Machine Learning Systems Detecting Illicit Drugs Based on Their ATR-FTIR Spectra
Abstract
1. Introduction
2. Related Work
3. Materials and Methods
3.1. Dataset Preparation
3.2. Exploratory Data Analysis
3.2.1. Statistical Measures
3.2.2. Principal Component Analysis (PCA)
3.2.3. Independent Component Analysis (ICA)
3.2.4. Autoencoders
3.3. Machine Learning Methods (MLM)
4. Results and Discussions
4.1. Exploratory Data Analysis
4.1.1. Statistical Measures
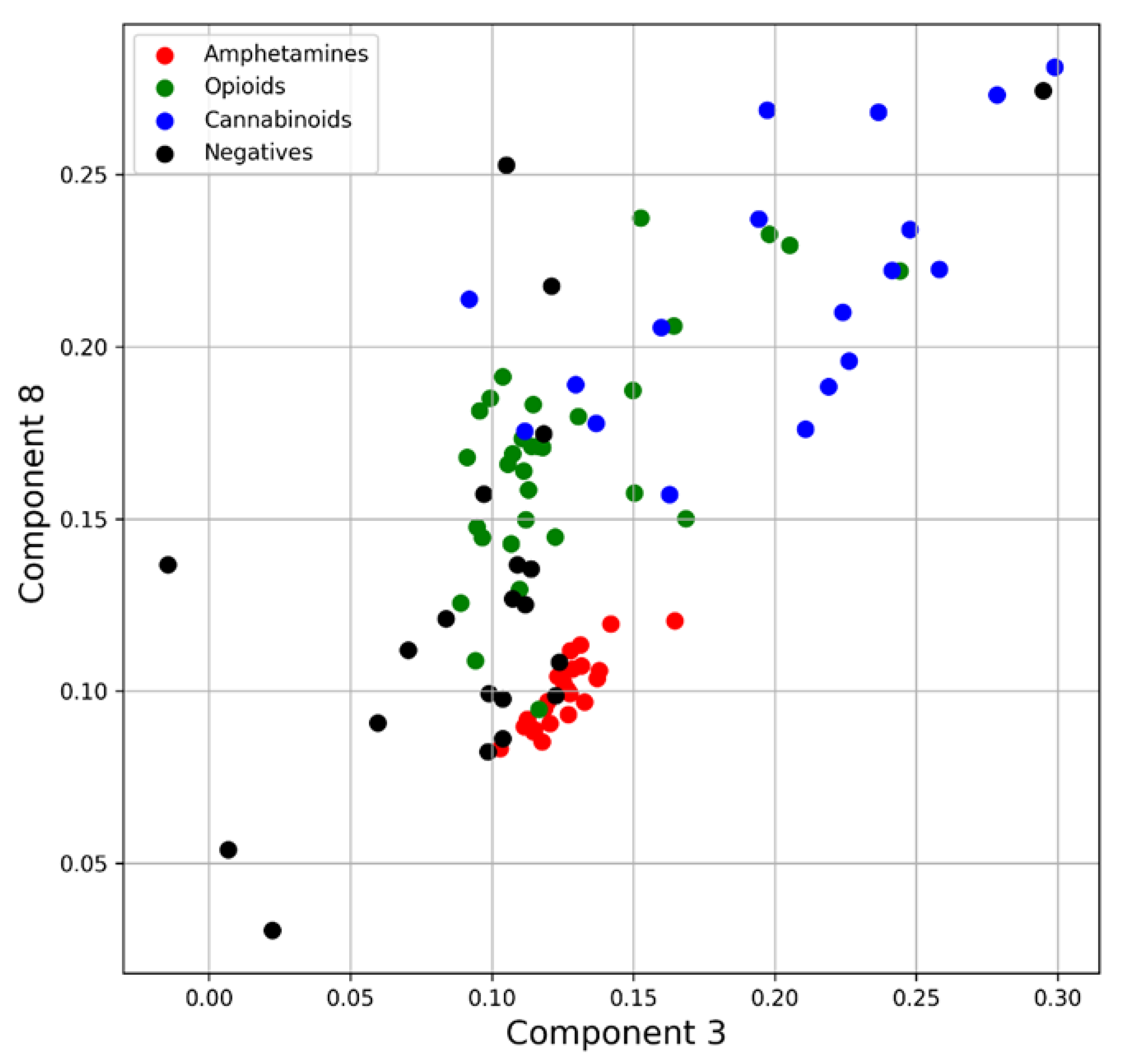
4.1.2. Principal Component Analysis
4.1.3. Independent Component Analysis
4.1.4. Transformers
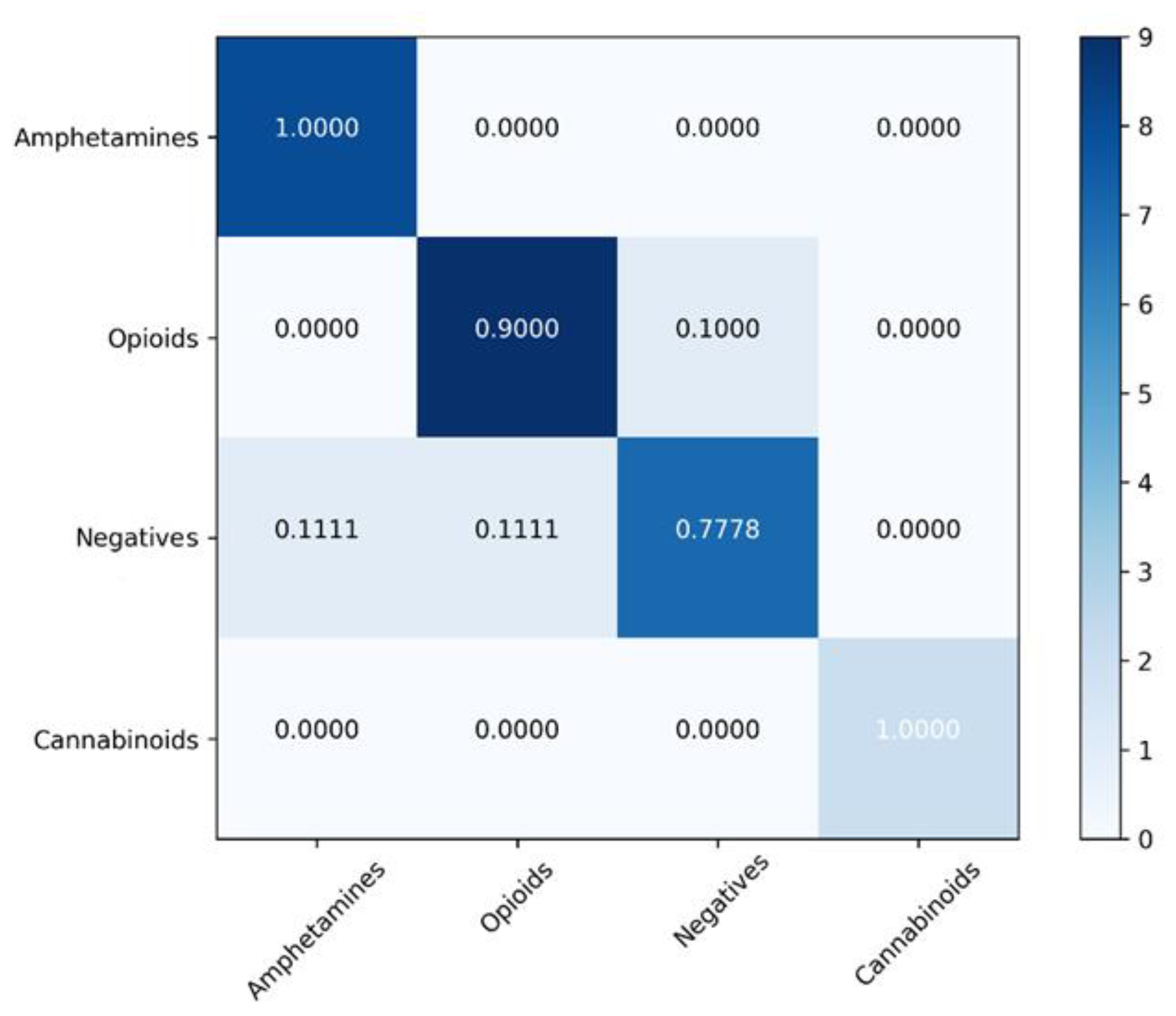
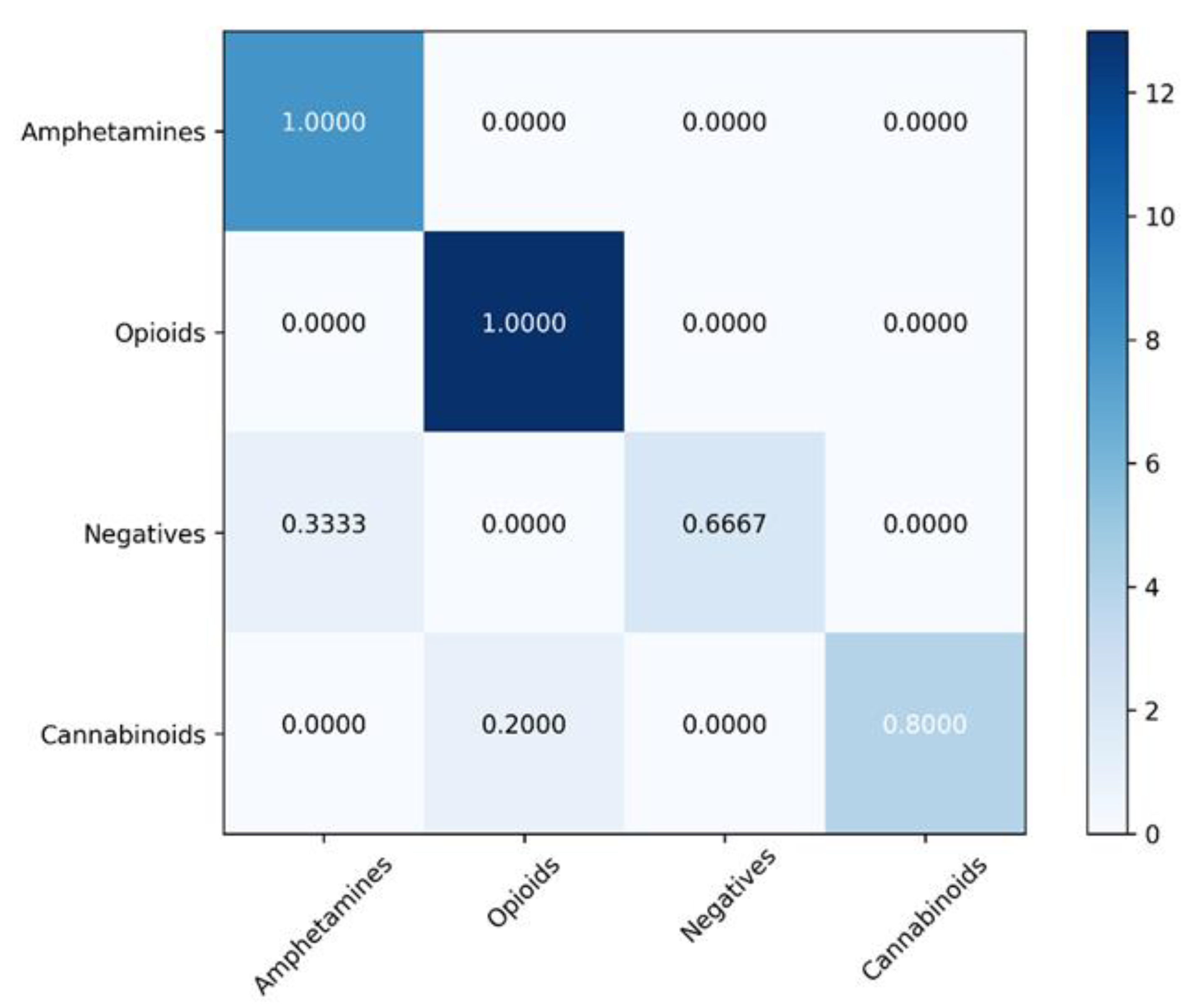
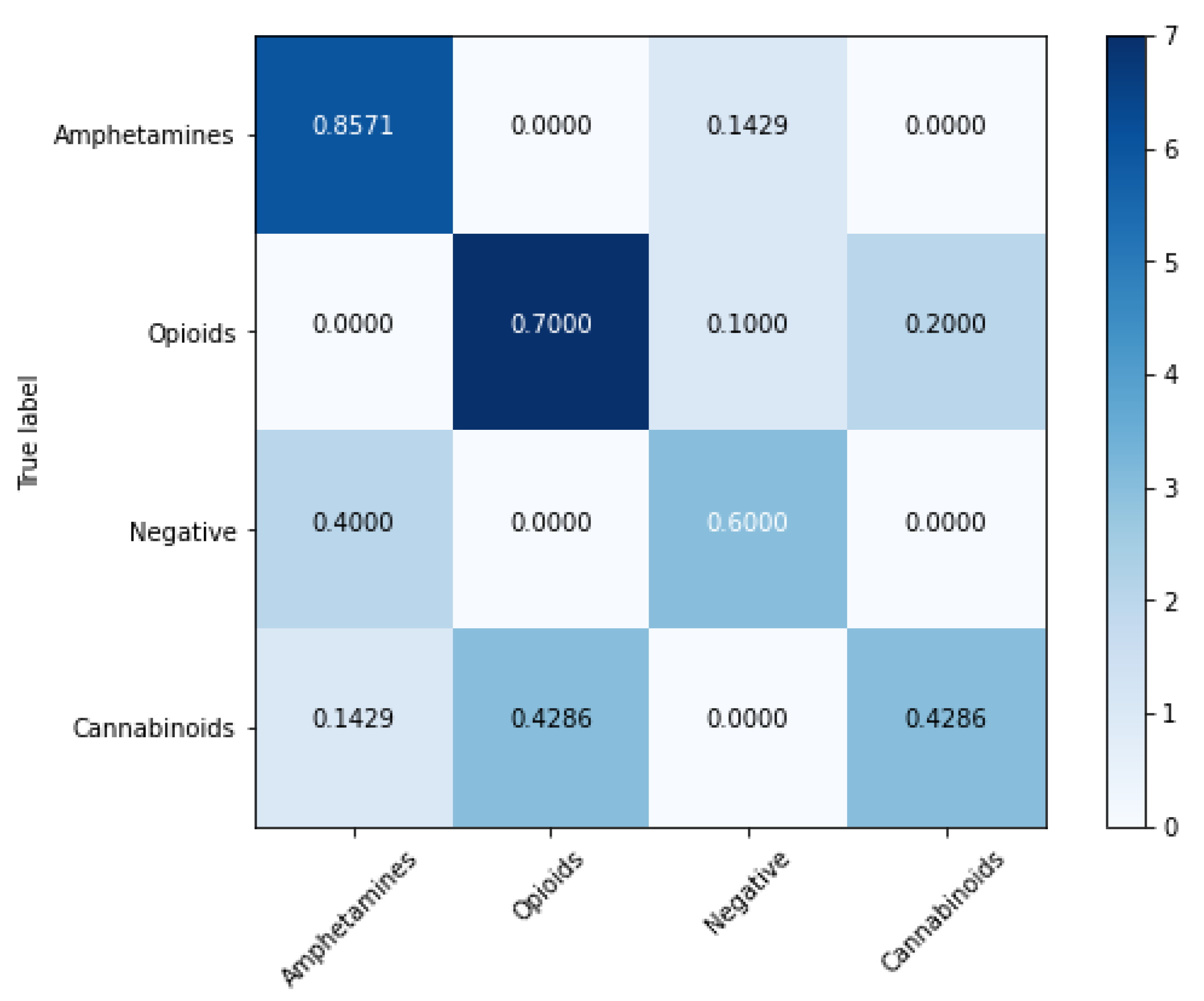
4.2. Classification Models
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Carvalho, M.; Carmo, H.; Costa, V.M.; Capela, J.P.; Pontes, H.; Remião, F.; Carvalho, F.; Bastos, M.d.L. Toxicity of amphetamines: An update. Arch. Toxicol. 2012, 86, 1167–1231. [Google Scholar] [CrossRef]
- Dean, B.V.; Stellpflug, S.J.; Burnett, A.M.; Engebretsen, K.M. 2C or not 2C: Phenethylamine designer drug review. J. Med. Toxicol. 2013, 9, 172–178. [Google Scholar] [CrossRef] [PubMed]
- Trachsel, D.; Lehmann, D.; Enzensperger, C. Phenethylamine: Von der Struktur zur Funktion; Nachtschatten-Verlag: Solothurn, Switzerland, 2013. [Google Scholar]
- Herrmann, E.S.; Johnson, P.S.; Johnson, M.W.; Vandrey, R. Novel drugs of abuse: Cannabinoids, stimulants, and hallucinogens. In Neuropathology of Drug Addictions and Substance Misuse; Elsevier: Amsterdam, The Netherlands, 2016; pp. 893–902. [Google Scholar]
- Zawilska, J.B.; Kacela, M.; Adamowicz, P. NBOMes–highly potent and toxic alternatives of LSD. Front. Neurosci. 2020, 14, 78. [Google Scholar] [CrossRef] [PubMed]
- Shi, V.Y.J.; Hsiao, M.; Loves, I.; Hamsavi, A. Comprehensive Guide to Hidradenitis Suppurativa; Elsevier: Amsterdam, The Netherlands, 2021; pp. 273–282. [Google Scholar]
- Kerrigan, S.; Goldberger, B.A. Opioids. In Principles of Forensic Toxicology; Springer: Berlin/Heidelberg, Germany, 2020; pp. 347–369. [Google Scholar]
- Zöllner, C.; Stein, C. Opioids. In Analgesia. Handbook of Experimental Pharmacology; Stein, C., Ed.; Springer: Berlin/Heidelberg, Germany, 2006; pp. 31–63. [Google Scholar]
- Pereira, L.S.; Lisboa, F.L.; Neto, J.C.; Valladão, F.N.; Sena, M.M. Screening method for rapid classification of psychoactive substances in illicit tablets using mid infrared spectroscopy and PLS-DA. Forensic Sci. Int. 2018, 288, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Koshute, P.; Hagan, N.; Jameson, N.J. Machine learning model for detecting fentanyl analogs from mass spectra. Forensic Chem. 2022, 27, 100379. [Google Scholar] [CrossRef]
- Lee, S.Y.; Lee, S.T.; Suh, S.; Ko, B.J.; Oh, H.B. Revealing unknown controlled substances and new psychoactive substances using high-resolution LC–MS-MS machine learning models and the hybrid similarity search algorithm. J. Anal. Toxicol. 2022, 46, 732–742. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.L.; Tan, J.; Ng, L.T.; Pan, J. Screening Unknown Novel Psychoactive Substances Using GC-MS Based Machine Learning. ChemRxiv 2022. [Google Scholar] [CrossRef]
- Piorunska-Sedlak, K.; Stypulkowska, K. Strategy for identification of new psychoactive substances in illicit samples using attenuated total reflectance infrared spectroscopy. Forensic Sci. Int. 2020, 312, 110262. [Google Scholar] [CrossRef] [PubMed]
- Scientific Working Group for the Analysis of Seized Drugs (SWGDRUG). Available online: www.swgdrug.org (accessed on 20 January 2023).
- Grigoletto, M.; Lisi, F. Looking for skewness in financial time series. Econom. J. 2009, 12, 310–323. [Google Scholar] [CrossRef]
- Loperfido, N. Kurtosis-based projection pursuit for outlier detection in financial time series. Eur. J. Financ. 2020, 26, 142–164. [Google Scholar] [CrossRef]
- McAlevey, L.G.; Stent, A.F. Kurtosis: A forgotten moment. Int. J. Math. Educ. Sci. Technol. 2018, 49, 120–130. [Google Scholar] [CrossRef]
- Deconinck, E.; Duchateau, C.; Balcaen, M.; Gremeaux, L.; Courselle, P. Chemometrics and infrared spectroscopy—A winning team for the analysis of illicit drug products. Rev. Anal. Chem. 2022, 41, 228–255. [Google Scholar] [CrossRef]
- Stone, J.V. Independent component analysis: An introduction. Trends Cogn. Sci. 2002, 6, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Himberg, J.; Mantyjarvi, J.; Korpipaa, P. Using PCA and ICA for exploratory data analysis in situation awareness. In Proceedings of the Conference Documentation International Conference on Multisensor Fusion and Integration for Intelligent Systems. MFI 2001 (Cat. No. 01TH8590), Baden-Baden, Germany, 20–22 August 2001; pp. 127–131. [Google Scholar]
- Kunapuli, S.S.; Bhallamudi, P.C. A review of deep learning models for medical diagnosis. In Machine Learning, Big Data, and IoT for Medical Informatics; Elsevier: Amsterdam, The Netherlands, 2021; pp. 389–404. [Google Scholar]
- Rodríguez-Pérez, R.; Bajorath, J. Evolution of Support Vector Machine and Regression Modeling in Chemoinformatics and Drug Discovery. J. Comput. Aided Mol. Des. 2022, 36, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Wade, C. Hands-On Gradient Boosting with XGBoost and Scikit-Learn: Perform Accessible Machine Learning and Extreme Gradient Boosting with Python; Packt Publishing: Birmingham, UK, 2020. [Google Scholar]
- Smith, C.; Koning, M. Decision Trees and Random Forests: A Visual Introduction For Beginners; Blue Windmill Media: Chicago, IL, USA, 2017. [Google Scholar]
- Bentéjac, C.; Csörgő, A.; Martínez-Muñoz, G. A comparative analysis of gradient boosting algorithms. Artif. Intell. Rev. 2021, 54, 1937–1967. [Google Scholar] [CrossRef]
- Vitola, J.; Pozo, F.; Tibaduiza, D.A.; Anaya, M. A Sensor Data Fusion System Based on K-Nearest Neighbor Pattern Classification for Structural Health Monitoring Applications. Sensors 2017, 17, 417. [Google Scholar] [CrossRef]
- Scafi, S.H.F.; Pasquini, C. Identification of counterfeit drugs using near-infrared spectroscopy. Analyst 2001, 126, 2218–2224. [Google Scholar] [CrossRef]
- Burlacu, C.M.; Burlacu, A.C.; Praisler, M. Sensitivity analysis of artificial neural networks identifying JWH synthetic cannabinoids built with alternative training strategies and methods. Inventions 2022, 7, 82. [Google Scholar] [CrossRef]
- Negoita, C.; Praisler, M.; Ion, A. Artificial intelligence application designed to screen for new psychoactive drugs based on their ATR-FTIR spectra. In AIP Conference Proceedings; Mishonov, T.M., Varonov, A.M., Eds.; AIP Publishing: Melville, NY, USA, 2019. [Google Scholar]
- United Nations Office on Drugs and Crime. Recommended Methods for the Identification and Analysis of Amphetamine, Methamphetamine and Their Ring-Substituted Analogues in Seized Materials; United Nations Publications: New York, NY, USA, 2006. [Google Scholar]
- United Nations Office on Drugs and Crime. Recommended Methods for the Identification and Analysis of Synthetic Cannabinoid Receptor Agonists in Seized Materials; United Nations Publications: New York, NY, USA, 2013. [Google Scholar]
- Gosav, S.; Praisler, M.; Birsa, M.L. Principal Component Analysis Coupled with Artificial Neural Networks—A Combined Technique Classifying Small Molecular Structures Using a Concatenated Spectral Database. Int. J. Mol. Sci. Spec. Issue Adv. Comput. Toxicol. 2011, 12, 6668–6684. [Google Scholar] [CrossRef]
- Ciochina, S.; Praisler, M.; Coman, M.M. Choosing Between Quantum Cascade Lasers (QCL) Equipping a New Hollow Fiber Infrared Scanner Designed to Detect New Psychoactive Substances (NPS). In AIP Conference Proceedings; Mishonov, T.M., Varonov, A.M., Eds.; AIP Publishing: Melville, NY, USA, 2019. [Google Scholar]





| Nr. crt. | Amphetamines | Opioids | Cannabinoids | Negatives |
|---|---|---|---|---|
| 1 | 2C-B HCl | 4’-Methyl acetyl fentanyl HCl | JWH-018 N-(5- chloropentyl) analog | 4-Acetoxy-N,N- Dimethyltrypt- amine oxalate |
| 2 | 2C-C HCl | para-Methyl acetyl fentanyl HCl | JWH-203 | Cocaine base |
| 3 | 2C-E HCl | Benzylfentanyl HCl | JWH-250 | Sertraline HCl |
| 4 | 2C-T-7 HCl | Acryl fentanyl HCl | JWH-122 | Trenbolone Hexahydro benzylcarbonate |
| 5 | 2C-T-2 HCl | 2-Furanylbenzyl fentanyl | JWH-018 adamantyl- carboxamide | 4-estren-3beta, 17beta-diol |
| 6 | 2C-I HCl | 2R,4S-2-Methyl fentanyl HCl | JWH-018 | Butalbital |
| 7 | 2,5-Dimethoxy-4-Chloro-amphetamine HCl | Despropionyl para-fluorofentanyl | JWH-307 | Boldenone Acetate |
| 8 | 2,5-Dimethoxy phenethylamine HCl | Despropionyl ortho-fluorofentanyl | JWH-081 | Cocaine HCl |
| 9 | 3,4-Dimethoxy amphetamine HCl | cis-3-Methyl fentanyl HCl | JWH-022 | Safrole |
| 10 | 2,5-Dimethoxyamphetamine HCl | Norfentanyl | JWH-210 | Phenazepam |
| 11 | DOI HCl | trans-3-Methyl fentanyl HCl | JWH-019 | Methenolone |
| 12 | d,l-4-Bromo-2,5-dimethoxyamphetamine HCl | para-Methoxy fentanyl HCl | JWH-073 | Methaqualone base |
| 13 | 4-Chloro-2,5-dimethoxyamphetamine HCl (DOC) | para-Chloroisobutyryl fentanyl HCl | JWH-018 Benzimidazole | MBZP HCl |
| 14 | 25B-NBOMe HCl | ortho-Methylacetyl fentanyl HCl | FUB-JWH-018 | Diazepam |
| 15 | 25C-NBOMe HCl | Heptanoyl fentanyl HCl | JWH-249 | Etaqualone HCl |
| 16 | 25I-NBOMe Base | beta-Hydroxy fentanyl HCl | JWH-018 indazole | Oxazepam |
| 17 | 25E-NBOMe HCl | 3-Methyl butyryl fentanyl HCl | AB-FUBICA | |
| 18 | 25D-NBOMe HCl | beta’-Phenyl fentanyl | ADB-PINACA | |
| 19 | 25H-NBOMe HCl | ortho-Fluoroisobutyryl fentanyl HCl | ||
| 20 | 25N-NBOMe HCl | para-Fluoroacetyl fentanyl HCl | ||
| 21 | 25C-NB3OMe HCl | meta-Fluoroisobutyryl fentanyl HCl | ||
| 22 | 25C-NB4OMe HCl | Tetrahydrofuran fentanyl 3-tetrahydrofurancarboxamide HCl | ||
| 23 | 25I-NBOMe HCl | para-Methyl cyclopropyl fentanyl HCl | ||
| 24 | 25I-NB3OMe HCl | para-Methoxy furanyl fentanyl HCl | ||
| 25 | 25I-NB4OMe HCl | ortho-Methyl cyclopropyl fentanyl HCl | ||
| 26 | ortho-Fluoro furanyl fentanyl HCl | |||
| 27 | N-benzyl para-fluoro norfentanyl HCl | |||
| 28 | N-Benzyl para-fluoro cyclopropyl norfentanyl HCl | |||
| 29 | Despropionyl meta- Fluorofentanyl | |||
| 30 | para-Fluoro fentanyl HCl | |||
| 31 | ortho-Methoxy furanyl fentanyl | |||
| 32 | Heroin Hydrochloride Monohydrate | |||
| 33 | W-18 | |||
| 34 | W-15 | |||
| 35 | 06-Monoacetyl morphine HCl | |||
| 36 | Morphine HCl trihydrate |
| Amphetamines | Opioids | Cannabinoids | Negatives | |
|---|---|---|---|---|
| Mean | 0.0338 | 0.0393 | 0.0452 | 0.0341 |
| Standard Deviation | 0.0263 | 0.0126 | 0.0128 | 0.0105 |
| Skewness | 1.877 | 1.261 | 2.567 | 0.7721 |
| Excess Kurtosis | 2.760 | 1.276 | 8.098 | 0.2311 |
| Minimum | 0.0126 | 0.0254 | 0.0258 | 0.0186 |
| Maximum | 0.135 | 0.0744 | 0.1072 | 0.0678 |
| Model | Balanced Accuracy (%) | Sensitivity (%) | Specificity (%) | Matthews Correlation Coefficient | ROC AUC |
|---|---|---|---|---|---|
| SVM | 92.08 ± 5.41 | 87.91 ± 5.16 | 96.25 ± 4.13 | 0.86 ± 0.04 | 0.91 |
| XGBoost | 91.99 ± 7.33 | 95.29 ± 7.59 | 88.69 ± 6.11 | 0.81 ± 0.05 | 0.91 |
| Random forest | 81.57 ± 8.66 | 71.15 ± 7.55 | 92.00 ± 8.74 | 0.67 ± 0.09 | 0.81 |
| Gradient Boosting | 76.46 ± 5.86 | 64.64 ± 4.95 | 88.28 ± 5.47 | 0.53 ± 0.05 | 0.76 |
| K-Nearest Neighbors | 66.88 ± 10.20 | 69.84 ± 10.20 | 90.64 ± 9.66 | 0.49 ± 0.12 | 0.80 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Darie, I.-F.; Anton, S.R.; Praisler, M. Machine Learning Systems Detecting Illicit Drugs Based on Their ATR-FTIR Spectra. Inventions 2023, 8, 56. https://doi.org/10.3390/inventions8020056
Darie I-F, Anton SR, Praisler M. Machine Learning Systems Detecting Illicit Drugs Based on Their ATR-FTIR Spectra. Inventions. 2023; 8(2):56. https://doi.org/10.3390/inventions8020056
Chicago/Turabian StyleDarie, Iulia-Florentina, Stefan Razvan Anton, and Mirela Praisler. 2023. "Machine Learning Systems Detecting Illicit Drugs Based on Their ATR-FTIR Spectra" Inventions 8, no. 2: 56. https://doi.org/10.3390/inventions8020056
APA StyleDarie, I.-F., Anton, S. R., & Praisler, M. (2023). Machine Learning Systems Detecting Illicit Drugs Based on Their ATR-FTIR Spectra. Inventions, 8(2), 56. https://doi.org/10.3390/inventions8020056





