Phylogenetic Analyses of Pristipomoides (Perciformes: Lutjanidae) Based on New Mitochondrial Genomes
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples, DNA Extraction, Amplification, and Sequencing
2.2. Sequence Assembly, Annotation, and Analysis
2.3. Phylogenetic Analysis
3. Results and Discussion
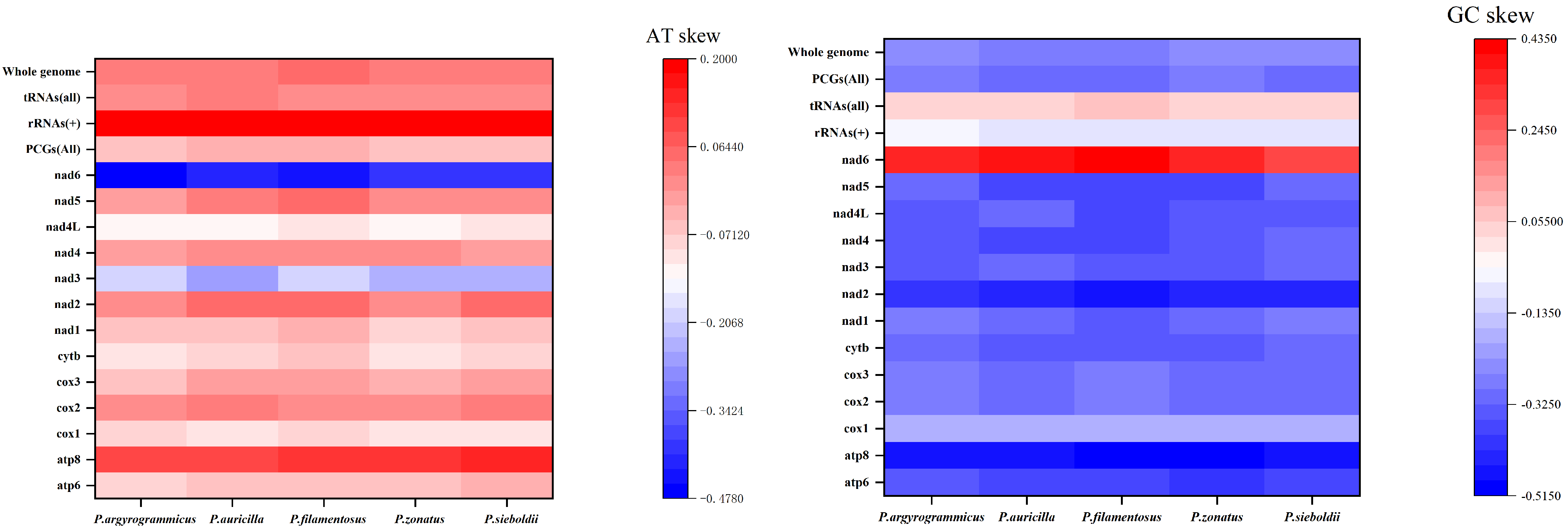
3.1. Genome Organization and Base Composition
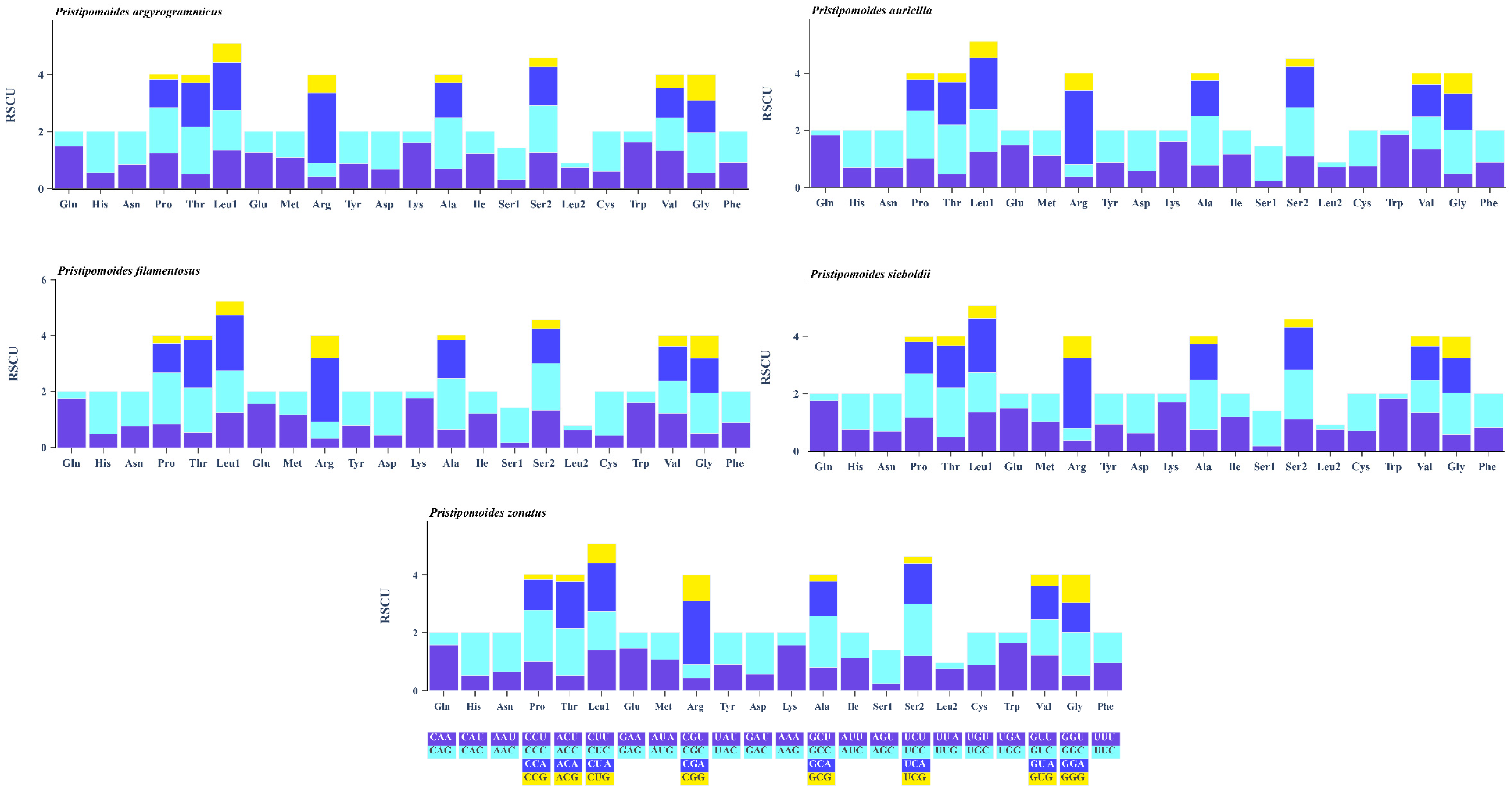
3.2. Protein Coding Genes and Codon Usage
3.3. Ka/Ks, Nucleotide Diversity
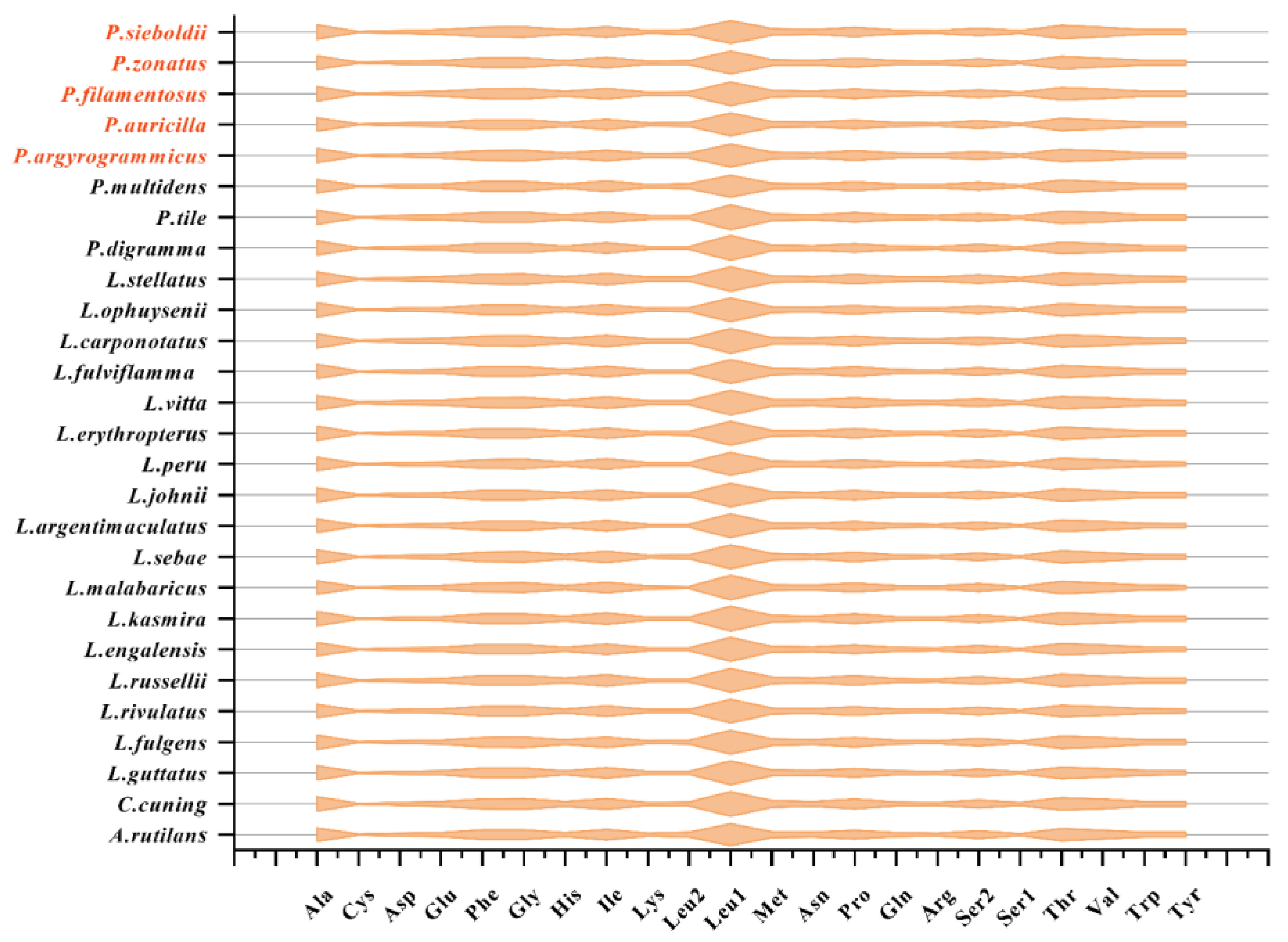
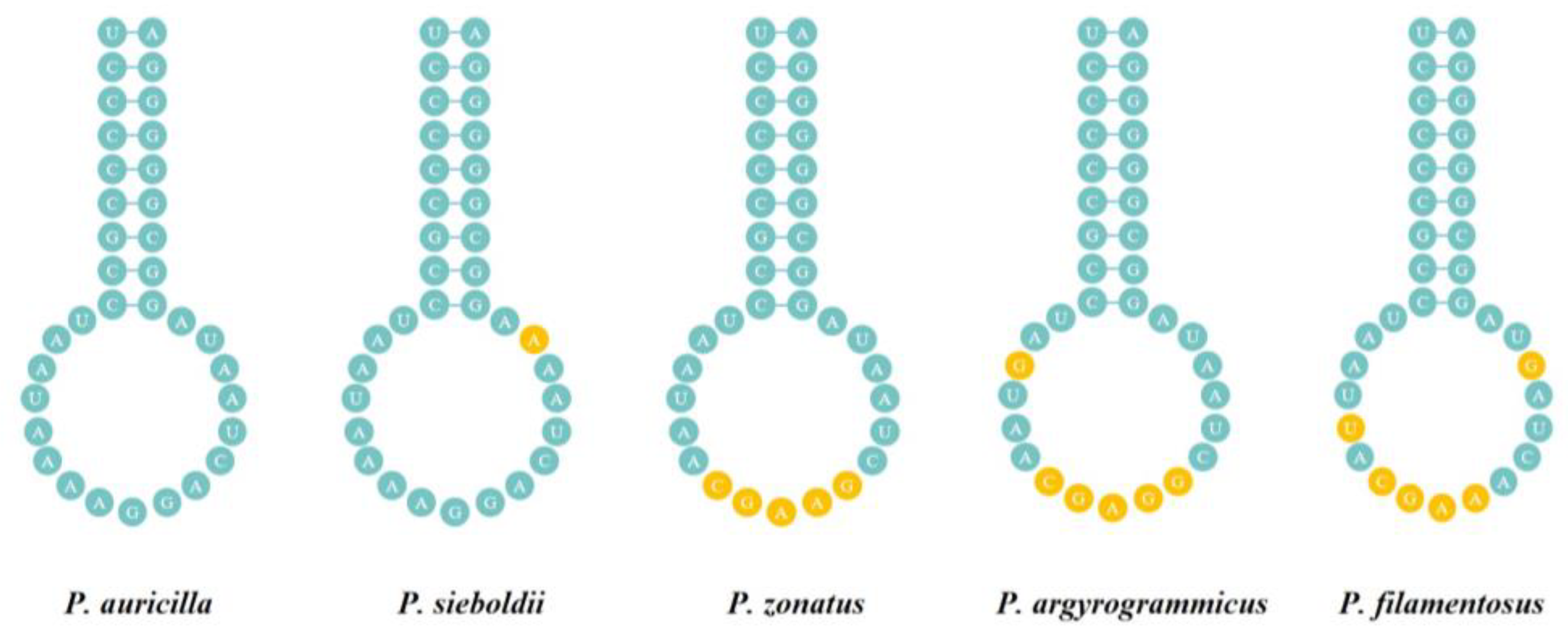
3.4. Transfer RNA and Ribosomal RNA Genes
3.5. Non-Coding Region and Overlap
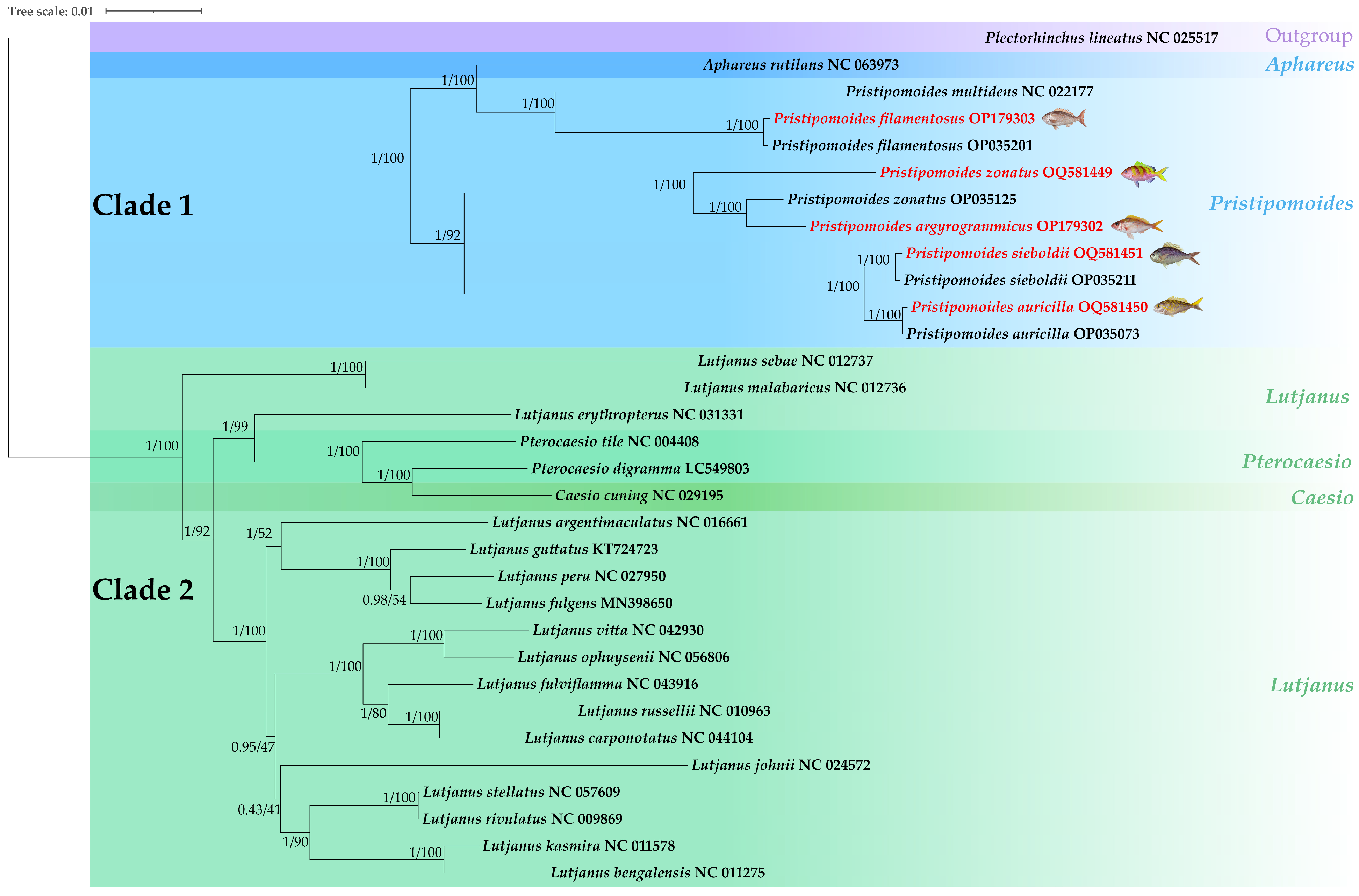
3.6. Phylogenetic Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Anderson, S.; Bankier, A.T.; Barrell, B.G.; de Bruijn, M.H.L.; Coulson, A.R.; Drouin, J.; Eperon, I.C.; Nierlich, D.P.; Roe, B.A.; Sanger, F.; et al. Sequence and Organization of the Human Mitochondrial Genome. Nature 1981, 290, 457–465. [Google Scholar] [CrossRef]
- Boore, J.L.; Brown, W.M. Big Trees from Little Genomes: Mitochondrial Gene Order as a Phylogenetic Tool. Curr. Opin. Genet. Dev. 1998, 8, 668–674. [Google Scholar] [CrossRef]
- Nagl, S.; Tichy, H.; Mayer, W.E.; Samonte, I.E.; McAndrew, B.J.; Klein, J. Classification and Phylogenetic Relationships of African Tilapiine Fishes Inferred from Mitochondrial DNA Sequences. Mol. Phylogenet. Evol. 2001, 20, 361–374. [Google Scholar] [CrossRef]
- Last, P.R.; Naylor, G.J.P.; Manjaji-Matsumoto, B.M. A Revised Classification of the Family Dasyatidae (Chondrichthyes: Myliobatiformes) Based on New Morphological and Molecular Insights. Zootaxa 2016, 4139, 345. [Google Scholar] [CrossRef]
- Satoh, T.P.; Miya, M.; Mabuchi, K.; Nishida, M. Structure and Variation of the Mitochondrial Genome of Fishes. BMC Genom. 2016, 17, 719. [Google Scholar] [CrossRef]
- Nelson, J.S.; Grande, T.C.; Wilson, M.V.H. Fishes of the World; John Wiley & Sons: Hoboken, NJ, USA, 2016; ISBN 978-1-119-22082-4. [Google Scholar]
- Allen, G.-R. FAO Species Catalogue: Volume 6—Snappers of the world: An annotated and illustrated catalogue of Lutjanid species known to date. In FAO Fisheries Synopsis; Food & Agriculture Organization: Kanagawa, Japan, 1985. [Google Scholar]
- Chu, C.; Rizman-Idid, M.; Chong, V.C. Phylogenetic Relationships of Selected Genera of Lutjanidae Inferred from Mitochondrial Regions, with a Note on the Taxonomic Status of Pinjalo pinjalo. Cienc. Mar. 2013, 39, 349–361. [Google Scholar] [CrossRef]
- Truong, O.; Vu, Q.; Dang, B.T. Phylogenetic Relationships of Emperors (Lethrinidae) and Snappers (Lutjanidae) in Vietnam Based on Mitochondrial DNA Sequences. In Proceedings of the International Conference on Biological, Environment and Food Engineering, Singapore, 15–16 May 2015. [Google Scholar]
- Kami, H.T. The Pristipomoides (Pisces: Lutjanidae) of Guam with Notes on Their Biology. Micronesica 1973, 9, 97–118. [Google Scholar]
- Lalitha, S. Primer Premier 5. Biotech Softw. Internet Rep. 2000, 1, 270–272. [Google Scholar] [CrossRef]
- Burland, T.G. DNASTAR’s Lasergene Sequence Analysis Software. In Bioinformatics Methods and Protocols; Misener, S., Krawetz, S.A., Eds.; Methods in Molecular BiologyTM; Humana Press: Totowa, NJ, USA, 1999; pp. 71–91. ISBN 978-1-59259-192-3. [Google Scholar]
- Chan, P.P.; Lin, B.Y.; Mak, A.J.; Lowe, T.M. TRNAscan-SE 2.0: Improved Detection and Functional Classification of Transfer RNA Genes. Nucleic Acids Res. 2021, 49, 9077–9096. [Google Scholar] [CrossRef]
- Laslett, D.; Canback, B. ARWEN: A program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics 2008, 24, 172–175. Available online: https://academic.oup.com/bioinformatics/article/24/2/172/228155 (accessed on 12 July 2022). [CrossRef]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.; Graham, M.; Van Domselaar, G.; Stothard, P. Proksee: In-Depth Characterization and Visualization of Bacterial Genomes. Nucleic Acids Res. 2023, 4, gkad326. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhang, Y.; Zhang, Z.; Zhu, J.; Yu, J. KaKs_Calculator 2.0: A Toolkit Incorporating Gamma-Series Methods and Sliding Window Strategies. Genom. Proteom. Bioinform. 2010, 8, 77–80. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Ferrer-Mata, A.; Sanchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sanchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. Available online: https://academic.oup.com/mbe/article/34/12/3299/4161815 (accessed on 12 March 2023). [CrossRef] [PubMed]
- Xiang, C.-Y.; Gao, F.; Jakovlić, I.; Lei, H.-P.; Hu, Y.; Zhang, H.; Zou, H.; Wang, G.-T.; Zhang, D. Using PhyloSuite for Molecular Phylogeny and Tree-Based Analyses. iMeta 2023, 2, e87. [Google Scholar] [CrossRef]
- Perna, N.T.; Kocher, T.D. Patterns of Nucleotide Composition at Fourfold Degenerate Sites of Animal Mitochondrial Genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Ranwez, V.; Douzery, E.J.P.; Cambon, C.; Chantret, N.; Delsuc, F. MACSE v2: Toolkit for the Alignment of Coding Sequences Accounting for Frameshifts and Stop Codons. Mol. Biol. Evol. 2018, 35, 2582–2584. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Talavera, G.; Castresana, J. Improvement of Phylogenies after Removing Divergent and Ambiguously Aligned Blocks from Protein Sequence Alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Nylander, J.A.A.; Ronquist, F.; Huelsenbeck, J.P.; Nieves-Aldrey, J. Bayesian Phylogenetic Analysis of Combined Data. Syst. Biol. 2004, 53, 47–67. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (ITOL) v3: An Online Tool for the Display and Annotation of Phylogenetic and Other Trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef] [PubMed]
- Boore, J.L. Animal Mitochondrial Genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Wang, R.; Sun, Y.; Xu, T. The Complete Mitochondrial Genome of the Small Yellow Croaker and Partitioned Bayesian Analysis of Sciaenidae Fish Phylogeny. Genet. Mol. Biol. 2012, 35, 191–199. [Google Scholar] [CrossRef]
- Kim, G.; Lee, J.-H.; Alam, M.J.; Lee, S.R.; Andriyono, S. Complete Mitochondrial Genome of Spanish Flag Snapper, Lutjanus carponotatus (Perciformes: Lutjanidae). Mitochondrial DNA Part B 2019, 4, 568–569. [Google Scholar] [CrossRef]
- Ojala, D.; Montoya, J.; Attardi, G. TRNA Punctuation Model of RNA Processing in Human Mitochondria. Nature 1981, 290, 470–474. [Google Scholar] [CrossRef]
- Osawa, S.; Ohama, T.; Jukes, T.H.; Watanabe, K. Evolution of the Mitochondrial Genetic Code I. Origin of AGR Serine and Stop Codons in Metazoan Mitochondria. J. Mol. Evol. 1989, 29, 202–207. [Google Scholar] [CrossRef]
- Sun, P.; Jiang, Y.; Yuan, X.; Zhang, H. The Complete Mitochondrial Genome of Lutjanus ophuysenii and Phylogenetic Analysis. Mitochondrial DNA Part B 2021, 6, 2396–2397. [Google Scholar] [CrossRef]
- Afriyie, G.; Wang, Z.; Dong, Z.; Ayisi Larbi, C.; Asiedu, B.; Guo, Y. Complete Mitochondrial Genome and Assembled DNA Barcoding Analysis of Lutjanus fulgens (Valenciennes, 1830) and Its Comparison with Other Lutjanus Species. Ecol. Evol. 2020, 10, 7971–7980. [Google Scholar] [CrossRef]
- Sun, C.-H.; Liu, H.-Y.; Xu, N.; Zhang, X.-L.; Zhang, Q.; Han, B.-P. Mitochondrial Genome Structures and Phylogenetic Analyses of Two Tropical Characidae Fishes. Front. Genet. 2021, 12, 627402. [Google Scholar] [CrossRef]
- Hurst, L.D. The Ka/Ks Ratio: Diagnosing the Form of Sequence Evolution. Trends Genet. 2002, 18, 486–487. [Google Scholar] [CrossRef]
- Zhang, Z.; Cheng, Q.; Ge, Y. The Complete Mitochondrial Genome of Rhynchocypris oxycephalus (Teleostei: Cyprinidae) and Its Phylogenetic Implications. Ecol. Evol. 2019, 9, 7819–7837. [Google Scholar] [CrossRef]
- Cai, X.; Qu, M.; Ding, S.; Wang, H.; Wang, H.; Hu, L.; Su, Y. Differentiation of Coral Trout (Plectropomus Leopardus) Based on an Analysis of Morphology and Complete Mitochondrial DNA: Are Cryptic Species Present? Acta Oceanol. Sin. 2013, 32, 40–46. [Google Scholar] [CrossRef]
- Wang, C.; Ye, P.; Liu, M.; Zhang, Y.; Feng, H.; Liu, J.; Zhou, H.; Wang, J.; Chen, X. Comparative Analysis of Four Complete Mitochondrial Genomes of Epinephelidae (Perciformes). Genes 2022, 13, 660. [Google Scholar] [CrossRef]
- Cornish-Bowden, A. Nomenclature for Incompletely Specified Bases in Nucleic Acid Sequences: Recommendations 1984. Nucleic Acids Res. 1985, 13, 3021–3030. [Google Scholar] [CrossRef]
- Campbell, N.J.; Barker, S.C. The Novel Mitochondrial Gene Arrangement of the Cattle Tick, Boophilus Microplus: Fivefold Tandem Repetition of a Coding Region. Mol. Biol. Evol. 1999, 16, 732–740. [Google Scholar] [CrossRef]
- Zhuang, X.; Qu, M.; Zhang, X.; Ding, S. A Comprehensive Description and Evolutionary Analysis of 22 Grouper (Perciformes, Epinephelidae) Mitochondrial Genomes with Emphasis on Two Novel Genome Organizations. PLoS ONE 2013, 8, e73561. [Google Scholar] [CrossRef]
- Sala, R.; Kusuma, A.B.; Pranata, B. Phylogenetic of Red Snapper (Lutjanidae) in Yapen Island Waters, Papua, Indonesia. Biodiversitas J. Biol. Divers. 2023, 24, 716–723. [Google Scholar] [CrossRef]
- Miller, T.L.; Cribb, T.H. Phylogenetic Relationships of Some Common Indo-Pacific Snappers (Perciformes: Lutjanidae) Based on Mitochondrial DNA Sequences, with Comments on the Taxonomic Position of the Caesioninae. Mol. Phylogenet. Evol. 2007, 44, 450–460. [Google Scholar] [CrossRef]
- Song, H.Y.; Jung, Y.-H.; Kim, B.; Choi, Y.J.; Nguyen, T.V.; Lee, D.-S. Complete Mitochondrial Genome of the Double-Lined Fusileer, Pterocaesio Digramma (Perciformes, Caesionidae): Mitogenome Characterization and Phylogenetic Analysis. Mitochondrial DNA Part B 2020, 5, 2617–2618. [Google Scholar] [CrossRef]
- Betancur-R, R.; Wiley, E.O.; Arratia, G.; Acero, A.; Bailly, N.; Miya, M.; Lecointre, G.; Ortí, G. Phylogenetic Classification of Bony Fishes. BMC Evol. Biol. 2017, 17, 162. [Google Scholar] [CrossRef]
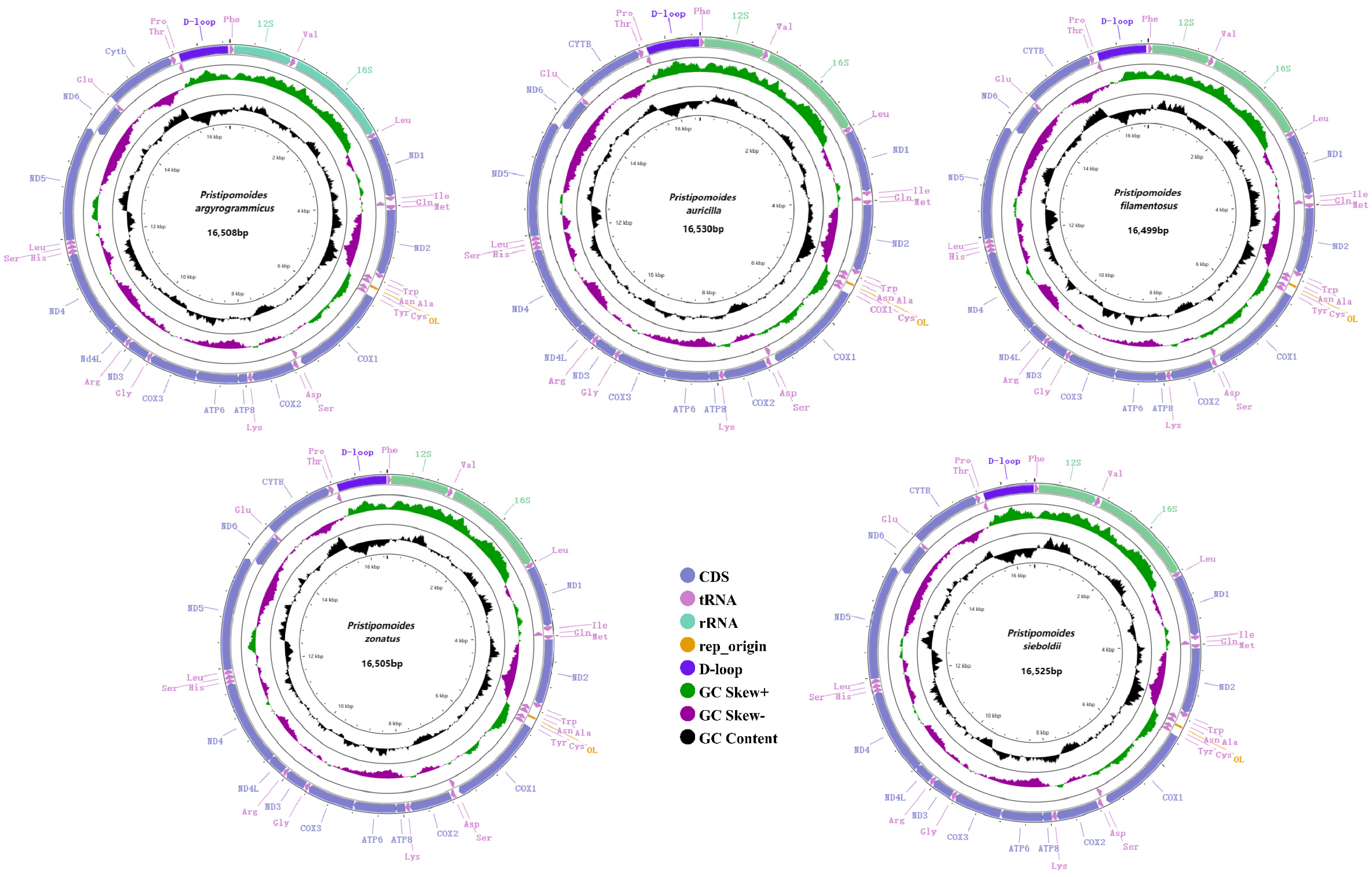



| Species | Length (bp) | GenBank Accession Number | Sampling Location |
|---|---|---|---|
| P. argyrogrammicus | 16,508 | OP179302 | Howard Shoal |
| P. auricilla | 16,530 | OQ581450 | Howard Shoal |
| P. filamentosus | 16,499 | OP179303 | Penguin Shoal |
| P. zonatus | 16,505 | OQ581449 | Dongsha Islands |
| P. sieboldii | 16,525 | OQ581451 | Shenhu Shoal |
| Gene | Strand | Length (bp) | Start Codons | Stop Codons | Anticodon | Intergenic Nucleotide * (bp) |
|---|---|---|---|---|---|---|
| P. argyrogrammicus/P. filamentosus/P. auricilla/P. sieboldii/P. zonatus | ||||||
| trnF | H | 68/68/69/69/68 | GAA | 0 | ||
| rrnS | H | 953/949/953/955/953 | 0/1 | |||
| trnV | H | 73/73/73/73/73 | TAC | 24/28 | ||
| rrnL | H | 1673/1673/1679/1679/1672 | 0 | |||
| trnL2 | H | 74/74/74/74/74 | TAA | 0/2 | ||
| nad1 | H | 975/975/975/975/975 | ATG/ATG/ATG/ATG/ATG | TAA/TAA/TAA/TAA/TAA | 2/3 | |
| trnI | H | 70/70/70/70/70 | GAT | −1/0 | ||
| trnQ | L | 71/71/71/71/71 | TTG | −1 | ||
| trnM | H | 69/69/69/69/69 | CAT | 0 | ||
| nad2 | H | 1047/1047/1047/1047/1047 | ATG/ATG/ATG/ATG/ATG | TAA/TAA/TAA/TAA/TAA | −1 | |
| trnW | H | 73/73/73/73/73 | TCA | 0 | ||
| trnA | L | 69/69/69/69/69 | TGC | 1 | ||
| trnN | L | 73/73/73/73/73 | GTT | 4 | ||
| OL | H | 35/35/35/35/35 | 0 | |||
| trnC | L | 67/67/67/67/67 | GCA | 0 | ||
| trnY | L | 70/71/71/71/70 | GTA | 1 | ||
| cox1 | H | 1560/1551/1551/1551/1560 | GTG/GTG/GTG/GTG/GTG | AGG/TAA/TAA/TAA/AGG | −9/1 | |
| trnS2 | L | 71/71/72/72/71 | TGA | 3 | ||
| trnD | H | 72/72/73/73/72 | GTC | 8 | ||
| cox2 | H | 691/691/691/691/691 | ATG/ATG/ATG/ATG/ATG | T/T/T/T/T | 0 | |
| trnK | H | 75/75/75/75/75 | TTT | 1 | ||
| atp8 | H | 168/168/168/168/168 | ATG/ATG/ATG/ATG/ATG | TAA/TAA/TAA/TAA/TAA | −10 | |
| atp6 | H | 684/684/684/684/684 | ATG/ATG/ATG/ATG/ATG | TAA/TAA/TAA/TAA/TAA | −1 | |
| cox3 | H | 786/786/786/786/786 | ATG/ATG/ATG/ATG/ATG | TAA/TAA/TAA/TAA/TAA | −1 | |
| trnG | H | 72/72/72/72/72 | TCC | 0 | ||
| nad3 | H | 351/351/351/351/351 | ATG/ATG/ATG/ATG/ATG | TAG/TAG/TAG/TAG/TAG | −2 | |
| trnR | H | 70/69/70/71/70 | TCG | 0/1 | ||
| nad4L | H | 297/297/297/297/297 | ATG/ATG/ATG/ATG/ATG | TAA/TAA/TAA/TAA/TAA | −7 | |
| nad4 | H | 1381/1381/1381/1381/1381 | ATG/ATG/ATG/ATG/ATG | T/T/T/T/T | 0 | |
| trnH | H | 69/69/69/69/69 | GTG | 0/2 | ||
| trnS1 | H | 67/68/68/68/68 | GCT | 4/5 | ||
| trnL1 | H | 73/73/73/73/73 | TAG | 0 | ||
| nad5 | H | 1839/1839/1839/1839/1839 | ATG/ATG/ATG/ATG/ATG | TAA/TAA/TAA/TAA/TAA | −4 | |
| nad6 | L | 522/522/522/522/522 | ATG/ATG/ATG/ATG/ATG | TAG/TAG/TAG/TAA/TAG | 0 | |
| trnE | L | 69/69/69/69/69 | TTC | 4/5 | ||
| cytb | H | 1141/1143/1143/1143/1141 | ATG/ATG/ATG/ATG/ATG | T/TAG/TAG/TAG/T | −1/0 | |
| trnT | H | 72/72/72/72/72 | TGT | −1 | ||
| trnP | L | 71/70/70/70/71 | TGG | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, C.; Li, D.; Zhang, Y.; Péré, M.; Zhuang, Z.; Liu, J.; Zhou, H.; Chen, X. Phylogenetic Analyses of Pristipomoides (Perciformes: Lutjanidae) Based on New Mitochondrial Genomes. Fishes 2023, 8, 343. https://doi.org/10.3390/fishes8070343
Liu C, Li D, Zhang Y, Péré M, Zhuang Z, Liu J, Zhou H, Chen X. Phylogenetic Analyses of Pristipomoides (Perciformes: Lutjanidae) Based on New Mitochondrial Genomes. Fishes. 2023; 8(7):343. https://doi.org/10.3390/fishes8070343
Chicago/Turabian StyleLiu, Chunhui, Dezhao Li, Yue Zhang, Maxime Péré, Zhibo Zhuang, Jingyu Liu, Haolang Zhou, and Xiao Chen. 2023. "Phylogenetic Analyses of Pristipomoides (Perciformes: Lutjanidae) Based on New Mitochondrial Genomes" Fishes 8, no. 7: 343. https://doi.org/10.3390/fishes8070343
APA StyleLiu, C., Li, D., Zhang, Y., Péré, M., Zhuang, Z., Liu, J., Zhou, H., & Chen, X. (2023). Phylogenetic Analyses of Pristipomoides (Perciformes: Lutjanidae) Based on New Mitochondrial Genomes. Fishes, 8(7), 343. https://doi.org/10.3390/fishes8070343






