Screening and Identification of Interacting Proteins of Mitfa in Red Tilapia
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. RNA Extraction and cDNA Synthesis
2.3. Expression and Purification of the Mitfa Recombinant Protein
2.4. His Pull-Down Assay
2.5. Western Blotting
2.6. Liquid Chromatography with Tandem Mass Spectrometry (LC-MS/MS) Analysis
2.7. Bioinformatics Analysis
2.8. Administration of the Mitfa Recombinant Protein for In Vivo Assays
2.9. Quantitative Real-Time PCR Analysis
3. Results
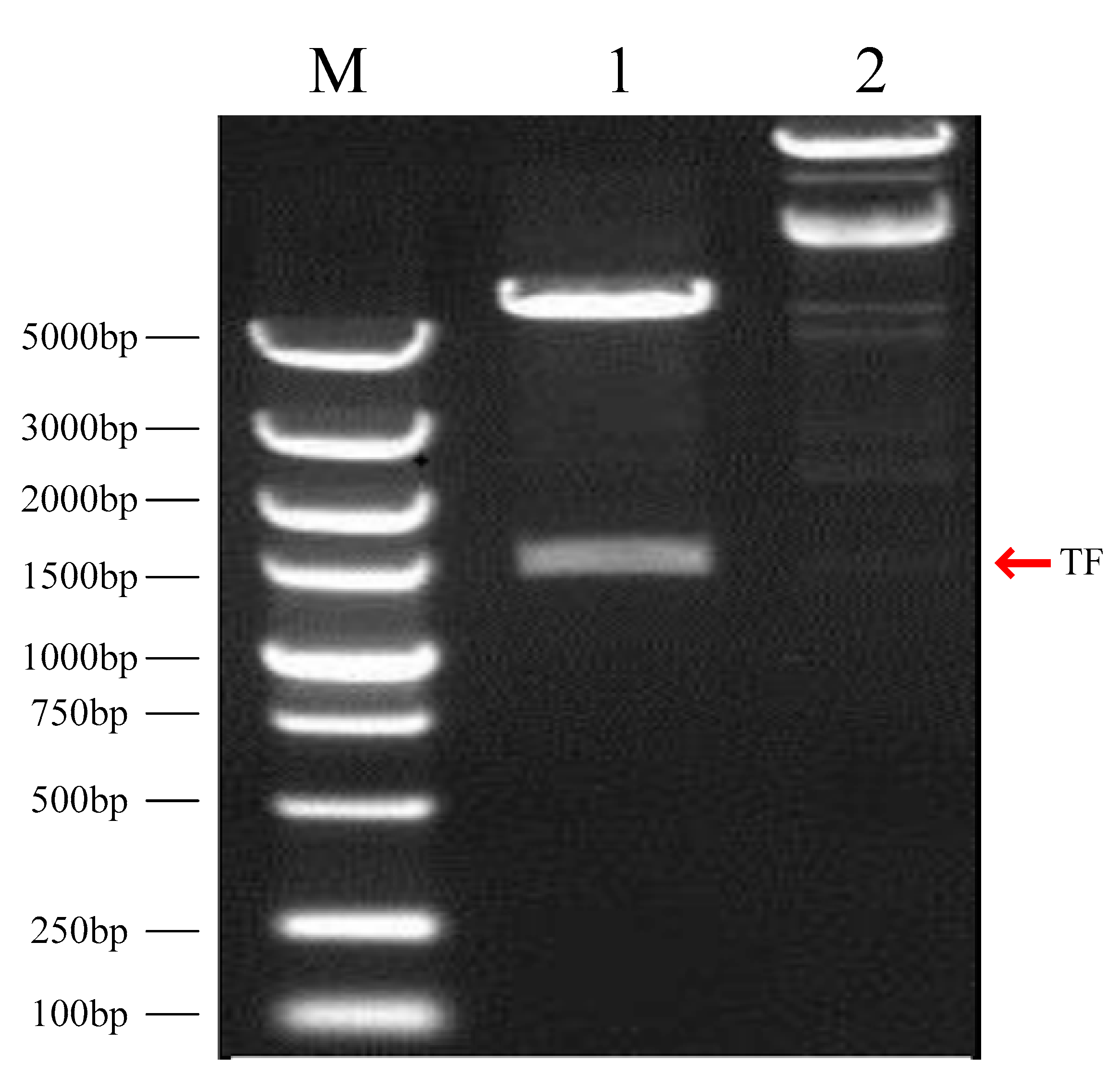
3.1. Construction of the pET28a (+)-Mitfa Prokaryotic Expression Vector
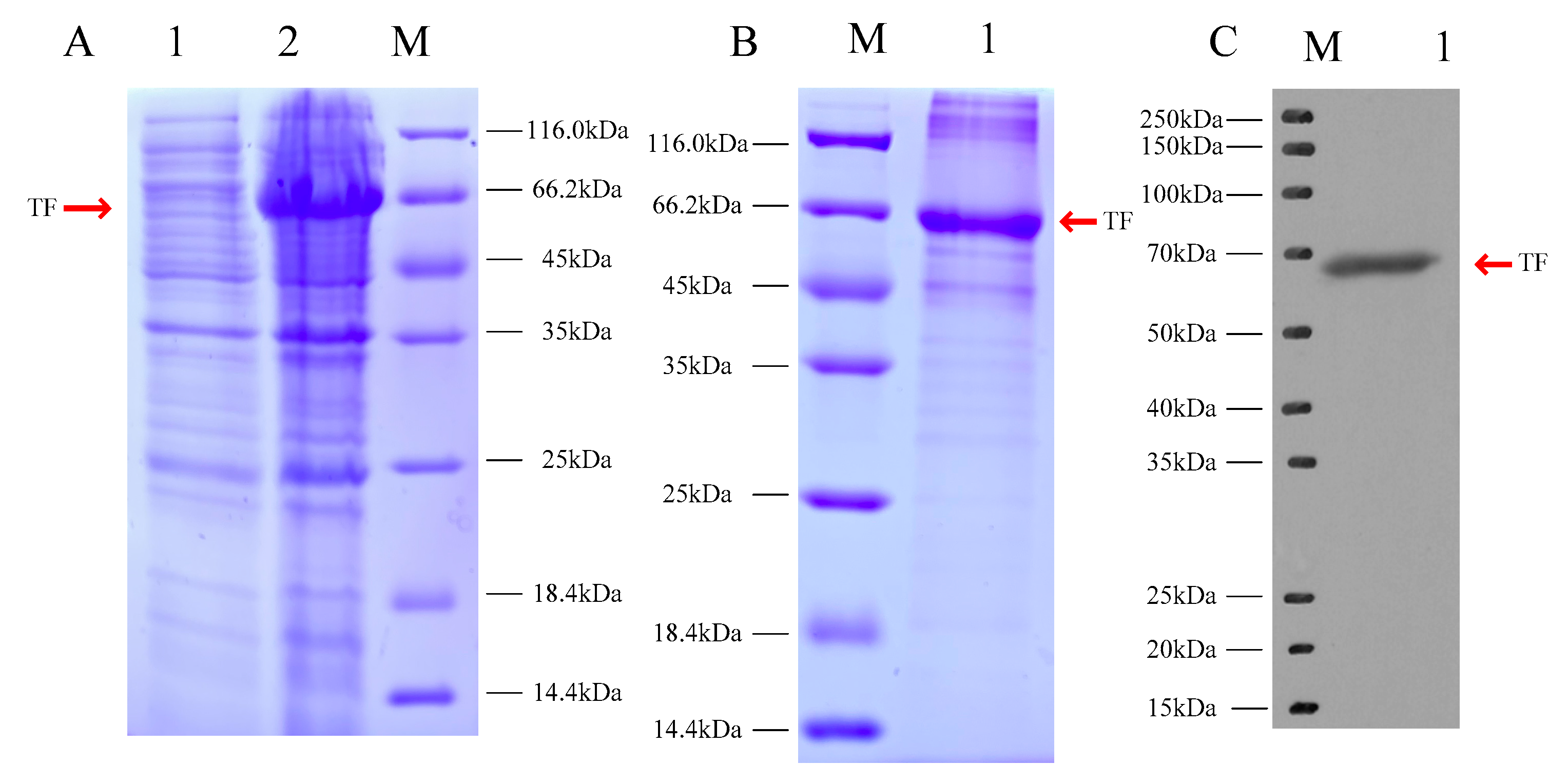
3.2. Expression and Purification of the Mitfa Fusion Protein
3.3. Screening and Identification of Proteins Interacting with Mitfa
3.4. Enrichment Analysis of the Mitfa Interaction Proteins
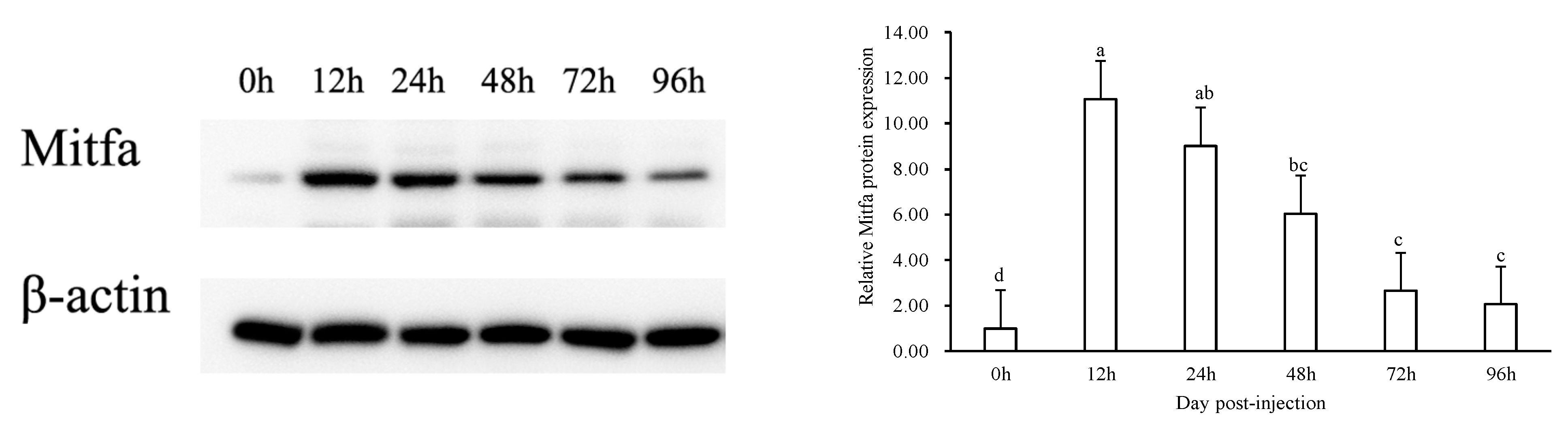
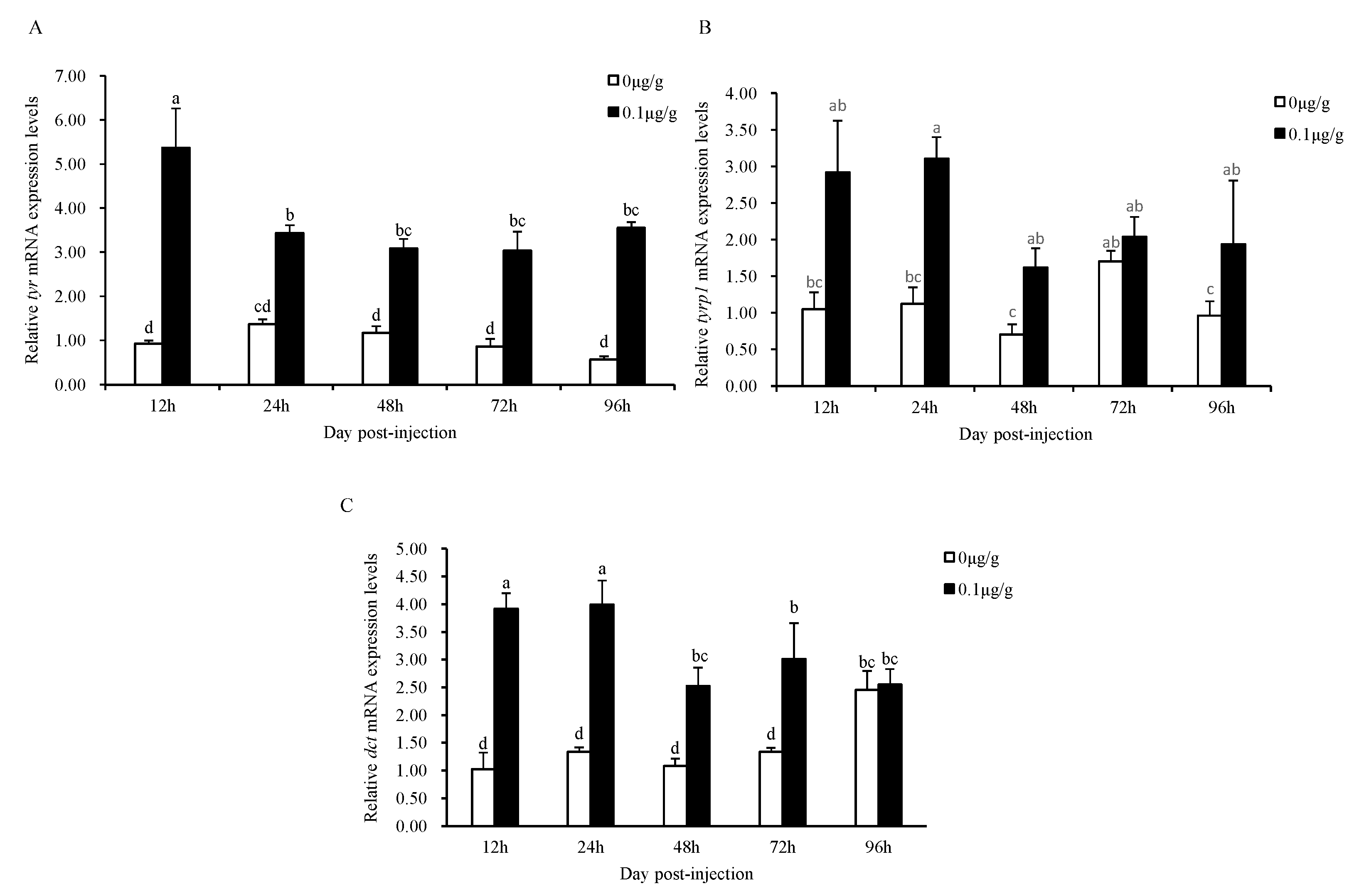
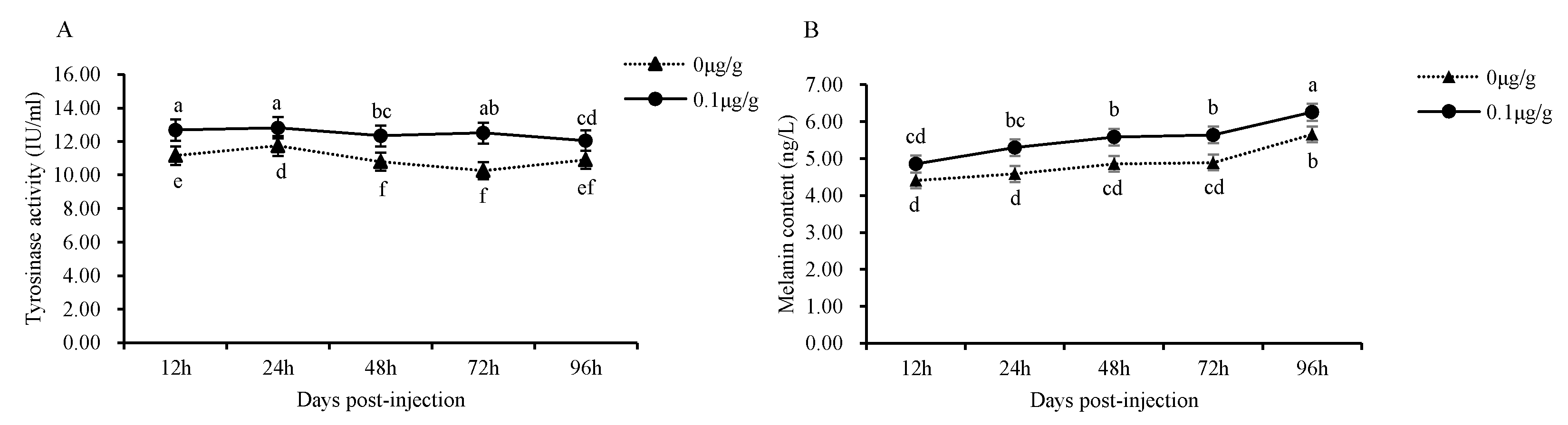
3.5. Administration of the Recombinant Mitfa Protein
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bauer, G.L.; Praetorius, C.; Bergsteinsdottir, K.; Hallsson, J.H.; Gisladottir, B.K.; Schepsky, A.; Steingrimsson, E. The role of mitf phosphorylation sites during coat color and eye development in mice analyzed by bacterial artificial chromosome transgene rescue. Genetics 2009, 183, 581–594. [Google Scholar] [CrossRef] [PubMed]
- Abrahamian, C.; Grimm, C. Endolysosomal cation channels and mitf in melanocytes and melanoma. Biomolecules 2021, 11, 1021. [Google Scholar] [CrossRef] [PubMed]
- Levy, C.; Khaled, M.; Fisher, D.E. Mitf master regulator of melanocyte development and melanoma oncogene. Trends. Mol. Med. 2006, 12, 406–414. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Zhu, W.Y. Mitf and melanocyte development, differentiation and functional regulation. J. Cell. Biol. 2002, 24, 346–351. [Google Scholar]
- Ma, X.Y.; Li, H.R.; Chen, Y.; Yang, J.; Chen, H.C.; Arnheiter, H.; Hou, L. The transcription factor mitf in RPE function and dysfunction. Prog. Retin. Eye Res. 2019, 73, 100766. [Google Scholar] [CrossRef]
- Minvielle, F.; Bed’hom, B.; Coville, J.L.; Ito, S.; Inoue-Murayama, M.; Gourichon, D. The “silver” Japanese quail and the mitf gene: Causal mutation, associated traits and homology with the “blue” chicken plumage. BMC Genet. 2010, 11, 15. [Google Scholar] [CrossRef]
- Wang, Y.; Li, S.M.; Jing, H.; Chen, S.Y.; Liu, P.Y. Mutations of tyr and mitf genes are associated with plumage colour phenotypes in geese. Asian Austral. J. Anim. 2014, 27, 778–783. [Google Scholar] [CrossRef]
- Goding, C.R.; Arnheiter, H. Mitf-the first 25 years. Gene Dev. 2019, 33, 983–1007. [Google Scholar] [CrossRef]
- Lister, J.A.; Robertson, C.P.; Lepage, T.; Johnson, S.L.; Raible, D.W. Nacre encodes a zebrafish microphthalmia-related protein that regulates neural-crest-derived pigment cell fate. Development 1999, 126, 3757–3767. [Google Scholar] [CrossRef]
- Wang, L.; Sun, F.; Wan, Z.Y.; Ye, B.Q.; Wen, Y.F.; Liu, H.M.; Yang, Z.T.; Pang, H.Y.; Meng, Z.N.; Fan, B.; et al. Genomic basis of striking fin shapes and colors in the fighting fish. Mol. Biol. Evol. 2021, 38, 3383–3396. [Google Scholar] [CrossRef]
- Jiang, B.J.; Wang, L.M.; Luo, M.K.; Fu, J.J.; Dong, Z.J. Molecular and functional analysis of the microphthalmia-associated transcription factor (mitf) gene duplicates in red tilapia. Comp. Biochem. Phys. A 2022, 271, 111257. [Google Scholar] [CrossRef] [PubMed]
- Lister, J.A.; Close, J.; Raible, D.W. Duplicate mitf genes in zebrafish: Complementary expression and conservation of melanogenic potential. Dev. Biol. 2001, 237, 333–344. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.H.; Wen, S.; Luo, C.; Zhang, Y.Q.; Tao, M.; Wang, D.W.; Deng, S.M.; Xiao, Y.M. Involvement of the mitfa gene in the development of pigment cell in Japanese ornamental koi carp (Cyprinus carpio L.). Genet. Mol. Res. 2015, 14, 2775–2784. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.Q.; Liu, J.H.; Peng, L.Y.; Ren, L.; Zhang, H.Q.; Zou, L.J.; Liu, W.B.; Xiao, Y.M. Comparative transcriptome analysis of molecular mechanism underlying gray-to-red body color formation in red crucian carp (Carassius auratus, red var.). Fish Physiol. Biochem. 2017, 43, 1387–1398. [Google Scholar] [CrossRef]
- Chen, X.D.; WU, G.D.; Song, H.M.; Wang, X.J.; Mou, X.D.; Liu, Y.; Liu, C.; Hu, Y.C. Structure and expression analysis of body color-related mitf gene in Amphilophus citrinellus. Prog. Fish. Sci. 2022, 43, 107–118. [Google Scholar]
- Jiang, B.J.; Fu, J.J.; Dong, Z.J.; Fang, M.; Zhu, W.B.; Wang, L.M. Maternal ancestry analyses of red tilapia strains based on D-loop sequences of seven tilapia populations. PeerJ 2019, 7, e7007. [Google Scholar] [CrossRef]
- Zhu, W.B.; Wang, L.M.; Dong, Z.J.; Chen, X.T.; Song, F.B.; Liu, N.; Yang, H.; Fu, J.J. Comparative transcriptome analysis identifies candidate genes related to skin color differentiation in red tilapia. Sci. Rep. 2016, 6, 31347. [Google Scholar] [CrossRef]
- Jiang, B.J.; Wang, L.M.; Luo, M.K.; Fu, J.J.; Dong, Z.J. Transcriptome analysis of skin color variation during and after overwintering of Malaysian red tilapia. Fish Physiol. Biochem. 2022, 48, 669–682. [Google Scholar] [CrossRef]
- Wang, L.M.; Jiang, B.J.; Zhu, W.B.; Fu, J.J.; Luo, M.K.; Liu, W.; Dong, Z.J. The role of melanocortin 1 receptor on melanogenesis pathway in skin color differentiation of red tilapia. Aquacult. Rep. 2022, 22, 100946. [Google Scholar] [CrossRef]
- Chen, Z.; Xue, H.F.; Zhao, H.Y.; Liu, K.; Li, H.; He, J.Z.; Wei, L.J. Cloning and expression of melanin-concentrating hormone 1 gene in Florida red tilapia (Oreochromis sp.). Nanfang Shuichan Kexue 2018, 14, 73–82. [Google Scholar]
- Yu, G.C.; Wang, L.G.; Han, Y.Y.; He, Q.Y. Clusterprofler: An R package for comparing biological themes among gene clusters. Omics 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Xie, C.; Mao, X.Z.; Huang, J.J.; Ding, Y.; Wu, J.M.; Dong, S.; Kong, L.; Gao, G.; Li, C.Y.; Wei, L.P. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39 (Suppl. S2), 316–322. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhang, Y.H.; Tang, Z.G.; Mao, J.W.; Kuang, Z.L.; Qin, C.B.; Li, W.S. Production of recombinant orange-spotted grouper (Epinephelus coioides) follicle-stimulating hormone (FSH) in single-chain form and dimer form by Pichia pastoris and their biological activities. Gen. Comp. Endocr. 2012, 178, 237–249. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.X.; Wang, B.; Liu, X.Z.; Xu, Y.J.; Shi, B.; Jiang, Y. Recombinant expression and bioactivity analysis of two leptin genes of Cynoglossus semilaevis. Shui Chan Xue Bao 2019, 43, 2279–2289. [Google Scholar]
- Tang, W.H.; Zhang, J.L.; Wang, Z.Y.; Hong, M.M. The cause of deviation made in determining the molecular weight of His-tag fusion proteins by SDS-PAGE. Sheng Li Xue Bao 2000, 26, 64–68. [Google Scholar]
- Cui, Y.C.; Song, Y.J.; Geng, Q.L.; Ding, Z.F.; Qin, Y.L.; Fan, R.W.; Dong, C.S.; Geng, J.J. The expression of krt2 and its effect on melanogenesis in alpaca skins. Acta Histochem. 2016, 118, 505–512. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.C.; Geng, J.J.; Geng, Q.L.; Ding, Z.F.; Dong, C.S. Human krt2 promotes melanogenesis in alpaca skin melanocytes in culture. Zhongguo Shengwu Huaxue Yu Fenzi Shengwu Xuebao 2017, 33, 303–310. [Google Scholar]
- Fan, R.; Xie, J.S.; Bai, J.M.; Wang, H.D.; Tian, X.; Bai, R.; Jia, X.Y.; Yang, L.; Song, Y.F.; Herrid, M.; et al. Skin transcriptome profiles associated with coat color in sheep. BMC Genet. 2013, 14, 389. [Google Scholar] [CrossRef]
- Wei, L.J.; Lv, Y.J.; Zou, H.; Wen, Y.H.; Gan, B.J.; Zhang, G.J.; Zhang, S.; Yan, X.Y.; Ye, X.C. Skin melanin synthesis of Jinbian carp and expression analysis of transport related genes. Nanfang Nongye Xuebao 2020, 51, 453–460. [Google Scholar]
- Chao, M.; Xu, H.; Huang, C.; Li, M.M.; Gong, X.T.; Tian, G.H.; Su, Y.; Li, Y.F.; Li, H.; Su, Y.Y. Research progress on color-related genes of fish. Hunan Wenli Xueyuan Xuebao Ziran Kexueban 2020, 32, 30–35. [Google Scholar]
- Si, Z.X.; Chen, H.L.; Xu, X.L.; Wang, J.; Wang, C.H. Effect of dct on pigmentation patterns in Oujiang color common carp. Zhongguo Shuichan Kexue 2020, 27, 605–612. [Google Scholar]
- Imes, D.L.; Geary, L.A.; Grahn, R.A.; Lyons, L.A. Albinism in the domestic cat (Felis catus) is associated with a tyrosinase (tyr) mutation. Anim. Genet. 2006, 37, 175–178. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, T.; Silversides, D.W.; Waymire, K.G.; Kwon, B.S.; Takeuchi, T.; Overbeek, P.A. Conserved cysteine to serine mutation in tyrosinase is responsible for the classical albino mutation in laboratory mice. Nucleic Acids Res. 1990, 18, 7293–7298. [Google Scholar] [CrossRef] [PubMed]
- Aigner, B.; Besenfelder, U.; Muller, M.; Brem, G. Tyrosinase gene variants in different rabbit strains. Mamm. Genome 2000, 11, 700–702. [Google Scholar] [CrossRef]
- Tobita-Teramoto, T.; Jang, G.Y.; Kino, K.; Salter, D.W.; Brumbaugh, J.; Akiyama, T. Autosomal albino chicken mutation (ca/ca) deletes hexanucleotide (-Δ GACTGG817) at a copper-binding site of the tyrosinase gene. Poultry Sci. 2000, 79, 46–50. [Google Scholar] [CrossRef]
- Bian, F.F.; Yang, X.F.; Ou, Z.J.; Luo, J.Z.; Tan, B.Z.; Yuan, M.R.; Chen, T.S.; Yang, R.B. Morphological characteristics and comparative transcriptome analysis of three different phenotypes of Pristella maxillaris. Front. Genet. 2013, 10, 698. [Google Scholar] [CrossRef]
- Chen, S.Y.; Chen, K.C.; Xu, J.M.; Li, F.W.; Ding, J.L.; Ma, Z.; Li, G.; Li, H. Insights into mRNA and long non-coding RNA profiling RNA sequencing in uterus of chickens with pink and blue eggshell colors. Front. Vet. Sci. 2021, 8, 736387. [Google Scholar] [CrossRef]
- Murashita, K.; Uji, S.; Yamamoto, T.; RøNnestad, I.; Kurokawa, T. Production of recombinant leptin and its effects on food intake in rainbow trout (Oncorhynchus mykiss). Comp. Biochem. Phys. B 2008, 150, 377–384. [Google Scholar] [CrossRef]
- Leedom, T.A.; Uchida, K.; Yada, T.; Iii, N.; Byatt, J.C.; Collier, R.J.; Tetsuya, H.; Gordon, G.E. Recombinant bovine growth hormone treatment of tilapia: Growth response, metabolic clearance, receptor binding and immunoglobulin production. Aquaculture 2002, 207, 359–380. [Google Scholar] [CrossRef]
- Wang, C.X.; Lu, B.Y.; Li, T.; Liang, G.Y.; Xu, M.M.; Liu, X.Y.; Tao, W.J.; Zhou, L.Y.; Kocher, T.D.; Wang, D.S. Nile tilapia: A model for studying teleost color patterns. J. Hered. 2021, 112, 469–484. [Google Scholar] [CrossRef] [PubMed]



| Primer | Sequences (5′-3′) |
|---|---|
| mitfa-his-F | ACCAAGTTTCTAACACATGA |
| mitfa-his -R | GTGGCATTATAAAGAGAGAC |
| mitfa F | AGGAAGCTGGAGCACGCA |
| mitfa R | GGGCAGTCACCAAGGACG |
| tyr F | TGTAATGCCACAGGGGAGGG |
| tyr R | TCCTGTCTCGTACTCGGGGA |
| tyrp1 F | TGCTCCACACCTTTACCGAC |
| tyrp1 R | TAACCGAGGTTTTCAGGGGC |
| dct F | TTCGAGTCGACACCAAACCC |
| dct R | TCAGGGGTCAAGGAGTGGAT |
| β-actin F | GTACCACCATGTACCCTGGC |
| β-actin R | TGAAGTTGTTGGGCGTTTGG |
| Sample ID | Spectrum | Number of Identification Spectra | Spectrum Resolution Rate | Number of Peptides | Number of Proteins | Unique-2 ** |
|---|---|---|---|---|---|---|
| His control protein | 468 | 1 | 0.21 | 1 | 1 | 0 |
| His-Mitfa fusion protein | 526 | 59 | 11.22 | 44 | 10 | 8 |
| Uniprot ID | Description | Gene Name | Coverage (%) | Unique Peptide | Score |
|---|---|---|---|---|---|
| I3JXH8_ORENI | Troponin T type 3a | tnnt3 | 3.29 | 1 | 1.46 |
| I3JKR2_ORENI | 2-phospho-D-glycerate hydro-lyase | eno3 | 2.39 | 1 | 2.00 |
| A0A669E6N4_ORENI | Decorin | dcn | 7.52 | 3 | 7.31 |
| XP_003447332.1 | Dopachrome tautomerase | dct | 13.78 | 7 | 13.93 |
| I3KRD3_ORENI | Keratin 15 | krt12 | 3.59 | 2 | 4.00 |
| A0A669B2B8_ORENI | Uncharacterized protein | tpm4 | 5.92 | 2 | 4.06 |
| A0A669BWG1_ORENI | IF rod domain-containing protein | krt4 | 4.26 | 2 | 4.34 |
| tr|I3JC76|I3JC76_ORENI | Tyrosinase | tyr | 11.45 | 1 | 2.00 |
| A0A669BU68_ORENI | Uncharacterized protein | myh1 | 3.58 | 7 | 14.45 |
| I3KFU7_ORENI | IF rod domain-containing protein | ina | 1.49 | 2 | 4.00 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, B.; Wang, L.; Fu, J.; Zhu, W.; Luo, M.; Dong, Z. Screening and Identification of Interacting Proteins of Mitfa in Red Tilapia. Fishes 2023, 8, 241. https://doi.org/10.3390/fishes8050241
Jiang B, Wang L, Fu J, Zhu W, Luo M, Dong Z. Screening and Identification of Interacting Proteins of Mitfa in Red Tilapia. Fishes. 2023; 8(5):241. https://doi.org/10.3390/fishes8050241
Chicago/Turabian StyleJiang, Bingjie, Lanmei Wang, Jianjun Fu, Wenbin Zhu, Mingkun Luo, and Zaijie Dong. 2023. "Screening and Identification of Interacting Proteins of Mitfa in Red Tilapia" Fishes 8, no. 5: 241. https://doi.org/10.3390/fishes8050241
APA StyleJiang, B., Wang, L., Fu, J., Zhu, W., Luo, M., & Dong, Z. (2023). Screening and Identification of Interacting Proteins of Mitfa in Red Tilapia. Fishes, 8(5), 241. https://doi.org/10.3390/fishes8050241







