Effect of the Flagellar Gene fliL on the Virulence of Pseudomonas plecoglossicida to Hybrid Grouper (Epinephelus fuscoguttatus ♀ × E. lanceolatus ♂)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. Construction of Deletion and Complement Strains
2.2.1. Construct the ΔfliL Strain
2.2.2. Construction of the Complement Strain C-ΔfliL
2.3. Validation of mRNA Levels of Deletion and Complement Strains
2.4. Determination of Growth Curve
2.5. Motility Measurement
2.6. Determination of Bacterial Adhesion Ability
2.6.1. Preparation of Surface Mucus
2.6.2. In Vitro Adhesion Experiment
2.7. Biofilm Determination
2.8. Measurement of the Chemotactic Response
2.9. Half Lethal Dose (LD50) Test for P. plecoglossicida
2.10. Transcriptomic Analysis
2.10.1. Library Preparation and Sequencing
2.10.2. Raw Data Statistics and Quality Control
2.10.3. Comparison with Reference Genome
2.10.4. Differentially Expressed Genes (DEGs) and Enrichment Analysis
2.10.5. Validation of Transcriptome Data
2.11. Data Analysis
2.12. Data Access
3. Results
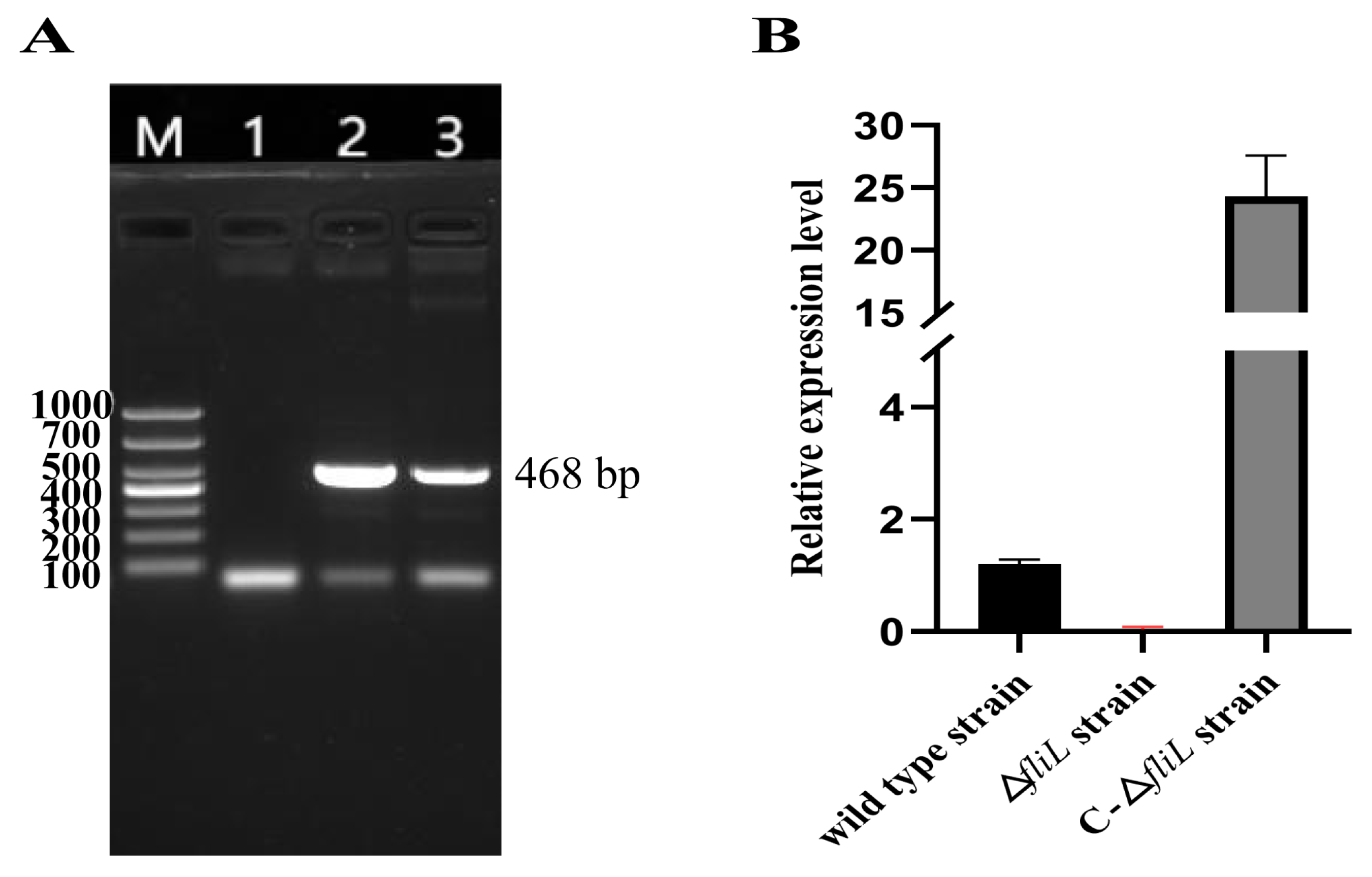
3.1. Construction of fliL Gene Deletion and Replacement Strain of P. plecoglossicida
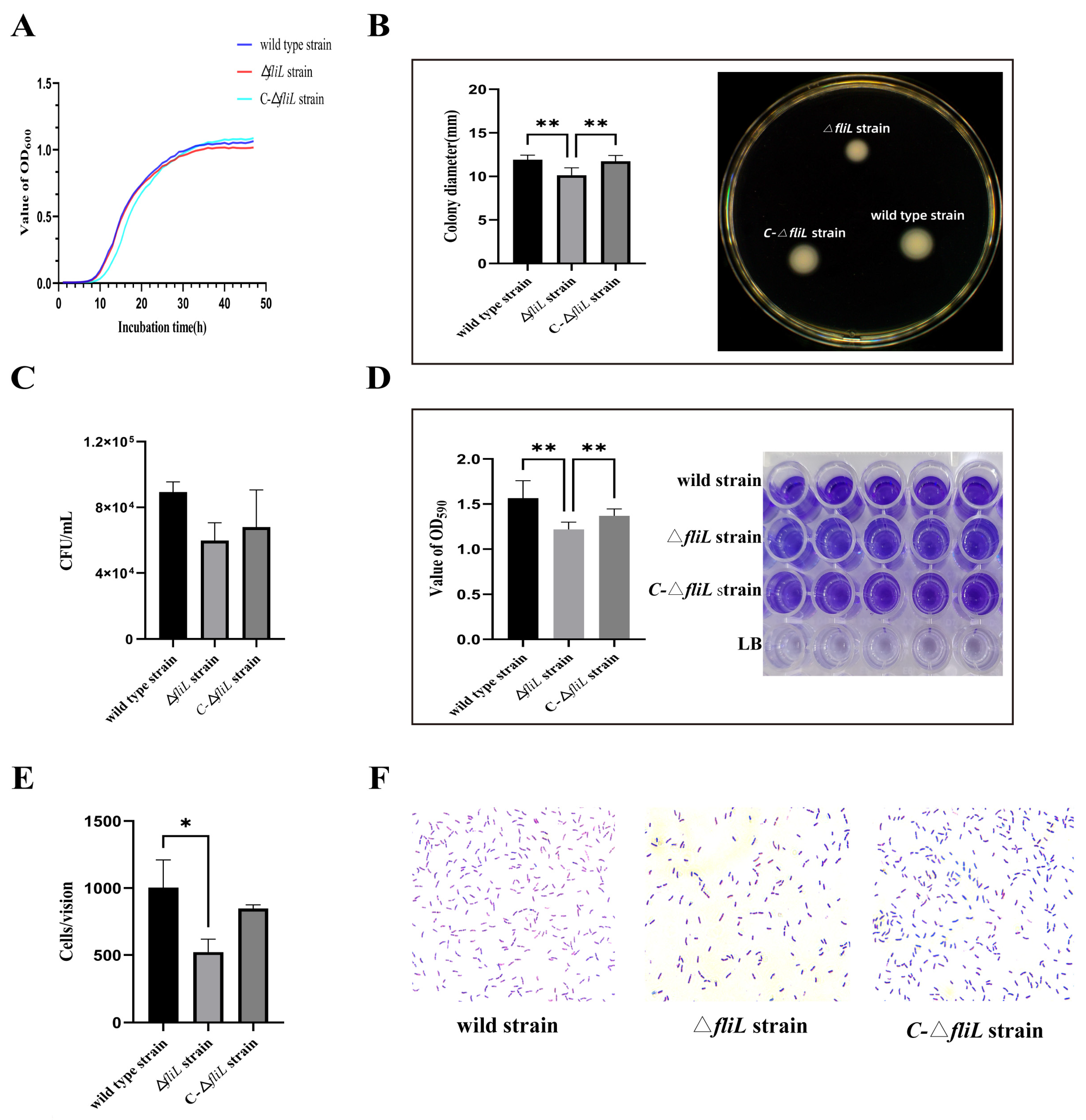
3.2. Effect of the fliL Gene on the Phenotype of P. plecoglossicida
3.3. Effect of the fliL Gene on the Virulence of P. plecoglossicida
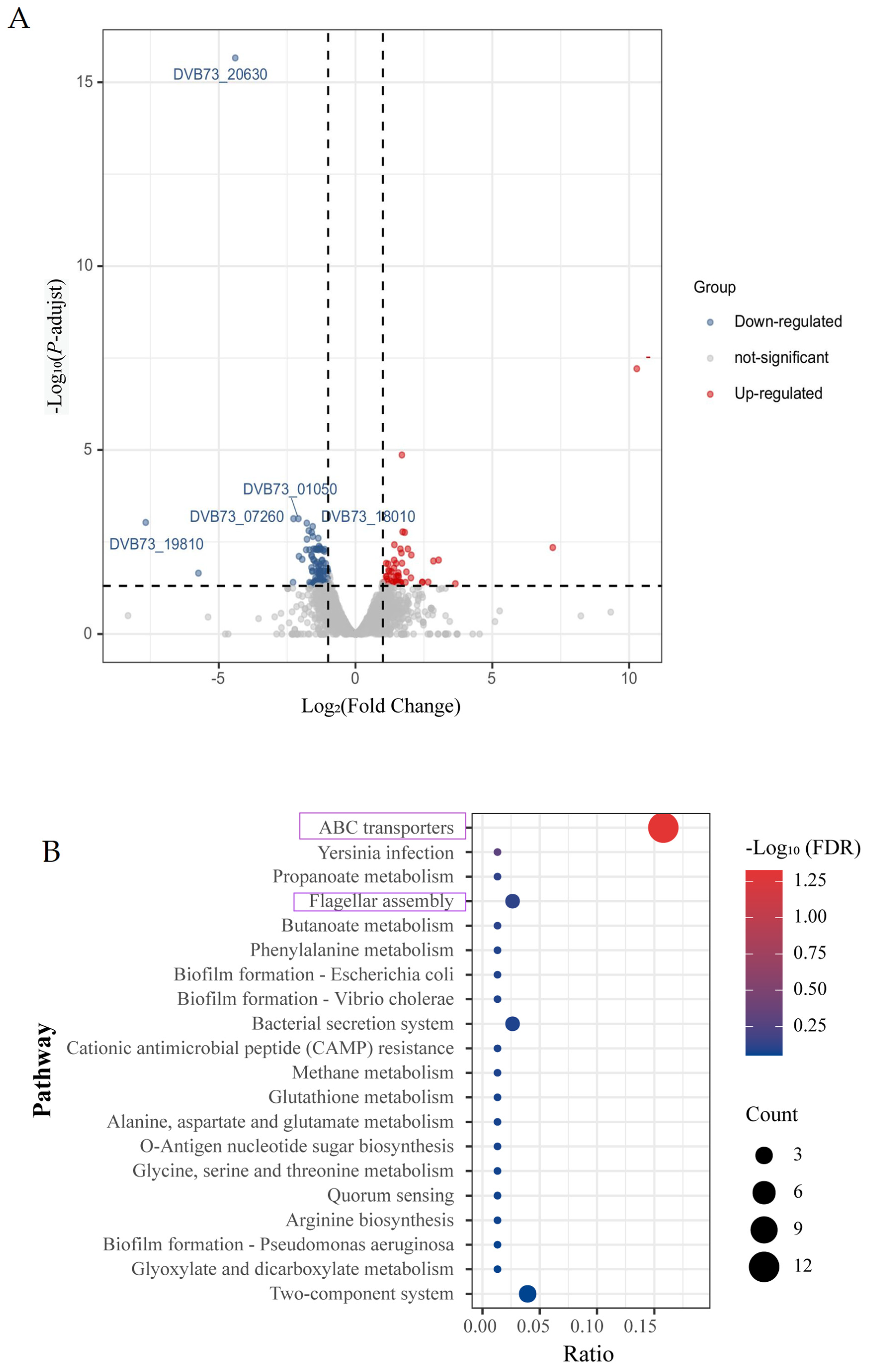
3.4. Effect of Knockout of fliL Gene on Transcriptome of Wild Strain of P. plecoglossicida
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Mzula, A.; Wambura, P.N.; Mdegela, R.H.; Shirima, G.M. Present Status of Aquaculture and the Challenge of Bacterial Diseases in Freshwater Farmed Fish in Tanzania; A Call for Sustainable Strategies. Aquac. Fish. 2021, 6, 247–253. [Google Scholar] [CrossRef]
- Rodger, H.D. Fish Disease Causing Economic Impact in Global Aquaculture. In Fish Vaccines; Adams, A., Ed.; Birkhäuser Advances in Infectious Diseases; Springer: Basel, Switzerland, 2016; pp. 1–34. ISBN 978-3-0348-0980-1. [Google Scholar]
- Wu, X.Y.; Xiong, J.B.; Fei, C.J.; Dai, T.; Zhu, T.F.; Zhao, Z.Y.; Pan, J.; Nie, L.; Chen, J. Prior exposure to ciprofloxacin disrupts intestinal homeostasis and predisposes ayu (Plecoglossus altivelis) to subsequent Pseudomonas plecoglossicida-induced infection. Zool. Res. 2022, 43, 648–665. [Google Scholar] [CrossRef]
- Cai, H.Y.; Ma, Y.; Qin, Y.X.; Zhao, L.M.; Yan, Q.P.; Huang, L.X. Vvrr2: A New Vibrio NcRNA Involved in Dynamic Synthesis of Multiple Biofilm Matrix Exopolusaccharides, Biofilm Structuring and Virulence. Aquaculture 2023, 563, 738925. [Google Scholar] [CrossRef]
- Du, Z.Y.; Zhang, M.M.; Qin, Y.X.; Zhao, L.M.; Huang, L.X.; Xu, X.J.; Yan, Q.P. The Role and Mechanisms of the Two-Component System EnvZ/OmpR on the Intracellular Survival of Aeromonas hydrophila. J. Fish Dis. 2022, 45, 1609–1621. [Google Scholar] [CrossRef] [PubMed]
- Park, S.C.; Shimamura, I.; Fukunaga, M.; Mori, K.I.; Nakai, T. Isolation of Bacteriophages Specific to a Fish Pathogen, Pseudomonas plecoglossicida, as a Candidate for Disease Control. Appl. Environ. Microbiol. 2000, 66, 1416–1422. [Google Scholar] [CrossRef] [Green Version]
- Xu, K.P.; Wang, Y.S.; Yang, W.X.H.; Cai, H.Y.; Zhang, Y.Y.; Huang, L.X. Strategies for Prevention and Control of Vibriosis in Asian Fish Culture. Vaccines 2022, 11, 98. [Google Scholar] [CrossRef]
- Nishimori, E.; Kita-Tsukamoto, K.; Wakabayashi, H. Pseudomonas plecoglossicida Sp. Nov., the Causative Agent of Bacterial Haemorrhagic Ascites of Ayu, Plecoglossus altivelis. Int. J. Syst. Evol. Microbiol. 2000, 50 Pt 1, 83–89. [Google Scholar] [CrossRef] [Green Version]
- Akayli, T.; Çanak, Ö.; Başaran, B. A New Pseudomonas Species Observed in Cultured Young Rainbow Trout (Oncorhynchus mykis Walbaum, 1792): Pseudomonas plecoglossicida. Biyol. Bilim. Araştırma Derg. 2011, 4, 107–111. [Google Scholar]
- Yuan, B.; Zhao, L.M.; Zhuang, Z.X.; Wang, X.R.; Fu, Q.; Huang, H.B.; Huang, L.X.; Qin, Y.X.; Yan, Q.P. Transcriptomic and Metabolomic Insights into the Role of the flgK Gene in the Pathogenicity of Pseudomonas plecoglossicida to Orange-Spotted Grouper (Epinephelus coioides). Zool. Res. 2022, 43, 952–965. [Google Scholar] [CrossRef]
- Zhang, J.T.; Zhou, S.M.; An, S.W.; Chen, L.; Wang, G.L. Visceral Granulomas in Farmed Large Yellow Croaker, Larimichthys crocea (Richardson), Caused by a Bacterial Pathogen, Pseudomonas plecoglossicida. J. Fish Dis. 2014, 37, 113–121. [Google Scholar] [CrossRef]
- Xin, G.; Zhao, L.M.; Zhuang, Z.X.; Wang, X.; Fu, Q.; Huang, H.B.; Huang, L.X.; Qin, Y.X.; Zhang, J.N.; Zhang, J.L.; et al. Function of the rpoD Gene in Pseudomonas plecoglossicida Pathogenicity and Epinephelus coioides Immune Response. Fish Shellfish Immunol. 2022, 127, 427–436. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Jiao, J.P.; Zhao, L.M.; Zhuang, Z.X.; Wang, X.R.; Fu, Q.; Huang, H.B.; Huang, L.X.; Qin, Y.X.; Zhang, J.N.; et al. The Contribution of exbB Gene to Pathogenicity of Pseudomonas plecoglossicida and Its Interactions with Epinephelus coioides. Fish Shellfish Immunol. 2022, 120, 610–619. [Google Scholar] [CrossRef] [PubMed]
- Jiao, J.P.; Zhao, L.M.; Huang, L.X.; Qin, Y.X.; Su, Y.Q.; Zheng, W.Q.; Zhang, J.N.; Yan, Q.P. The Contributions of fliG Gene to the Pathogenicity of Pseudomonas plecoglossicida and Pathogen-Host Interactions with Epinephelus coioides. Fish Shellfish Immunol. 2021, 119, 238–248. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Zhao, L.M.; Li, Q.; Huang, L.X.; Qin, Y.X.; Wang, P.; Zhu, C.Z.; Yan, Q.P. flgC Gene Is Involved in the Virulence Regulation of Pseudomonas plecoglossicida and Affects the Immune Response of Epinephelus coioides. Fish Shellfish Immunol. 2023, 132, 108512. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.W.; Kumar, A.; Kojima, S.; Homma, M. FliL Associates with the Stator to Support Torque Generation of the Sodium-Driven Polar Flagellar Motor of Vibrio. Mol. Microbiol. 2015, 98, 101–110. [Google Scholar] [CrossRef]
- Tachiyama, S.; Chan, K.L.; Liu, X.L.; Hathroubi, S.; Peterson, B.; Khan, M.F.; Ottemann, K.M.; Liu, J.; Roujeinikova, A. The Flagellar Motor Protein FliL Forms a Scaffold of Circumferentially Positioned Rings Required for Stator Activation. Proc. Natl. Acad. Sci. USA 2022, 119, e2118401119. [Google Scholar] [CrossRef]
- Lin, T.S.; Zhu, S.W.; Kojima, S.; Homma, M.; Lo, C.J. FliL Association with Flagellar Stator in the Sodium-Driven Vibrio Motor Characterized by the Fluorescent Microscopy. Sci. Rep. 2018, 8, 11172. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.Y.; Patellis, J.; Belas, R. Activity of Proteus Mirabilis FliL Is Viscosity Dependent and Requires Extragenic DNA. J. Bacteriol. 2013, 195, 823–832. [Google Scholar] [CrossRef] [Green Version]
- Mengucci, F.; Dardis, C.; Mongiardini, E.J.; Althabegoiti, M.J.; Partridge, J.D.; Kojima, S.; Homma, M.; Quelas, J.I.; Lodeiro, A.R. Characterization of FliL Proteins in Bradyrhizobium diazoefficiens: Lateral FliL Supports Swimming Motility, and Subpolar FliL Modulates the Lateral Flagellar System. J. Bacteriol. 2020, 202, e00708-19. [Google Scholar] [CrossRef]
- Partridge, J.D.; Nieto, V.; Harshey, R.M. A New Player at the Flagellar Motor: FliL Controls Both Motor Output and Bias. mBio 2015, 6, e02367. [Google Scholar] [CrossRef] [Green Version]
- Takekawa, N.; Isumi, M.; Terashima, H.; Zhu, S.W.; Nishino, Y.; Sakuma, M.; Kojima, S.; Homma, M.; Imada, K. Structure of Vibrio FliL, a New Stomatin-like Protein That Assists the Bacterial Flagellar Motor Function. mBio 2019, 10, e00292-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Josenhans, C.; Suerbaum, S. The Role of Motility as a Virulence Factor in Bacteria. Int. J. Med. Microbiol. 2002, 291, 605–614. [Google Scholar] [CrossRef] [PubMed]
- Suaste-Olmos, F.; Domenzain, C.; Mireles-Rodríguez, J.C.; Poggio, S.; Osorio, A.; Dreyfus, G.; Camarena, L. The Flagellar Protein FliL Is Essential for Swimming in Rhodobacter sphaeroides. J. Bacteriol. 2010, 192, 6230–6239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, H.D.; Xu, Z.J.; Tao, Z.; Li, W.Y.; Li, Y.; Yang, A.; Wang, W.; Yin, X.L.; Yan, X.J. Efficacy and Safety of Pseudomonas plecoglossicida Mutant ΔtssD-1 as a Live Attenuated Vaccine for the Large Yellow Croaker (Larimichthys crocea). Aquaculture 2021, 531, 735976. [Google Scholar] [CrossRef]
- Howery, K.E.; Rather, P.N. Allelic Exchange Mutagenesis in Proteus mirabilis. Methods Mol. Biol. 2019, 2021, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Vijayan, S.P.; Rekha, P.D.; Dinesh, U.; Arun, A.B. Determination of Acute Lethal Dose 50 (LD50) of Uranyl Nitrate in Male Swiss Albino Mice. Res. J. Pharm. Technol. 2018, 11, 1086. [Google Scholar] [CrossRef]
- Melo Ferreira, R.; Sabo, A.R.; Winfree, S.; Collins, K.S.; Janosevic, D.; Gulbronson, C.J.; Cheng, Y.-H.; Casbon, L.; Barwinska, D.; Ferkowicz, M.J.; et al. Integration of Spatial and Single-Cell Transcriptomics Localizes Epithelial Cell-Immune Cross-Talk in Kidney Injury. JCI Insight 2021, 6, e147703. [Google Scholar] [CrossRef]
- Rong, Y.; Tang, L.; Yin, H.F.; Sun, D.Y. A Modified Coriolis Method Was Used to Calculate the Number of BALB/cA-Nu Mice Treated with 1,2. LD50 of Dichloropropane. Chin. J. Occup. Dis. Labor Hyg. 2019, 37, 53–55. [Google Scholar]
- Dong, Y.; Bai, H.T.; Dong, F.; Zhang, X.B.; Ema, H. Gene Knockout in Highly Purified Mouse Hematopoietic Stem Cells by CRISPR/Cas9 Technology. J. Immunol. Methods 2021, 495, 113070. [Google Scholar] [CrossRef]
- Sasidharan, S.; Saudagar, P. Knockout of Tyrosine Aminotransferase Gene by Homologous Recombination Arrests Growth and Disrupts Redox Homeostasis in Leishmania Parasite. Parasitol. Res. 2022, 121, 3229–3241. [Google Scholar] [CrossRef]
- Zhang, Y.X.; Zhang, Y.; Qi, Y.P. Plant Gene Knockout and Knockdown by CRISPR-Cpf1 (Cas12a) Systems. Methods Mol. Biol. 2019, 1917, 245–256. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.R.; Cho, J.S.; Cho, I.J.; Park, D.; Lee, S.Y. Markerless Gene Knockout and Integration to Express Heterologous Biosynthetic Gene Clusters in Pseudomonas putida. Metab. Eng. 2018, 47, 463–474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, X.; Tu, Y.Q.; Wu, H.; Zhang, L.J.; Zhao, S.N. Gene Knockout Revealed the Role of Gene FeoA in Cell Growth and Division of Lactobacillus delbrueckii Subsp. Bulgaricus. Arch. Microbiol. 2021, 203, 3541–3549. [Google Scholar] [CrossRef]
- Zhu, X.W.; Wu, Y.K.; Lv, X.Q.; Liu, Y.F.; Du, G.C.; Li, J.H.; Liu, L. Combining CRISPR-Cpf1 and Recombineering Facilitates Fast and Efficient Genome Editing in Escherichia coli. ACS Synth. Biol. 2022, 11, 1897–1907. [Google Scholar] [CrossRef]
- Carroll, D. Genome Engineering with Zinc-Finger Nucleases. Genetics 2011, 188, 773–782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dalvie, N.C.; Lorgeree, T.; Biedermann, A.M.; Love, K.R.; Love, J.C. Simplified Gene Knockout by CRISPR-Cas9-Induced Homologous Recombination. ACS Synth. Biol. 2022, 11, 497–501. [Google Scholar] [CrossRef]
- Kim, S.E.; Kim, J.W.; Kim, Y.J.; Kwon, D.N.; Kim, J.H.; Kang, M.J. Generation of Fibroblasts Lacking the Sal-like 1 Gene by Using Transcription Activator-like Effector Nuclease-Mediated Homologous Recombination. Asian-Australas. J. Anim. Sci. 2016, 29, 564–570. [Google Scholar] [CrossRef]
- Sakaguchi, K.; He, J.L.; Tani, S.; Kano, Y.; Suzuki, T. A Targeted Gene Knockout Method Using a Newly Constructed Temperature-Sensitive Plasmid Mediated Homologous Recombination in Bifidobacterium longum. Appl. Microbiol. Biotechnol. 2012, 95, 499–509. [Google Scholar] [CrossRef]
- Langford, M.L.; Zabaleta, J.; Ochoa, A.C.; Testerman, T.L.; McGee, D.J. In Vitro and in Vivo Complementation of the Helicobacter Pylori Arginase Mutant Using an Intergenic Chromosomal Site. Helicobacter 2006, 11, 477–493. [Google Scholar] [CrossRef] [Green Version]
- Alonso, M.N.; Malaga, W.; Mc Neil, M.; Jackson, M.; Romano, M.I.; Guilhot, C.; Santangelo, M.P. Efficient Method for Targeted Gene Disruption by Homologous Recombination in Mycobacterium avium Subspecie paratuberculosis. Res. Microbiol. 2020, 171, 203–210. [Google Scholar] [CrossRef]
- Chaussé, A.M.; Roche, S.M.; Moroldo, M.; Hennequet-Antier, C.; Holbert, S.; Kempf, F.; Barilleau, E.; Trotereau, J.; Velge, P. Epithelial Cell Invasion by Salmonella typhimurium Induces Modulation of Genes Controlled by Aryl Hydrocarbon Receptor Signaling and Involved in Extracellular Matrix Biogenesis. Virulence 2023, 14, 2158663. [Google Scholar] [CrossRef]
- Prada, C.F.; Casadiego, M.A.; Freire, C.C. Evolution of helicobacter Spp: Variability of Virulence Factors and Their Relationship to Pathogenicity. PeerJ 2022, 10, e13120. [Google Scholar] [CrossRef] [PubMed]
- Sano, G.; Takada, Y.; Goto, S.; Maruyama, K.; Shindo, Y.; Oka, K.; Matsui, H.; Matsuo, K. Flagella Facilitate Escape of salmonella from Oncotic Macrophages. J. Bacteriol. 2007, 189, 8224–8232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, J.X.; Zhang, X.; Wang, X.F.; Xu, C.H.; Chang, S.; Wu, H.J.; Wang, T.; Liang, H.H.; Gao, H.C.; Zhou, Y.; et al. Structural Basis of Assembly and Torque Transmission of the Bacterial Flagellar Motor. Cell 2021, 184, 2665–2679.e19. [Google Scholar] [CrossRef] [PubMed]
- Chilcott, G.S.; Hughes, K.T. Coupling of Flagellar Gene Expression to Flagellar Assembly in Salmonella enterica Serovar Typhimurium and Escherichia coli. Microbiol. Mol. Biol. Rev. 2000, 64, 694–708. [Google Scholar] [CrossRef] [Green Version]
- Attieh, Z.; Mouawad, C.; Rejasse, A.; Jehanno, I.; Perchat, S.; Hegna, I.K.; Økstad, O.A.; Kallassy Awad, M.; Sanchis-Borja, V.; El Chamy, L. The fliK Gene Is Required for the Resistance of Bacillus thuringiensis to Antimicrobial Peptides and Virulence in Drosophila melanogaster. Front. Microbiol. 2020, 11, 611220. [Google Scholar] [CrossRef]
- Zhou, G.; Wang, Y.S.; Peng, H.; Liu, H.Z.; Feng, J.; Li, S.J.; Sun, T.L.; Li, C.L.; Shi, Q.S.; Xie, X.B. Outer Membrane Protein of OmpF Contributes to Swimming Motility, Biofilm Formation, Osmotic Response as Well as the Transcription of Maltose Metabolic Genes in Citrobacter werkmanii. World J. Microbiol. Biotechnol. 2023, 39, 15. [Google Scholar] [CrossRef]
- Zhou, G.; Wang, Y.S.; Peng, H.; Li, S.J.; Sun, T.L.; Shen, P.F.; Xie, X.B.; Shi, Q.S. Roles of OmpA of Citrobacter werkmanii in Bacterial Growth, Biocide Resistance, Biofilm Formation and Swimming Motility. Appl. Microbiol. Biotechnol. 2021, 105, 2841–2854. [Google Scholar] [CrossRef]
- Dong, Z.C.; Chen, Y. Transcriptomics: Advances and Approaches. Sci. China Life Sci. 2013, 56, 960–967. [Google Scholar] [CrossRef] [Green Version]
- Wirka, R.C.; Pjanic, M.; Quertermous, T. Advances in Transcriptomics: Investigating Cardiovascular Disease at Unprecedented Resolution. Circ. Res. 2018, 122, 1200–1220. [Google Scholar] [CrossRef]
- Creecy, J.P.; Conway, T. Quantitative Bacterial Transcriptomics with RNA-Seq. Curr. Opin. Microbiol. 2015, 23, 133–140. [Google Scholar] [CrossRef] [Green Version]
- Siezen, R.J.; Wilson, G.; Todt, T. Prokaryotic Whole-Transcriptome Analysis: Deep Sequencing and Tiling Arrays. Microb. Biotechnol. 2010, 3, 125–130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- James, K.; Cockell, S.J.; Zenkin, N. Deep Sequencing Approaches for the Analysis of Prokaryotic Transcriptional Boundaries and Dynamics. Methods 2017, 120, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Ahn, A.C.; Cavalca, L.; Colombo, M.; Schuurmans, J.M.; Sorokin, D.Y.; Muyzer, G. Transcriptomic Analysis of Two Thioalkalivibrio Species Under Arsenite Stress Revealed a Potential Candidate Gene for an Alternative Arsenite Oxidation Pathway. Front. Microbiol. 2019, 10, 1514. [Google Scholar] [CrossRef] [Green Version]
- Roe, C.; Williamson, C.H.D.; Vazquez, A.J.; Kyger, K.; Valentine, M.; Bowers, J.R.; Phillips, P.D.; Harrison, V.; Driebe, E.; Engelthaler, D.M.; et al. Bacterial Genome Wide Association Studies (BGWAS) and Transcriptomics Identifies Cryptic Antimicrobial Resistance Mechanisms in Acinetobacter baumannii. Front. Public Health 2020, 8, 451. [Google Scholar] [CrossRef]
- Lebreton, A.; Cossart, P. RNA- and Protein-Mediated Control of Listeria monocytogenes Virulence Gene Expression. RNA Biol. 2017, 14, 460–470. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.W.; Zhao, H.Q.; Yang, H.; He, C.Y.; Shu, W.; Cui, Z.L.; Liu, Q.Z. Insights into the Impact of Small RNA SprC on the Metabolism and Virulence of Staphylococcus aureus. Front. Cell. Infect. Microbiol. 2022, 12, 746746. [Google Scholar] [CrossRef]
- Papp-Wallace, K.M.; Maguire, M.E. Manganese Transport and the Role of Manganese in Virulence. Annu. Rev. Microbiol. 2006, 60, 187–209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wandersman, C.; Delepelaire, P. Bacterial Iron Sources: From Siderophores to Hemophores. Annu. Rev. Microbiol. 2004, 58, 611–647. [Google Scholar] [CrossRef]
- Zheng, C.K.; Qiu, J.; Zhai, Y.M.; Wei, M.; Zhou, X.H.; Jiao, X.N. ZrgA Contributes to Zinc Acquisition in Vibrio Parahaemolyticus. Virulence 2023, 14, 2156196. [Google Scholar] [CrossRef]
- Kieuvongngam, V.; Chen, J. Structures of the Peptidase-Containing ABC Transporter PCAT1 under Equilibrium and Nonequilibrium Conditions. Proc. Natl. Acad. Sci. USA 2022, 119, e2120534119. [Google Scholar] [CrossRef] [PubMed]
- Beis, K. Structural Basis for the Mechanism of ABC Transporters. Biochem. Soc. Trans. 2015, 43, 889–893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lewis, V.G.; Ween, M.P.; McDevitt, C.A. The Role of ATP-Binding Cassette Transporters in Bacterial Pathogenicity. Protoplasma 2012, 249, 919–942. [Google Scholar] [CrossRef] [PubMed]

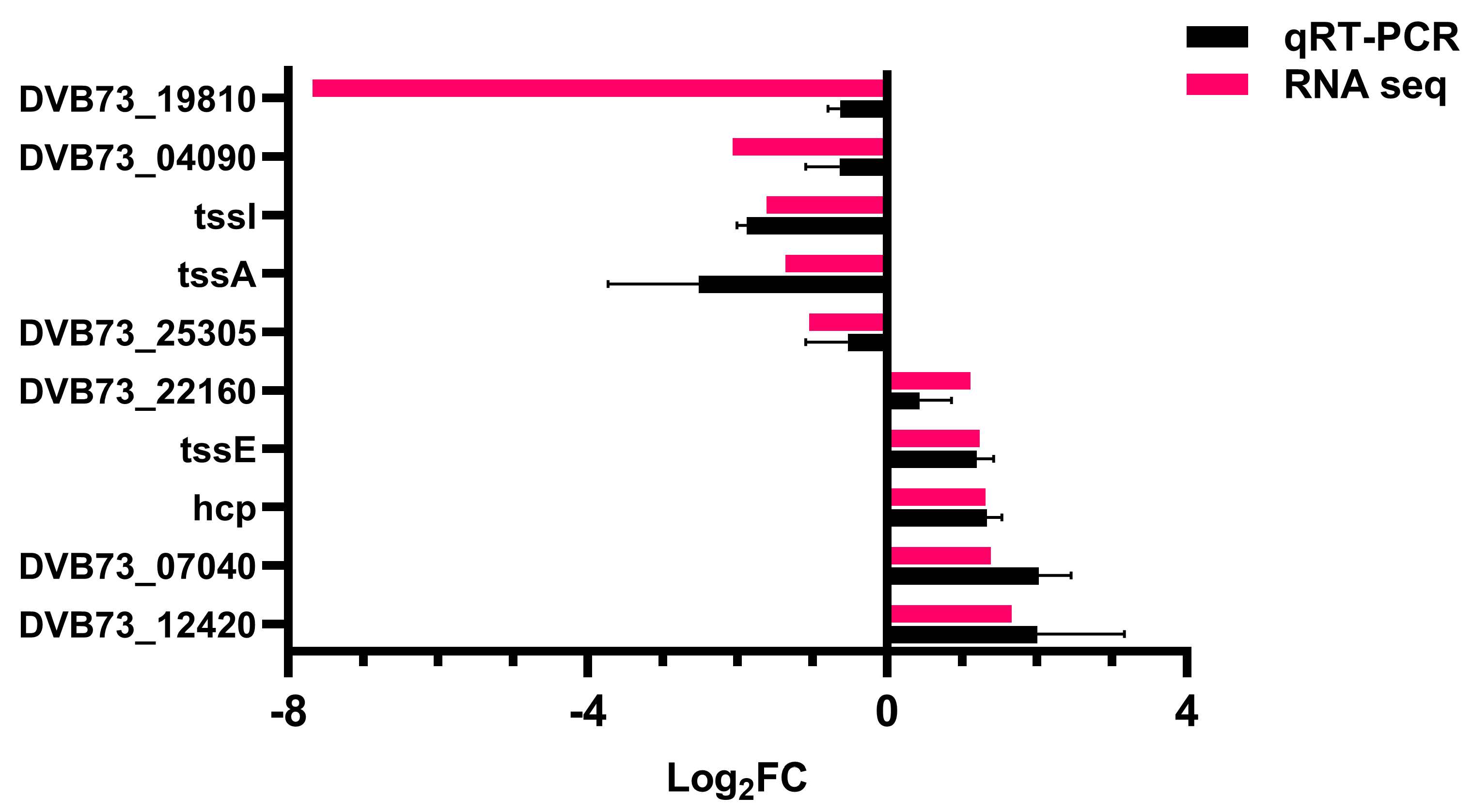

| Dose (CFU/Fish) | The Number of Test Animals | Wild Type Strain | ΔfliL Strain | C-ΔfliL Strain | |||
|---|---|---|---|---|---|---|---|
| The Number of Dead | Mortality/% | The Number of Dead | Mortality/% | The Number of Dead | Mortality/% | ||
| 5 × 106 | 10 | 10 | 100 | 10 | 100 | 10 | 100 |
| 5 × 105 | 10 | 10 | 100 | 7 | 70 | 10 | 100 |
| 5 × 104 | 10 | 9 | 90 | 4 | 40 | 9 | 90 |
| 5 × 103 | 10 | 6 | 60 | 3 | 30 | 8 | 80 |
| 5 × 102 | 10 | 0 | 0 | 0 | 0 | 3 | 30 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, L.; Zhang, J.; Zhao, L.; Li, Q.; Huang, L.; Qin, Y.; Yan, Q. Effect of the Flagellar Gene fliL on the Virulence of Pseudomonas plecoglossicida to Hybrid Grouper (Epinephelus fuscoguttatus ♀ × E. lanceolatus ♂). Fishes 2023, 8, 397. https://doi.org/10.3390/fishes8080397
Shi L, Zhang J, Zhao L, Li Q, Huang L, Qin Y, Yan Q. Effect of the Flagellar Gene fliL on the Virulence of Pseudomonas plecoglossicida to Hybrid Grouper (Epinephelus fuscoguttatus ♀ × E. lanceolatus ♂). Fishes. 2023; 8(8):397. https://doi.org/10.3390/fishes8080397
Chicago/Turabian StyleShi, Lian, Junjie Zhang, Lingmin Zhao, Qi Li, Lixing Huang, Yingxue Qin, and Qingpi Yan. 2023. "Effect of the Flagellar Gene fliL on the Virulence of Pseudomonas plecoglossicida to Hybrid Grouper (Epinephelus fuscoguttatus ♀ × E. lanceolatus ♂)" Fishes 8, no. 8: 397. https://doi.org/10.3390/fishes8080397
APA StyleShi, L., Zhang, J., Zhao, L., Li, Q., Huang, L., Qin, Y., & Yan, Q. (2023). Effect of the Flagellar Gene fliL on the Virulence of Pseudomonas plecoglossicida to Hybrid Grouper (Epinephelus fuscoguttatus ♀ × E. lanceolatus ♂). Fishes, 8(8), 397. https://doi.org/10.3390/fishes8080397






