Anthropometric Measurements for Predicting Low Appendicular Lean Mass Index for the Diagnosis of Sarcopenia: A Machine Learning Model
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Participants
2.2. Instruments
2.2.1. Body Composition Assessment Using DXA
- Participant preparation: participants were asked to avoid intense physical activity.
- Positioning: participants were set in a supine position on the scanning table, free of external metal objects. The limbs were positioned alongside the body, palms facing downward, feet in a neutral or slightly inward position, and face facing upward in a neutral position.
- Post-processing: scans were performed by trained personnel following the manufacturer’s instructions [38].
2.2.2. Anthropometry
- Acromiale: upper border of the most lateral part of the acromion.
- Radiale: proximal–lateral border of the radial head.
- Mid acromiale–radiale: midpoint between the acromiale and radiale marks.
- Triceps skinfold: posterior midline of the upper arm, aligned with the mid acromiale–radiale point.
- Calf skinfold: medial aspect of the leg at its maximum girth.
- Body mass: measured using a calibrated digital scale, with the participants in anthropometric position (standing, feet shoulder-width apart, arms relaxed at the sides).
- Stretch stature: measured with a stadiometer, participants barefoot and upright, feet together, head in the Frankfurt plane.
- Triceps skinfold (TSF): taken with a skinfold caliper, vertically at the mid acromiale–radiale point, parallel to the arm’s longitudinal axis.
- Calf skinfold (CSF): participant standing with the right foot on the anthropometric box and right knee flexed at 90°. The skinfold was measured vertically at the designated point on the medial leg.
- Arm Relaxed Girth (ARG): measured at the mid acromiale–radiale point with a non-elastic tape, arm hanging naturally.
- Arm Flexed and Tensed Girth (AFTG): arm flexed at 90° in front of the body. Participants were asked to contract the biceps maximally, and the girth was measured at the point of greatest muscle prominence.
- Forearm Girth (FG): with the forearm slightly flexed and in a supine position, the maximum girth was measured.
- Calf Girth (CG): with the participants standing on the anthropometric box, measured at the level of the leg skinfold point.
2.3. Procedures
2.4. Data Analysis
2.5. Ethical and Biosafety Considerations
3. Results
3.1. General and Descriptive Result
Sarcopenia
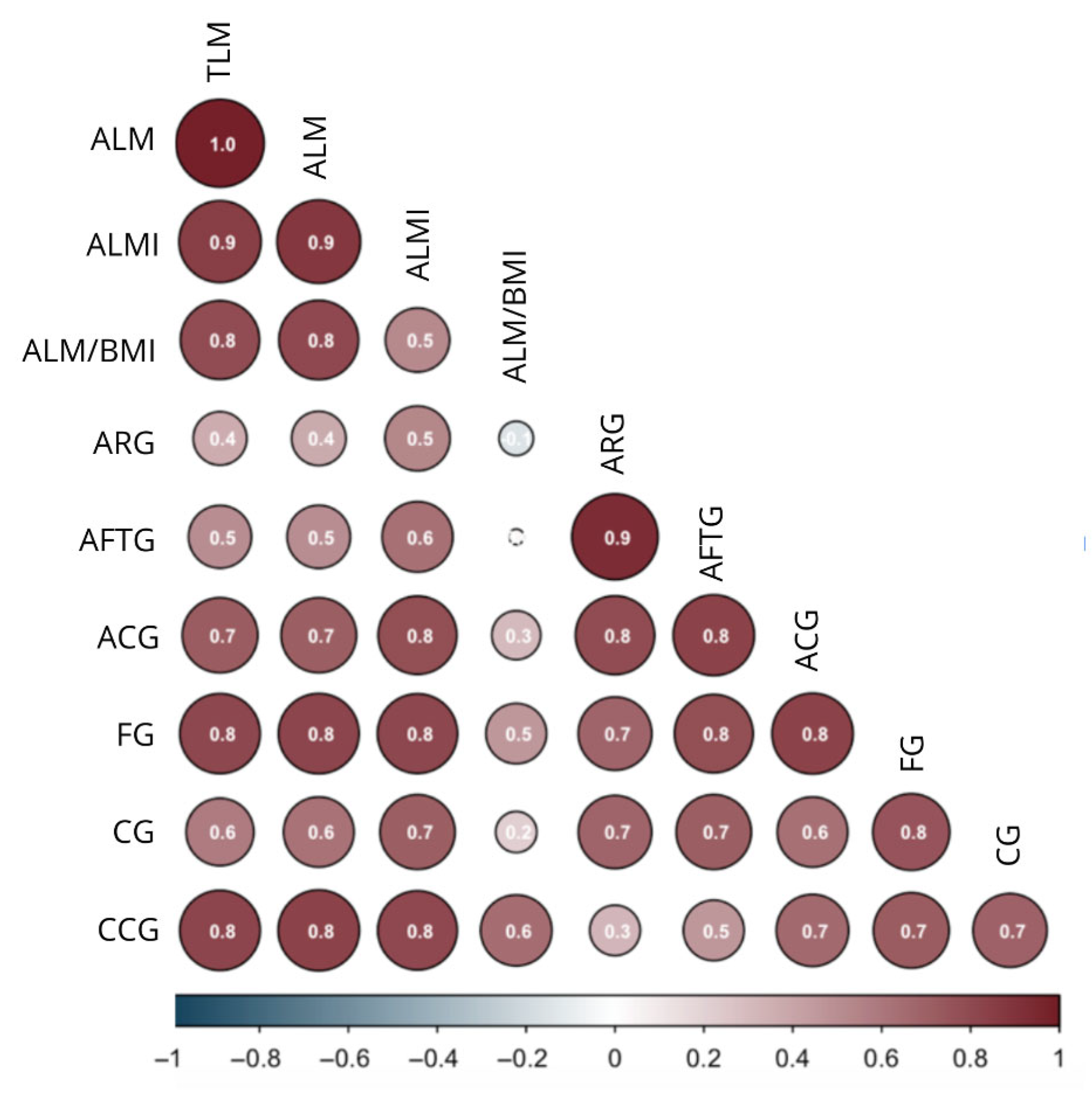
3.2. Body Composition Values Related to Anthropometry
3.3. Predictive Models for Appendicular Lean Mass Index (ALMI) Using Anthropometry
3.3.1. Logistic Regression Models
3.3.2. Decision Trees
| Female | Male | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Node | split | n | loss | yval | (yprob) | Node | split | n | loss | yval | (yprob) |
| (1) | root | 87 | 18 | 1 | (0.793–0.207) | (1) | root | 42 | 8 | 1 | (0.809–0.191) |
| (2) | BMI ≥ 25.83 | 61 | 1 | 1 | (0.984–0.016) ☨ | (2) | ARG ≥ 28.75 | 30 | 0 | 1 | (0.000–0.000) ☨ |
| (3) | BMI < 25.83 | 26 | 9 | 2 | (0.346–0.333) | (3) | ARG < 28.75 | 12 | 4 | 2 | (0.333–0.666) ☨ |
| (4) | CCG ≤ 26.89 | 12 | 4 | 1 | (0.666–0.333) ☨ | ||||||
| (5) | CCG < 26.89 | 14 | 1 | 2 | (0.071–0.929) ☨ | ||||||
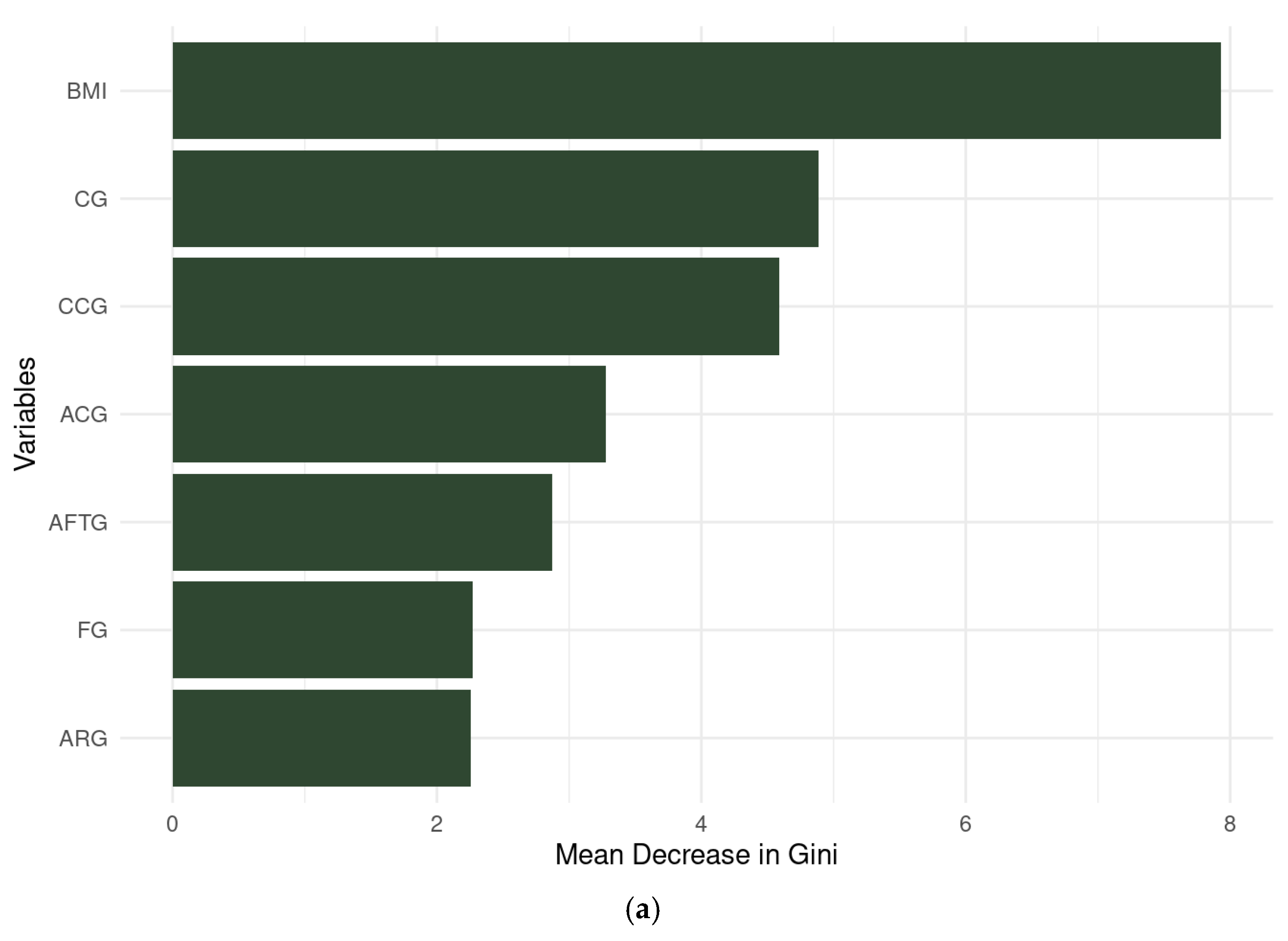
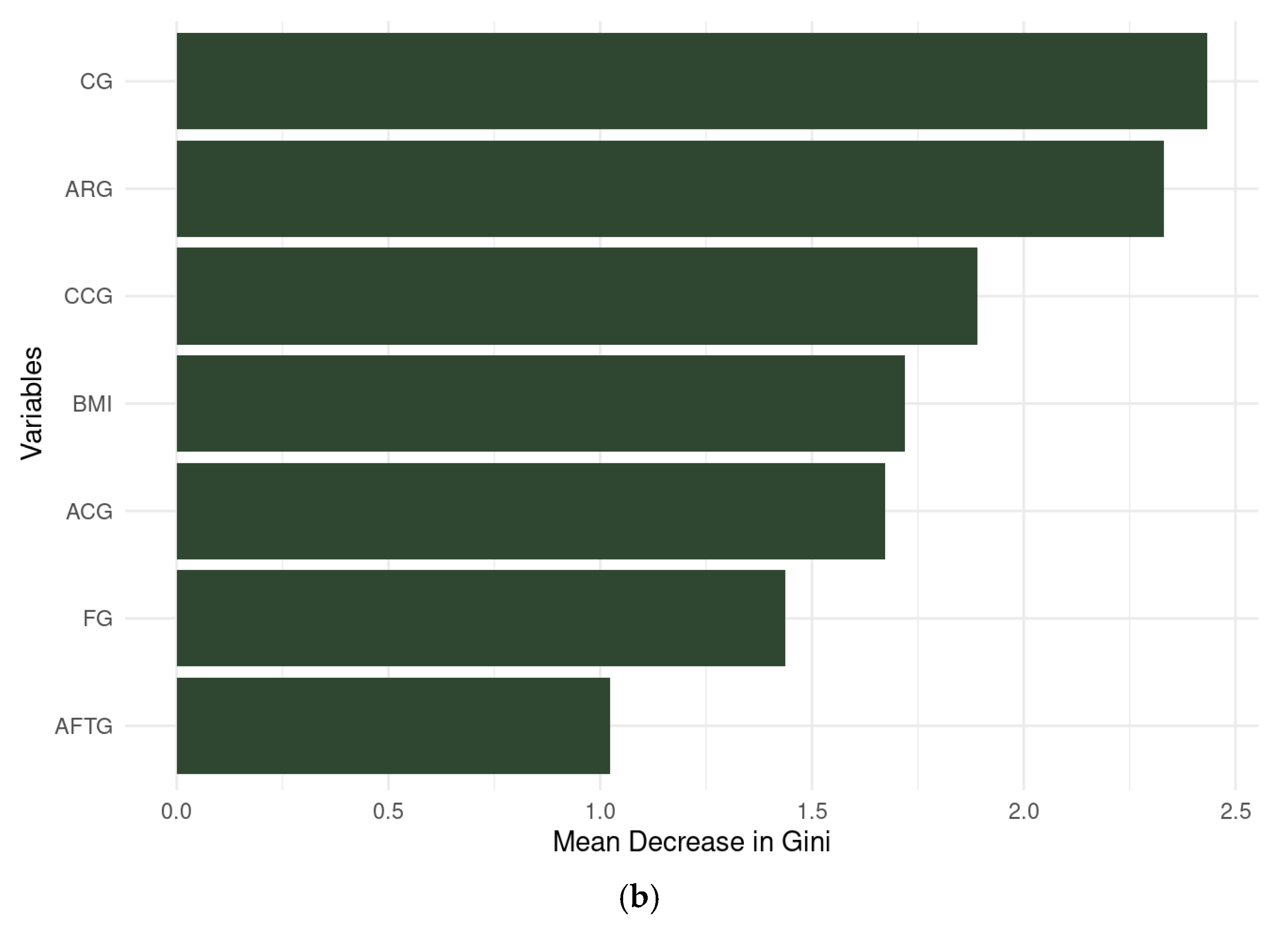
3.3.3. Random Forest
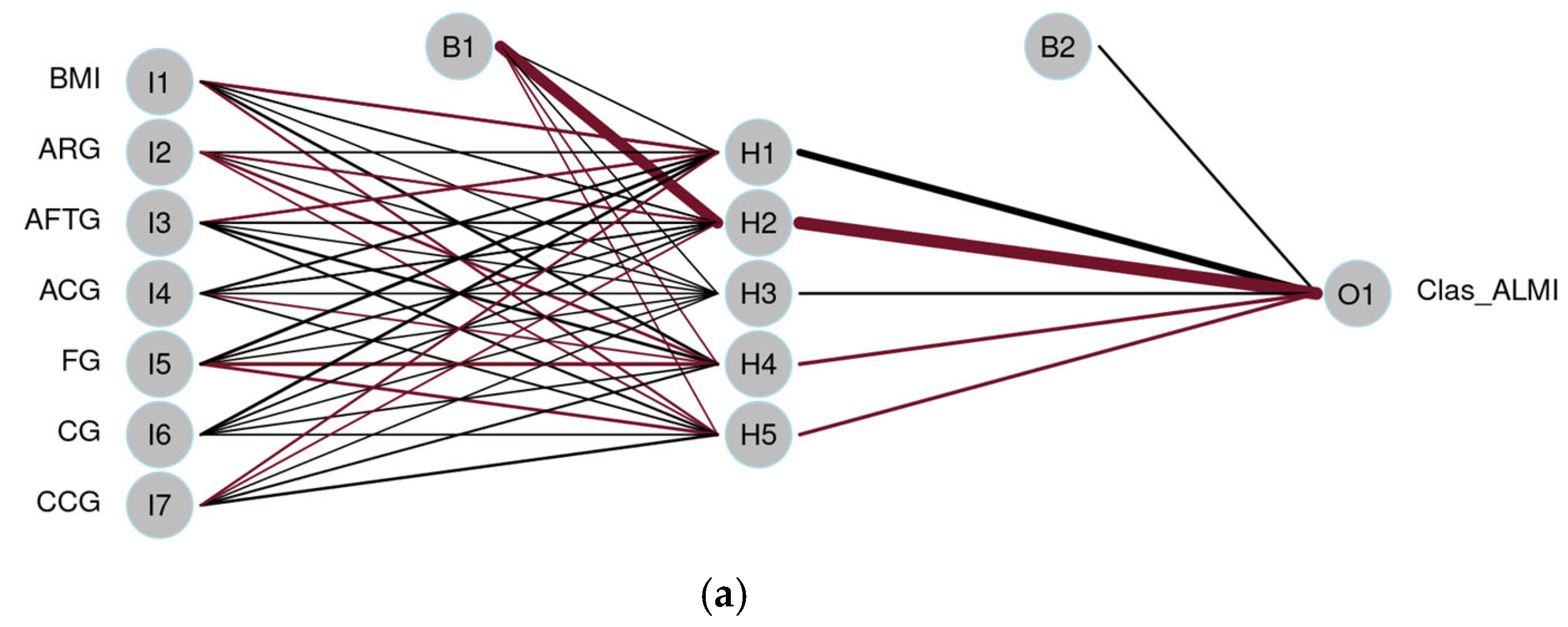
3.3.4. Artificial Neural Network
3.3.5. LASSO Regression
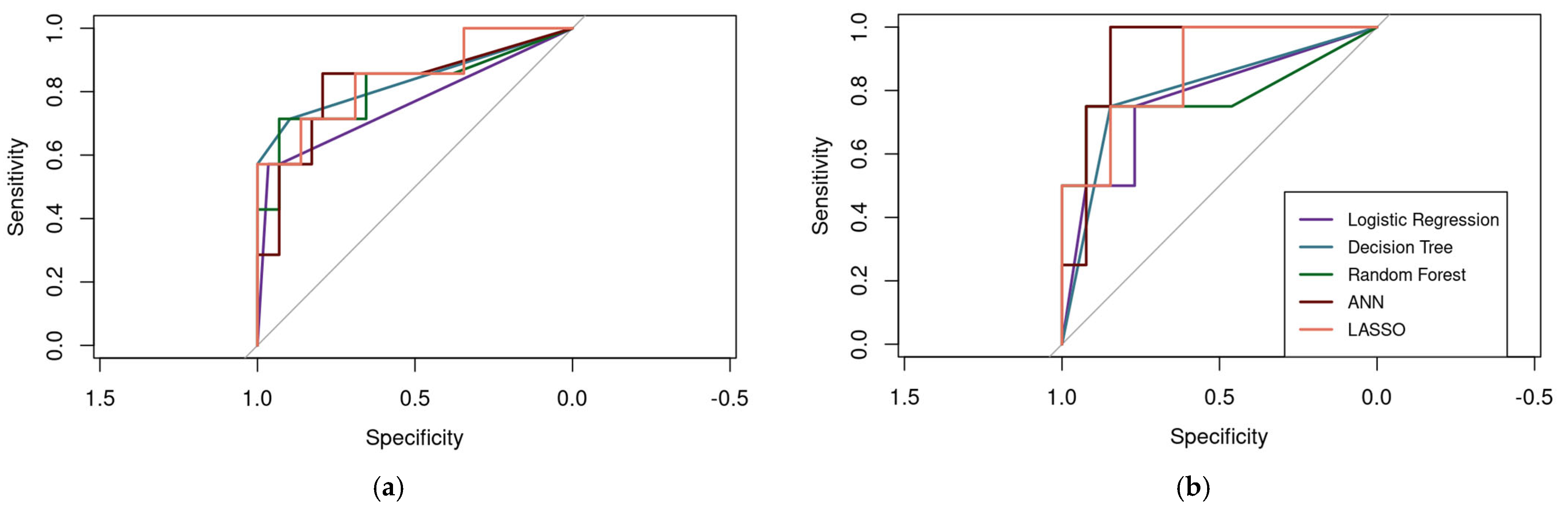
3.3.6. Performance Metrics and Model Comparison
| Female | Male | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| LR | DT | RF | ANN | LASSO | LR | DT | RF | ANN | LASSO | |
| Accuracy | 0.86 | 0.92 | 0.86 | 0.86 | 0.89 | 0.77 | 0.82 | 0.88 | 0.88 | 0.82 |
| Sensitivity | 0.57 | 0.57 | 0.57 | 0.57 | 0.57 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 |
| Specificity | 0.93 | 1.00 | 0.93 | 0.93 | 0.97 | 0.77 | 0.85 | 0.92 | 0.85 | 0.85 |
| Precision | 0.67 | 1.00 | 0.67 | 0.67 | 0.80 | 0.50 | 0.60 | 0.75 | 0.60 | 0.60 |
| AUC | 0.76 | 0.84 | 0.82 | 0.82 | 0.84 | 0.77 | 0.80 | 0.79 | 0.92 | 0.87 |
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ACG | Arm Corrected Girth |
| AFTG | Arm Flexed and Tensed Girth |
| AI | Artificial Intelligence |
| ALM | Appendicular Lean Mass |
| ALMI | Appendicular Lean Mass Index |
| ANN | Artificial Neural Network |
| ARG | Arm Relaxed Girth |
| AUC | ROC curve |
| AWGS | Asian Working Group for Sarcopenia |
| BAs | Boosted algorithms |
| BIA | Bioelectrical Impedance Analysis |
| BMI | Body Mass Index |
| BUN | Blood Urea Nitrogen |
| CAIAM | Centro de Atención Integral al Adulto Mayor |
| CCG | Calf Corrected Girth |
| CG | Calf Girth |
| CoGs | Corrected Girths |
| CoG | Corrected Girth |
| CSF | Calf Skinfold |
| CUTonalá | University of Guadalajara Tonalá Campus |
| DIF | Sistema Nacional para el Desarrollo Integral de la Familia |
| DT | Decision Trees |
| DXA | Dual-Energy X-ray Absorptiometry |
| EWGSOP | European Working Group on Sarcopenia in Older People |
| F | Female |
| FG | Forearm Girth |
| IQR | Interquartile Range |
| ISAK | International Society for the Advancement of Kinanthropometry |
| LASSO | Least Absolute Shrinkage and Selection Operator |
| LECEN | Laboratorio de Evaluación y Cuidado del Estado Nutricio |
| LR | Logistic Regression |
| M | Male |
| ML | Machine Learning |
| MS | Muscle Strength |
| n | number of cases |
| RBCs | Red Blood Cells |
| RFs | Random Forests |
| SAMJ | Sarcopenia in Older Adults from Jalisco |
| SPPB | Short Physical Performance Battery |
| SVMs | Support Vector Machines |
| TSF | Triceps Skinfold |
| WBCs | White Blood Cells |
| WHR | Waist-to-Hip Ratio |
Appendix A
Appendix A.1
| Variables | n (SD) | Female n (%) | Male n (%) | |
|---|---|---|---|---|
| Sex | 183 | 123 (67.2) | 60 (32.8) | |
| Age (years) | 70 (6.37) | 70.1 (6.28) | 70.1 (6.41) | |
| Monthly income (MNX) | 7023.1 (7974.0) | 6181.9 (5441.3) | 7028.3 (8033.0) | |
| Marital status | Single | 23 (12.6) | 20 (16.3) | 3 (5.0) |
| Married | 75 (41.0) | 33 (26.8) | 42 (70.0) | |
| Domestic partnership | 5 (2.7) | 5 (4.1) | 0 (0) | |
| Widow | 64 (35.0) | 54 (43.9) | 10 (16.7) | |
| Separated | 7 (3.8) | 5 (4.1) | 2 (3.3) | |
| Divorced | 9 (4.9) | 6 (4.9) | 3 (5.0) | |
| Living arrangement | Alone | 37 (20.2) | 27 (22.0) | 10 (16.7) |
| Family | 90 (49.2) | 76 (61.8) | 14 (23.3) | |
| Partner and family | 34 (18.6) | 12 (9.8) | 22 (36.7) | |
| Partner | 21 (11.5) | 7 (5.7) | 14 (23.3) | |
| Friend | 1 (0.5) | 1 (0.8) | 0 (0) | |
| Smoker | Yes | 18 (9.8) | 10 (8.1) | 8 (13.3) |
| No | 165 (90.2) | 113 (91.9) | 52 (86.7) | |
| Alcohol consumption | Yes | 28 (15.3) | 14 (11.4) | 14 (23.3) |
| No | 155 (84.7) | 109 (88.6) | 46 (76.7) | |
| Insulin resistance | Yes | 26 (14.2) | 16 (13.0) | 10 (16.7) |
| No | 157 (85.8) | 107 (87) | 50 (83.3) | |
| Diagnosed depression | Yes | 35 (19.1) | 30 (24.4) | 5 (8.3) |
| No | 148 (80.9) | 93 (75.6) | 55 (91.7) |
References
- Soto-Perez-de-Celis, E.; de Glas, N.A.; Hsu, T.; Kanesvaran, R.; Steer, C.; Navarrete-Reyes, A.P.; Battisti, N.M.L.; Chavarri-Guerra, Y.; O’Donovan, A.; Avila-Funes, J.A.; et al. Global geriatric oncology: Achievements and challenges. J. Geriatr. Oncol. 2017, 8, 374–386. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Decade of Healthy Ageing: Plan of Action 2021-2030; World Health Organization: Geneva, Switzerland, 2025. [Google Scholar]
- De Cabo, R.; Le Couteur, D. Kasper, D., Fauci, A., Hauser, S., Longo, D., Jameson, J., Loscalzo, J., Eds.; The biology of aging. In Harrison’s Principles of Internal Medicine; McGraw-Hill Education: New York, NY, USA, 2015; pp. 94e1–94e8. [Google Scholar]
- Manrique-Espinoza, B.; Salinas-Rodríguez, A.; Moreno-Tamayo, K.M.; Acosta-Castillo, I.; Sosa-Ortiz, A.L.; Gutiérrez-Robledo, L.M.; Téllez-Rojo, M.M. Condiciones de salud y estado funcional de los adultos mayores en México. Salud Public. Mex. 2013, 55, S323–S331. [Google Scholar] [CrossRef]
- Steffl, M.; Sima, J.; Shiells, K.; Holmerova, I. The increase in health care costs associated with muscle weakness in older people without long-term illnesses in the Czech Republic: Results from the Survey of Health, Ageing and Retirement in Europe (SHARE). Clin. Int. Aging 2017, 12, 2003–2007. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Jentoft, A.J.; Bahat, G.; Bauer, J.; Boirie, Y.; Bruyère, O.; Cederholm, T.; Cooper, C.; Landi, F.; Rolland, Y.; Sayer, A.A.; et al. Sarcopenia: Revised European consensus on definition and diagnosis. Age Ageing 2019, 48, 16–31. [Google Scholar] [CrossRef]
- Musumeci, G. Sarcopenia and Exercise “The State of the Art”. J. Funct. Morphol. Kinesiol. 2017, 2, 40. [Google Scholar] [CrossRef]
- Cruz-Jentoft, A.J.; Baeyens, J.P.; Bauer, J.M.; Boirie, Y.; Cederholm, T.; Landi, F.; Martin, F.C.; Michel, J.P.; Rolland, Y.; Schneider, S.M.; et al. Sarcopenia: European consensus on definition and diagnosis: Report of the European Working Group on Sarcopenia in Older People. Age Ageing 2010, 39, 412–423. [Google Scholar] [CrossRef]
- Chen, L.-K.; Woo, J.; Assantachai, P.; Auyeung, T.-W.; Chou, M.-Y.; Iijima, K.; Jang, H.C.; Kang, L.; Kim, M.; Kim, S.; et al. Asian Working Group for Sarcopenia: 2019 consensus update on sarcopenia diagnosis and treatment. JAMDA 2020, 21, 300–307.e2. [Google Scholar] [CrossRef]
- Morley, J.E.; Vellas, B.; Abellan van Kan, G.; Anker, S.D.; Bauer, J.M.; Bernabei, R.; Cesari, M.; Chumlea, W.C.; Doehner, W.; Evans, J.; et al. Frailty consensus: A call to action. JAMDA 2013, 14, 392–397. [Google Scholar] [CrossRef]
- Bauer, J.; Morley, J.E.; Schols, A.M.W.J.; Ferrucci, L.; Cruz-Jentoft, A.J.; Dent, E.; Baracos, V.E.; Crawford, J.A.; Doehner, W.; Heymsfield, S.B.; et al. Sarcopenia: A time for action. An SCWD position paper. J. Cachexia Sarcopenia Muscle 2019, 10, 956–961. [Google Scholar] [CrossRef]
- Bhasin, S.; Travison, T.G.; Manini, T.M.; Patel, S.; Pencina, K.M.; Fielding, R.A.; Magaziner, J.M.; Newman, A.B.; Kiel, D.P.; Cooper, C.; et al. Sarcopenia definition: The position statements of the sarcopenia definition and outcomes consortium. JAGS 2020, 68, 1410–1418. [Google Scholar] [CrossRef]
- Suzan, V.; Suzan, A.A. A bibliometric analysis of sarcopenia: Top 100 articles. Eur. Geriatr. Med. 2021, 12, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Kemmler, W.; von Stengel, S.; Kohl, M. Developing sarcopenia criteria and cutoffs for an older caucasian cohort—A strictly biometrical approach; a strictly biometrical approach. Clin. Int. Aging 2018, 13, 1365–1373. [Google Scholar] [CrossRef]
- Woo, J.; Leung, J. Anthropometric cut points for definition of sarcopenia based on incident mobility and physical limitation in older chinese people. J. Gerontol. A Biol. Sci. Med. Sci. 2016, 71, 935–940. [Google Scholar] [CrossRef][Green Version]
- Calvani, R.; Picca, A.; Marini, F.; Biancolillo, A.; Cesari, M.; Pesce, V.; Lezza, A.M.S.; Bossola, M.; Leeuwenburgh, C.; Bernabei, R.; et al. The “BIOmarkers associated with sarcopenia and physical frailty in elderly persons” (BIOSPHERE) study: Rationale, design and methods. Eur. J. Intern. Med. 2018, 56, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Confortin, S.C.; Ono, L.M.; Barbosa, A.R.; d’Orsi, E. Sarcopenia e sua associação com mudanças nos fatores socioeconômicos, comportamentais e de saúde: Estudo EpiFloripa Idoso. Cad. Saúde Pública 2018, 34. [Google Scholar] [CrossRef]
- Espinel-Bermúdez, M.C.; Sánchez-García, S.; García Peña, C.; Trujillo, X.; Huerta-Viera, M.; Granados-García, V.; Hernández-González, S.; Arias-Merino, E.D. Factores asociados a sarcopenia en adultos mayores mexicanos: Encuesta nacional de salud y nutrición 2012. Rev. Med. Inst. Mex. Seguro Soc. 2018, 56, 46–53. [Google Scholar]
- Martín Sierra, J.A.; Calderón Loeza, G.Y.; Zapata Vázquez, R.E.; Novelo Tec, J.F. Sarcopenia y factores asociados en los adultos mayores de una unidad de medicina familiar en Yucatán, México. Atención Fam. 2021, 28, 191. [Google Scholar] [CrossRef]
- Palus, S.; Springer, J.I.; Doehner, W.; von Haehling, S.; Anker, M.; Anker, S.D.; Springer, J. Models of sarcopenia: Short review. Int. J. Cardiol. 2017, 238, 19–21. [Google Scholar] [CrossRef]
- Alemán-Mateo, H.; Lee, S.Y.; Javed, F.; Thornton, J.; Heymsfield, S.B.; Pierson, R.N.; Pi-Sunyer, F.X.; Wang, Z.M.; Wang, J.; Gallagher, D. Elderly Mexicans have less muscle and greater total and truncal fat compared to African-Americans and Caucasians with the same BMI. J. Nutr. Health Aging 2009, 13, 919–923. [Google Scholar] [CrossRef]
- Alemán-Mateo, H.; López-Teros, M.T.; Ruiz-Valenzuela, R.E.; Ramírez-Torres, M.; Urquidez-Romero, R. Sarcopenia: Influence of regional skeletal muscle cutoff points and fat-free mass in older mexican people—A pilot study. Curr. Gerontol. Geriatr. 2020, 8037503, 1–9. [Google Scholar] [CrossRef]
- Alley, D.E.; Shardell, M.D.; Peters, K.W.; McLean, R.R.; Dam, T.-T.L.; Kenny, A.M.; Fragala, M.S.; Harris, T.B.; Kiel, D.P.; Guralnik, J.M.; et al. Grip strength cutpoints for the identification of clinically relevant weakness. J. Gerontol. A Biol. Sci. Med. Sci. 2014, 69, 559–566. [Google Scholar] [CrossRef]
- Pinheiro, P.A.; da Silva Coqueiro, R.; Carneiro, J.A.O.; Correia, T.M.L.; Pereira, R.; Fernandes, M.H. Anthropometric indicators as screening tools for sarcopenia in older adult women. Enferm 2020, 30, 269–274. [Google Scholar]
- Rech, C.R.; Dellagrana, R.A.; Marucci, M.D.F.N.; Petroski, E.L. Validade de equações antropométricas para estimar a massa muscular em idosos. Rev. Bras. Cineantropometria Desempenho Hum. 2012, 14, 23–31. [Google Scholar]
- Rangel Peniche, D.B.; Alemán Mateo, H.; de los Angeles Aguilera Barreiro, M.; Ruiz Valenzuela, R.E.; Ramírez-Torres, M.; Urquidez-Romero, R. Differences in body composition in older people from two regions of Mexico: Implications for diagnoses of sarcopenia and sarcopenic obesity. Biomed. Res. Int. 2018, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Greener, J.G.; Kandathil, S.M.; Moffat, L.; Jones, D.T. A guide to machine learning for biologists. Nat. Rev. Mol. Cell Biol. 2022, 23, 40–55. [Google Scholar] [CrossRef]
- Panch, T.; Szolovits, P.; Atun, R. Artificial intelligence, machine learning and health systems. J. Glob. Health 2018, 8, 020303. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Z.; Zhang, J.; Wang, C.; Wang, Y.; Chen, H.; Shan, L.; Huo, J.; Gu, J.; Ma, X. Deep learning model for classifying endometrial lesions. J. Transl. Med. 2021, 19, 10. [Google Scholar] [CrossRef]
- Ou Yang, W.-Y.; Lai, C.-C.; Tsou, M.-T.; Hwang, L.-C. Development of machine learning models for prediction of osteoporosis from clinical health examination data. Int. J. Environ. Res. Public. Health 2021, 18, 7635. [Google Scholar] [CrossRef]
- Albores-Mendez, E.M.; Aguilera Hernández, A.D.; Melo-González, A.; Vargas-Hernández, M.A.; Gutierrez de la Cruz, N.; Vazquez-Guzman, M.A.; Castro-Marín, M.; Romero-Morelos, P.; Winkler, R. A diagnostic model for overweight and obesity from untargeted urine metabolomics of soldiers. PeerJ 2022, 10, e13754. [Google Scholar] [CrossRef]
- Butler, É.M.; Derraik, J.G.B.; Taylor, R.W.; Cutfield, W.S. Prediction models for early childhood obesity: Applicability and existing issues. Horm. Res. Paediatr. 2018, 90, 358–367. [Google Scholar] [CrossRef]
- Chung, H.; Jo, Y.; Ryu, D.; Jeong, C.; Choe, S.; Lee, J. Artificial-intelligence-driven discovery of prognostic biomarker for sarcopenia. J. Cachexia Sarcopenia Muscle 2021, 12, 2220–2230. [Google Scholar] [CrossRef] [PubMed]
- Cook, N.R. Statistical evaluation of prognostic versus diagnostic models: Beyond the ROC curve. Clin. Chem. 2008, 54, 17–23. [Google Scholar] [CrossRef]
- Hendriksen, J.M.T.; Geersing, G.J.; Moons, K.G.M.; de Groot, J.A.H. Diagnostic and prognostic prediction models. J. Thromb. Haemost. 2013, 11, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Wolf, M.B.; DeLand, E.C. A comprehensive, computer-model-based approach for diagnosis and treatment of complex acid–base disorders in critically-ill patients. J. Clin. Monit. Comput. 2011, 25, 353–364. [Google Scholar] [CrossRef]
- von Elm, E.; Altman, D.G.; Egger, M.; Pocock, S.J.; Gøtzsche, P.C.; Vandenbroucke, J.P. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: Guidelines for reporting observational studies. Lancet 2007, 370, 1453–1457. [Google Scholar] [CrossRef]
- Messina, C.; Albano, D.; Gitto, S.; Tofanelli, L.; Bazzocchi, A.; Ulivieri, F.M.; Guglielmi, G.; Sconfienza, L.M. Body composition with dual energy X-ray absorptiometry: From basics to new tools. Quant. Imaging Med. Surg. 2020, 10, 1687–1698. [Google Scholar] [CrossRef]
- Esparza-Ros, F.; Vaquero-Cristóbal, R.; Marfell-Jones, M. International Standards for Anthropometric Assessment; International Society for the Advancement of Kinanthropometry (ISAK): Glasgow, Scotland, 2019. [Google Scholar]
- Barraza Gómez, F.; Hadler Garcés, A.; Jeria Fernández, J.; Riffo Esquivel, C. Cuantificación de la masa muscular de los miembros apendiculares, por medio ecuaciones antropométricas. J. Mov. Health. 2009, 10, 44–48. [Google Scholar] [CrossRef]
- Herrera-Amante, C.A.; Ramos-García, C.O.; Alacid, F.; Quiroga-Morales, L.A.; Martínez-Rubio, A.J.; Bonilla, D.A. Development of alternatives to estimate resting metabolic rate from anthropometric variables in paralympic swimmers. J. Sports Sci. 2021, 39, 2133–2143. [Google Scholar] [CrossRef]
- Bonilla, D.A.; De León, L.G.; Alexander-Cortez, P.; Odriozola-Martínez, A.; Herrera-Amante, C.A.; Vargas-Molina, S.; Petro, J.L. Simple anthropometry-based calculations to monitor body composition in athletes: Scoping review and reference values. Nutr. Health 2022, 28, 95–109. [Google Scholar] [CrossRef]
- Ge, S.; Du, Q.; Feng, X.; Liu, Y.; Wang, H.; Hai, S.; Shi, X.; Sun, W.; Ma, A.; Lv, T.; et al. Optimal cutoffs for the diagnosis of sarcopenia in older Chinese adults. Front. Nutr. 2022, 9, 853323. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-P.; Joh, J.-Y.; Kim, S.; Hwang, H.-S.; Shin, I.-S. The application of different appendicular skeletal muscle cutoff points and research definitions associated with health-related quality of life in Korean older people: Data from KNHANES 2008–2011. BMC Geriatr. 2014, 14, 144. [Google Scholar] [CrossRef] [PubMed]
- López-Teros, M.T.; Rosas-Carrasco, O.; Sánchez-García, S.; Castro-Porras, L.; Luna-López, A.; Agudelo-Botero, M. The association of osteosarcopenia with functional disability in community-dwelling Mexican adults 50 and older. Front. Med. 2021, 8, 674724. [Google Scholar] [CrossRef]
- Viana, J.U.; Dias, J.M.D.; Pereira, L.S.M.; Silva, S.L.A.D.; Hoelzle, L.F.; Dias, R.C. Pontos de corte alternativos para massa muscular apendicular para verificação da sarcopenia em idosos brasileiros: Dados da Rede Fibra—Belo Horizonte/Brasil. Fisioter. E Pesqui. 2018, 25, 166–172. [Google Scholar] [CrossRef]
- World Medical Association. World medical association declaration of Helsinki. Ethical principles for medical research involving human subjects. JAMA 2025, 333, 71–74. [Google Scholar] [CrossRef] [PubMed]
- Mukaka, M.M. Statistics corner: A guide to appropriate use of correlation coefficient in medical research. Malawi Med. J. 2012, 24, 69–71. [Google Scholar]
- Secretaría de Salud. In Reglamento de la Ley General de Salud en Materia de Investigación para la Salud; Diario Oficial de la Federación: Mexico City, Mexico, 6 January 1987. Available online: https://www.diputados.gob.mx/LeyesBiblio/regley/Reg_LGS_MIS.pdf (accessed on 1 June 2025).
- Ranstam, J.; Cook, J.A. LASSO regression. Br. J. Surg. 2018, 105, 1348. [Google Scholar] [CrossRef]
- Shaikhina, T.; Lowe, D.; Daga, S.; Briggs, D.; Higgings, R.; Khovanova, N. Decision tree and random forest models for outcome prediction in antibody incompatible kidney transplantation. Biomed. Signal Process. Control. 2019, 52, 456–462. [Google Scholar] [CrossRef]
- Podgorelec, V.; Kokol, P.; Stiglic, B.; Rozman, I. Decision trees: An overview and their use in medicine. J. Med. Syst. 2002, 26, 445–463. [Google Scholar] [CrossRef]
- Horning, N. Random Forests: An algorithm for image classification and generation of continuous fields data sets. In Proceedings of the International Conference on Geoinformatics for Spatial Infrastructure Development in Earth and Allied Sciences, Osaka, Japan, 9–11 December 2010; Volume 911, pp. 1–6. [Google Scholar]
- Olshvang, D.; Harris, C.; Chellappa, R.; Santhanam, P. Predictive modeling of lean body mass, appendicular lean mass, and appendicular skeletal muscle mass using machine learning techniques: A comprehensive analysis utilizing NHANES data and the Look AHEAD study. PLoS ONE 2024, 19, e0309830. [Google Scholar] [CrossRef]
- Cichosz, S.L.; Rasmussen, N.H.; Vestergaard, P.; Hejlesen, O. Precise prediction of total body lean and fat mass from anthropometric and demographic data: Development and validation of neural network models. J. Diabetes Sci. Technol. 2021, 15, 1337–1343. [Google Scholar] [CrossRef] [PubMed]
- Buccheri, E.; Dell’Aquila, D.; Russo, M.; Chiaramonte, R.; Musumeci, G.; Vecchio, M. Can artificial intelligence simplify the screening of muscle mass loss? Heliyon 2023, 9, e16323. [Google Scholar] [CrossRef]
- Marazzato, F.; McCarthy, C.; Field, R.H.; Nguyen, H.; Nguyen, T.; Shepherd, J.A.; Tinsley, G.M.; Heymsfield, S.B. Advances in digital anthropometric body composition assessment: Neural network algorithm prediction of appendicular lean mass. Eur. J. Clin. Nutr. 2024, 78, 452–454. [Google Scholar] [CrossRef] [PubMed]
- Birk, N.; Kulkarni, B.; Bhogadi, S.; Aggarwal, A.; Walia, G.K.; Gupta, V.; Rani, U.; Mahajan, H.; Kinra, S.; Mallinson, P.A. Machine learning-based equations for improved body composition estimation in Indian adults. medRxiv 2024. [Google Scholar] [CrossRef]
- Guo, J.; He, Q.; She, C.; Liu, H.; Li, Y. A machine learning–based online web calculator to aid in the diagnosis of sarcopenia in the US community. Digit. Health 2024, 10, 1–14. [Google Scholar] [CrossRef]
- Buccheri, E.; Dell’Aquila, D.; Russo, M.; Chiaramonte, R.; Vecchio, M. Appendicular skeletal muscle mass in older adults can be estimated with a simple equation using a few zero-cost variables. J. Geriatr. Phys. Ther. 2024, 47, E149–E158. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; He, Q.; Pan, Y.; Zhang, X.; Li, M.; Chen, S. Estimation of appendicular skeletal muscle mass for women aged 60-70 years using a machine learning approach. J. Am. Med. Dir. Assoc. 2022, 23, 1985.e1–1985.e7. [Google Scholar] [CrossRef]
- Kang, Y.-J.; Yoo, J.-I.; Ha, Y. Sarcopenia feature selection and risk prediction using machine learning: A cross-sectional study. Medicine 2019, 98, e17699. [Google Scholar] [CrossRef]
- Du, J.; Tao, X.; Zhu, L.; Wang, H.; Qi, W.; Min, X.; Wei, S.; Zhang, X.; Liu, Q. Development of a visualized risk prediction system for sarcopenia in older adults using machine learning: A cohort study based on CHARLS. Front. Public Health 2025, 13, 1–8. [Google Scholar] [CrossRef]
- Li, N.; Ou, J.; He, H.; He, J.; Zhang, L.; Peng, Z.; Zhong, J.; Jiang, N. Exploration of a machine learning approach for diagnosing sarcopenia among Chinese community-dwelling older adults using sEMG-based data. J. Neuroeng. Rehabil. 2024, 21, 69. [Google Scholar] [CrossRef]
- Yang, S.; Yu, J.; Chen, Q.; Sun, X.; Hu, Y.; Su, T.; Li, J.; Jin, L. Development and external validation of a machine-learning based model to predict pre-sarcopenia in MASLD population: Results from NHANES 2017–2018. Ann. Hepatol. 2024, 30, 101585. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.; Zhang, X.; Sun, G.; Zeng, P.; Zheng, C.; Wang, K. Sarcopenia prediction model based on machine learning and SHAP values for community-based older adults with cardiovascular disease in China. Front. Public Health 2025, 13, 1527304. [Google Scholar] [CrossRef]
- Johnson, T.; Su, J.; Andres, J.; Henning, A.; Ren, J. Sex differences in fat distribution and muscle fat infiltration in the lower extremity: A Retrospective Diverse-Ethnicity 7T MRI Study in a Research Institute Setting in the USA. Diagnostics 2024, 14, 2260. [Google Scholar] [CrossRef]
- Fragala, M.S.; Clark, M.H.; Walsh, S.J.; Kleppinger, A.; Judge, J.O.; Kuchel, G.A.; Kenny, A.M. Gender differences in anthropometric predictors of physical performance in older adults. Gend. Med. 2012, 9, 445–456. [Google Scholar] [CrossRef]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic minority over-sampling technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Rajput, D.; Wang, W.J.; Chen, C.C. Evaluation of a decided sample size in machine learning applications. BMC Bioinform. 2023, 24, 48. [Google Scholar] [CrossRef] [PubMed]
- Vabalas, A.; Gowen, E.; Poliakoff, E.; Casson, A.J. Machine learning algorithm validation with a limited sample size. PLoS ONE 2019, 14, e0224365. [Google Scholar] [CrossRef] [PubMed]
- Goldenholz, D.M.; Sun, H.; Ganglberger, W.; Westover, M.B. Sample Size Analysis for Machine Learning Clinical Validation Studies. Biomedicines 2023, 11, 685. [Google Scholar] [CrossRef]
- Castillo-Olea, C.; Garcia-Zapirain Soto, B.; Zuñiga, C. Evaluation of prevalence of the sarcopenia level using machine learning techniques: Case study in Tijuana Baja California, Mexico. Int. J. Environ. Res. Public Health 2020, 17, 1917. [Google Scholar] [CrossRef]
- Pineda-Zuluaga, M.C.; González-Correa, C.H.; Sepulveda-Gallego, L.E. Cut-off points for low skeletal muscle mass in older adults: Colombia versus other populations. F1000Res 2022, 11, 304. [Google Scholar] [CrossRef]
- Abdalla, P.P.; Silva, A.M.; Venturini, A.C.R.; Santos, A.P.D.; Carvalho, A.D.S.; Siqueira, V.A.A.A.; Gomide, E.B.; Alves, N.M.D.C.; Ferriolli, E.; Machado, D.R.L. Cut-off points of appendicular lean soft tissue for identifying sarcopenia in older adults in Brazil: A cross-sectional study. Nutr. Hosp. 2020, 37, 306–312. [Google Scholar] [CrossRef]
- Alwosheel, A.; van Cranenburgh, S.; Chorus, C.G. Is your dataset big enough? Sample size requirements when using artificial neural networks for discrete choice analysis. J. Choice Model. 2018, 167–182. [Google Scholar] [CrossRef]
- Adão, T.; Lameira, G.; dos Santos, J.; Palha, F. Technical error of measurement in anthropometry. Rev. Bras. Med. Esporte. 2005, 11, 81–85. [Google Scholar]
- Esparza-Ros, F. Manual de Cineantropometría; Grupo Español De Cineantropometría: Zaragoza, España, 1993. [Google Scholar]
- Padilla, C.J.; Ferreyro, F.A.; Arnold, W.D. Anthropometry as a readily accessible health assessment of older adults. Exp. Gerontol. 2021, 153, 111464. [Google Scholar] [CrossRef] [PubMed]
- Turimov Mustapoevich, D.; Kim, W. Machine learning applications in sarcopenia detection and management: A comprehensive survey. Healthcare 2023, 11, 2483. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Torriani, M. Artificial intelligence in the evaluation of body composition. Semin. Musculoskelet. Radiol. 2020, 24, 030–037. [Google Scholar] [CrossRef] [PubMed]
- Ozgur, S.; Altinok, Y.A.; Bozkurt, D.; Saraç, Z.F.; Akçiçek, S.F. Performance evaluation of machine learning algorithms for sarcopenia diagnosis in older adults. Healthcare 2023, 11, 2699. [Google Scholar] [CrossRef]
- Hui, A.T.; Ahn, S.S.; Lye, C.T.; Deng, J. Ethical challenges of artificial intelligence in health care: A narrative review. EBEM 2021, 12, 55–71. [Google Scholar] [CrossRef]


| Male Mean (SD) | Female Mean (SD) | p-Value | |
|---|---|---|---|
| Body composition | |||
| ALMI (kg/m2) | 8.13 (0.94) | 6.66 (0.85) | <0.001 |
| Anthropometry | |||
| BMI (kg/m2) | 28.6 (3.85) | 28.99 (5.05) | 0.542 |
| ARG (cm) | 30.94 (3.24) | 30.84 (4.05) | <0.001 |
| AFTG (cm) | 31.62 (3.03) | 30.04 (3.98) | <0.001 |
| FG (cm) | 27.03 (1.99) | 24.08 (2.12) | <0.001 |
| CG (cm) | 36.27 (2.9) | 34.59 (3.47) | <0.001 |
| CoG arm (cm) | 26.94 (2.35) | 24.07 (2.64) | <0.001 |
| CoG calf (cm) | 36.27 (2.55) | 28.06 (2.5) | <0.001 |
| Body Composition | Male Median (IQR) | Female Median (IQR) | p-Value |
|---|---|---|---|
| ALM (kg) | 22.13 (3.7) | 11.68 (3.37) | <0.001 |
| ALM/BMI | 0.71 (0.13) | 0.53 (0.1) | <0.001 |
| Sex | ALMI (kg/m2) | ALM/BMI |
|---|---|---|
| Male | <7.49 | <0.69 |
| Female | <5.93 | <0.46 |
| Anthropometry | DXA Fat Mass (kg) | DXA Lean Mass (kg) | ||||||
|---|---|---|---|---|---|---|---|---|
| Appendicular r | Arms r | Legs r | Appendicular r | Arms (kg) r | Legs (kg) r | ALMI (kg/m2) r | ALM/BMI r | |
| Body mass (kg) | 0.496 ** | 0.474 ** | 0.479 ** | 0.802 ** | 0.744 ** | 0.809 ** | 0.798 ** | 0.317 ** |
| BMI (kg/m2) | 0.731 ** | 0.702 ** | 0.704 ** | 0.731 ** | 0.287 ** | 0.352 ** | 0.589 ** | −0.287 ** |
| TSF (mm) | 0.772 ** | 0.754 ** | 0.740 ** | −0.235 ** | −0.283 ** | −0.209 * | −0.065 | −0.607 ** |
| CSF (mm) | 0.721 ** | 0.613 ** | 0.718 ** | −0.299 ** | −0.373 ** | −0.269 ** | −0.159 * | −0.579 ** |
| ARG (cm) | 0.698 ** | 0.733 ** | 0.641 ** | 0.400 ** | 0.400 ** | 0.396 ** | 0.547 | −0.137 ** |
| AFTG (cm) | 0.574 ** | 0.654 ** | 0.522 ** | 0.527 ** | 0.529 ** | 0.515 ** | 0.646 ** | 0.037 |
| FG (cm) | 0.303 ** | 0.359 ** | 0.271 ** | 0.825 ** | 0.824 ** | 0.818 ** | 0.809 ** | 0.461** |
| CG (cm) | 0.549 ** | 0.444 ** | 0.554 ** | 0.630 ** | 0.540 ** | 0.655 ** | 0.701 ** | 0.202 * |
| Calf CoG (cm) | −0.562 | −0.080 | −0.049 | 0.831 ** | 0.805 ** | 0.825 ** | 0.786 ** | 0.650 ** |
| Arm CoG (cm) | 0.317 ** | 0.430 ** | 0.267 ** | 0.710 ** | 0.685 ** | 0.773 ** | 0.293 ** | 0.202 * |
| Estimate | Standard Error | Z Value | p Value | |||||
|---|---|---|---|---|---|---|---|---|
| F | M | F | M | F | M | F | M | |
| Intercept | 7376.80 | 1802.22 | 706,862.64 | 973,842.13 | 0.010 | 0.002 | 0.992 | 0.999 |
| BMI | −185.10 | −3.13 | 15,623.58 | 14,513.51 | −0.012 | 0.000 | 0.991 | 1.000 |
| ARG | 506.38 | −56.27 | 40,247.17 | 99,777.97 | 0.013 | −0.001 | 0.990 | 1.000 |
| AFTG | −331.97 | 24.36 | 28,252.76 | 44,860.02 | −0.012 | 0.001 | 0.991 | 1.000 |
| Arm CoG | −222.99 | 23.28 | 19,574.73 | 45,842.11 | −0.011 | 0.001 | 0.991 | 1.000 |
| FG | −48.82 | −21.73 | 6982.93 | 33,221.02 | −0.007 | −0.001 | 0.994 | 0.999 |
| CG | −48.11 | 19.88 | 10,491.04 | 26,609.56 | −0.005 | 0.001 | 0.996 | 0.999 |
| Calf CoG | −14.11 | −47.95 | 3338.89 | 29,310.10 | −0.004 | −0.002 | 0.997 | 0.999 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
González-Martin, A.M.; Limón-Villegas, E.S.; Reyes-Castillo, Z.; Esparza-Ros, F.; Hernández-Palma, L.A.; Santillán-Rivera, M.S.; Herrera-Amante, C.A.; Ramos-García, C.O.; Righini, N. Anthropometric Measurements for Predicting Low Appendicular Lean Mass Index for the Diagnosis of Sarcopenia: A Machine Learning Model. J. Funct. Morphol. Kinesiol. 2025, 10, 276. https://doi.org/10.3390/jfmk10030276
González-Martin AM, Limón-Villegas ES, Reyes-Castillo Z, Esparza-Ros F, Hernández-Palma LA, Santillán-Rivera MS, Herrera-Amante CA, Ramos-García CO, Righini N. Anthropometric Measurements for Predicting Low Appendicular Lean Mass Index for the Diagnosis of Sarcopenia: A Machine Learning Model. Journal of Functional Morphology and Kinesiology. 2025; 10(3):276. https://doi.org/10.3390/jfmk10030276
Chicago/Turabian StyleGonzález-Martin, Ana M., Edgar Samid Limón-Villegas, Zyanya Reyes-Castillo, Francisco Esparza-Ros, Luis Alexis Hernández-Palma, Minerva Saraí Santillán-Rivera, Carlos Abraham Herrera-Amante, César Octavio Ramos-García, and Nicoletta Righini. 2025. "Anthropometric Measurements for Predicting Low Appendicular Lean Mass Index for the Diagnosis of Sarcopenia: A Machine Learning Model" Journal of Functional Morphology and Kinesiology 10, no. 3: 276. https://doi.org/10.3390/jfmk10030276
APA StyleGonzález-Martin, A. M., Limón-Villegas, E. S., Reyes-Castillo, Z., Esparza-Ros, F., Hernández-Palma, L. A., Santillán-Rivera, M. S., Herrera-Amante, C. A., Ramos-García, C. O., & Righini, N. (2025). Anthropometric Measurements for Predicting Low Appendicular Lean Mass Index for the Diagnosis of Sarcopenia: A Machine Learning Model. Journal of Functional Morphology and Kinesiology, 10(3), 276. https://doi.org/10.3390/jfmk10030276












