Y.; Qu, L.; Shao, W.; Zhou, Y. Unleashing the potential of sam for medical adaptation via hierarchical decoding. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 16–22 June 2024; pp. 3511–3522. [Google Scholar]
- Chen, T.; Zhu, L.; Deng, C.; Cao, R.; Wang, Y.; Zhang, S.; Li, Z.; Sun, L.; Zang, Y.; Mao, P. Sam-adapter: Adapting segment anything in underperformed scenes. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Paris, France, 2–3 October 2023; pp. 3367–3375. [Google Scholar]
- Killeen, B.D.; Wang, L.J.; Zhang, H.; Armand, M.; Taylor, R.H.; Dreizin, D.; Osgood, G.; Unberath, M. Fluorosam: A language-aligned foundation model for X-ray image segmentation. arXiv 2024, arXiv:2403.08059. [Google Scholar]
- Ma, J.; He, Y.; Li, F.; Han, L.; You, C.; Wang, B. Segment anything in medical images. Nat. Commun. 2024, 15, 654. [Google Scholar] [CrossRef]
- Cheng, J.; Ye, J.; Deng, Z.; Chen, J.; Li, T.; Wang, H.; Su, Y.; Huang, Z.; Chen, J.; Jiang, L.; et al. SAM-Med2D. arXiv 2023, arXiv:2308.16184. [Google Scholar] [CrossRef]
- Houlsby, N.; Giurgiu, A.; Jastrzebski, S.; Morrone, B.; De Laroussilhe, Q.; Gesmundo, A.; Attariyan, M.; Gelly, S. Parameter-efficient transfer learning for NLP. In Proceedings of the International Conference on Machine Learning, Long Beach, CA, USA, 10–15 June 2019; pp. 2790–2799. [Google Scholar]
- Zhang, K.; Liu, D. Customized segment anything model for medical image segmentation. arXiv 2023, arXiv:2304.13785. [Google Scholar] [CrossRef]
- Wu, J.; Wang, Z.; Hong, M.; Ji, W.; Fu, H.; Xu, Y.; Xu, M.; Jin, Y. Medical sam adapter: Adapting segment anything model for medical image segmentation. Med. Image Anal. 2025, 102, 103547. [Google Scholar] [CrossRef]
- Alayrac, J.B.; Donahue, J.; Luc, P.; Miech, A.; Barr, I.; Hasson, Y.; Lenc, K.; Mensch, A.; Millican, K.; Reynolds, M.; et al. Flamingo: A visual language model for few-shot learning. Adv. Neural Inf. Process. Syst. 2022, 35, 23716–23736. [Google Scholar]
- Wang, X.; Zhang, X.; Cao, Y.; Wang, W.; Shen, C.; Huang, T. Seggpt: Segmenting everything in context. arXiv 2023, arXiv:2304.03284. [Google Scholar] [CrossRef]
- Hu, J.; Shang, Y.; Yang, Y.; Guo, X.; Peng, H.; Ma, T. Icl-sam: Synergizing in-context learning model and sam in medical image segmentation. Med. Imaging Deep. Learn. 2024, 250, 641–656. [Google Scholar]
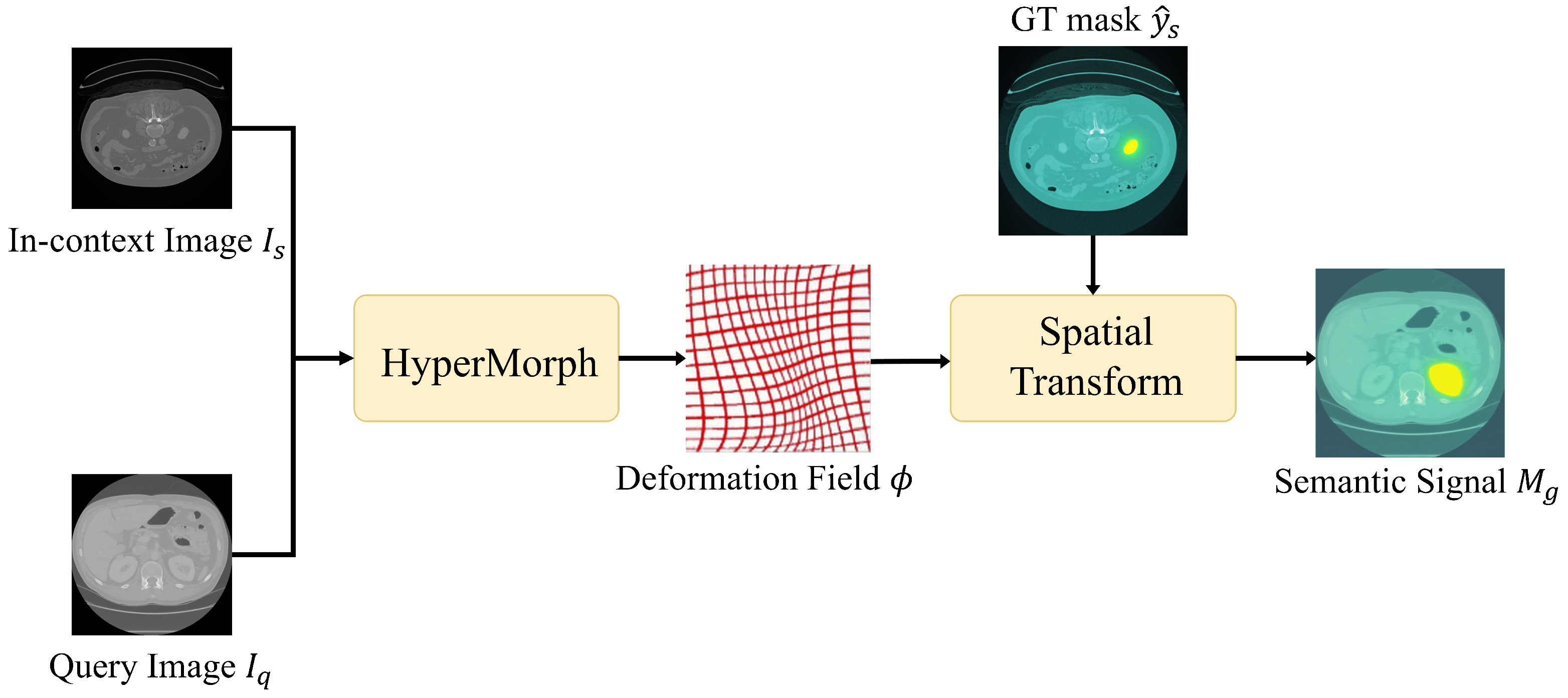
- Hoopes, A.; Hoffmann, M.; Fischl, B.; Guttag, J.; Dalca, A.V. Hypermorph: Amortized hyperparameter learning for image registration. In Proceedings of the International Conference on Information Processing in Medical Imaging, Virtual Event, 28–30 June 2021; pp. 3–17. [Google Scholar]
- Jaderberg, M.; Simonyan, K.; Zisserman, A.; Kavukcuoglu, K. Spatial transformer networks. In Proceedings of the NIPS’15: Proceedings of the 29th International Conference on Neural Information Processing Systems—Volume 2, Cambridge, MA, USA, 7–12 December 2015; Volume 28. [Google Scholar]
- Haralick, R.M.; Sternberg, S.R.; Zhuang, X. Image analysis using mathematical morphology. IEEE Trans. Pattern Anal. Mach. Intell. 1987, 4, 532–550. [Google Scholar] [CrossRef]
- Toussaint, G.T. Solving geometric problems with the rotating calipers. In Proceedings of the Proc. IEEE Melecon, Athens, Greece, 1–8 May 1983; Volume 83, p. A10. [Google Scholar]
- Orlando, J.I.; Fu, H.; Breda, J.B.; Van Keer, K.; Bathula, D.R.; Diaz-Pinto, A.; Fang, R.; Heng, P.A.; Kim, J.; Lee, J.; et al. Refuge challenge: A unified framework for evaluating automated methods for glaucoma assessment from fundus photographs. Med. Image Anal. 2020, 59, 101570. [Google Scholar] [CrossRef]
- Baid, U.; Ghodasara, S.; Mohan, S.; Bilello, M.; Calabrese, E.; Colak, E.; Farahani, K.; Kalpathy-Cramer, J.; Kitamura, F.C.; Pati, S.; et al. The rsna-asnr-miccai brats 2021 benchmark on brain tumor segmentation and radiogenomic classification. arXiv 2021, arXiv:2107.02314. [Google Scholar]
- Heller, N.; Isensee, F.; Trofimova, D.; Tejpaul, R.; Zhao, Z.; Chen, H.; Wang, L.; Golts, A.; Khapun, D.; Shats, D.; et al. The kits21 challenge: Automatic segmentation of kidneys, renal tumors, and renal cysts in corticomedullary-phase ct. arXiv 2023, arXiv:2307.01984. [Google Scholar]
- Yu, J.; Qin, J.; Xiang, J.; He, X.; Zhang, W.; Zhao, W. Trans-UNeter: A new Decoder of TransUNet for Medical Image Segmentation. In Proceedings of the IEEE International Conference on Bioinformatics and Biomedicine, Istanbul, Turkiye, 5–8 December 2023; pp. 2338–2341. [Google Scholar]
- Huang, S.; Liang, H.; Wang, Q.; Zhong, C.; Zhou, Z.; Shi, M. Seg-sam: Semantic-guided sam for unified medical image segmentation. arXiv 2024, arXiv:2412.12660. [Google Scholar]
- Choi, J.Y.; Yoo, T.K. Evaluating ChatGPT-4o for ophthalmic image interpretation: From in-context learning to code-free clinical tool generation. Inform. Health 2025, 2, 158–169. [Google Scholar] [CrossRef]
- Shi, L.; Li, X.; Hu, W.; Chen, H.; Chen, J.; Fan, Z.; Gao, M.; Jing, Y.; Lu, G.; Ma, D.; et al. EBHI-Seg: A novel enteroscope biopsy histopathological hematoxylin and eosin image dataset for image segmentation tasks. Front. Med. 2023, 10, 1114673. [Google Scholar] [CrossRef] [PubMed]
- Gamper, J.; Alemi Koohbanani, N.; Benet, K.; Khuram, A.; Rajpoot, N. Pannuke: An open pan-cancer histology dataset for nuclei instance segmentation and classification. In Proceedings of the European Congress on Digital Pathology, Warwick, UK, 10–13 April 2019; pp. 11–19. [Google Scholar]
- Da, Q.; Huang, X.; Li, Z.; Zuo, Y.; Zhang, C.; Liu, J.; Chen, W.; Li, J.; Xu, D.; Hu, Z.; et al. DigestPath: A benchmark dataset with challenge review for the pathological detection and segmentation of digestive-system. Med. Image Anal. 2022, 80, 102485. [Google Scholar] [CrossRef] [PubMed]
- Graham, S.; Chen, H.; Gamper, J.; Dou, Q.; Heng, P.A.; Snead, D.; Tsang, Y.W.; Rajpoot, N. MILD-Net: Minimal information loss dilated network for gland instance segmentation in colon histology images. Med. Image Anal. 2019, 52, 199–211. [Google Scholar] [CrossRef] [PubMed]



| Category | REFUGE | BraTS21 | kiTS23 |
|---|---|---|---|
| Segmentation task | Glaucoma | Brain tumor | Renal tumor |
| Imaging modality | 2D color fundus | 3D MP-MRI | 3D CE-CT |
| Target object | Optic disc/cup | Tumor subregion | Kidney, tumor, cyst |
| Sample number | 1200 | 4500 | 599 |
| Dataset | Method | Number of In-Context Samples | ||||
| 1 | 2 | 4 | 8 | 16 | ||
| REFUGE [41] | SegGPT [35] | 31.82 | 38.55 | 43.19 | 47.88 | 51.52 |
| UniverSeg [3] | 57.97 | 70.80 | 75.29 | 78.96 | 81.48 | |
| Neuralizer [10] | 67.74 | 71.09 | 73.14 | 73.25 | 75.31 | |
| ICL-SAM [36] | 73.91 | 78.89 | 80.57 | 82.64 | 83.91 | |
| Ours | 77.75 | 80.97 | 82.69 | 84.85 | 85.93 | |
| BraTS21 [42] | SegGPT [35] | 10.15 | 14.67 | 21.30 | 27.76 | 34.95 |
| UniverSeg [3] | 20.78 | 30.59 | 47.06 | 57.80 | 67.04 | |
| Neuralizer [10] | 21.61 | 22.84 | 25.40 | 25.62 | 32.02 | |
| ICL-SAM [36] | 33.87 | 45.84 | 58.79 | 65.79 | 71.91 | |
| Ours | 36.92 | 48.58 | 61.52 | 68.29 | 74.05 | |
| KiTS23 [43] | SegGPT [35] | 25.61 | 34.02 | 43.88 | 51.74 | 58.26 |
| UniverSeg [3] | 44.87 | 56.71 | 71.79 | 78.38 | 83.40 | |
| Neuralizer [10] | 37.15 | 52.78 | 55.10 | 63.84 | 65.15 | |
| ICL-SAM [36] | 64.55 | 71.81 | 80.65 | 84.39 | 86.29 | |
| Ours | 68.19 | 75.18 | 85.45 | 87.78 | 89.72 | |
| Method | REFUGE | BraTS21 | KiTS23 |
|---|---|---|---|
| in-context selection | 82.87 | 72.13 | 84.93 |
| semantic guidance | 82.28 | 70.47 | 84.52 |
| multi-granularity prompt | 83.81 | 72.21 | 86.37 |
| mask fusion | 84.42 | 72.38 | 87.08 |
| VG-SAM (Ours) | 85.93 | 74.05 | 89.72 |
| Method | REFUGE | BraTS21 | KiTS23 |
|---|---|---|---|
| Ours with SAM [13] | 79.69 | 68.26 | 84.83 |
| Ours with MedSAM [29] | 83.17 | 70.65 | 83.39 |
| Ours with SAM-Med2D [30] | 84.26 | 72.14 | 87.37 |
| Ours with SA-SAM [45] | 85.93 | 74.05 | 89.72 |
| Dataset | Method | Preference (100%) | DSC |
|---|---|---|---|
| KiTS23 [43] | HyperMorph | 21.95 | 86.58 |
| Ours (VG-SAM) | 78.05 | 89.72 |
| Method | Inference Time (s) | Peak Memory (MB) | DSC | ||
|---|---|---|---|---|---|
| Retrieval | Segmentation | Total | |||
| UniverSeg [3] | - | 0.020 | 0.020 | 2997 | 83.40 |
| ICL-SAM [36] | - | 0.167 | 0.167 | 4515 | 86.29 |
| Ours (VG-SAM) | 0.102 | 0.095 | 0.197 | 4115 | 89.72 |
| Segmentation Cost | Resolution of Query Images | ||
|---|---|---|---|
| Time (s) | 0.071 | 0.095 | 0.406 |
| GPU Memory (MB) | 4105 | 4115 | 4133 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dai, G.; Wang, Q.; Qin, Y.; Wei, G.; Huang, S. VG-SAM: Visual In-Context Guided SAM for Universal Medical Image Segmentation. Fractal Fract. 2025, 9, 722. https://doi.org/10.3390/fractalfract9110722
Dai G, Wang Q, Qin Y, Wei G, Huang S. VG-SAM: Visual In-Context Guided SAM for Universal Medical Image Segmentation. Fractal and Fractional. 2025; 9(11):722. https://doi.org/10.3390/fractalfract9110722
Chicago/Turabian StyleDai, Gang, Qingfeng Wang, Yutao Qin, Gang Wei, and Shuangping Huang. 2025. "VG-SAM: Visual In-Context Guided SAM for Universal Medical Image Segmentation" Fractal and Fractional 9, no. 11: 722. https://doi.org/10.3390/fractalfract9110722
APA StyleDai, G., Wang, Q., Qin, Y., Wei, G., & Huang, S. (2025). VG-SAM: Visual In-Context Guided SAM for Universal Medical Image Segmentation. Fractal and Fractional, 9(11), 722. https://doi.org/10.3390/fractalfract9110722









