Identification of a New Insulin-like Growth Factor 3 (igf3) in Turbot (Scophthalmus maximus): Comparison and Expression Analysis of IGF System Genes during Gonadal Development
Abstract
1. Introduction
2. Materials and Methods
2.1. Fish Sampling
2.2. Histological Observation
2.3. igf3 Isolation
2.4. Phylogenetic and Bioinformatics Analyses
2.5. Quantitative Real-Time PCR (qPCR)
2.6. Statistical Analysis
3. Results
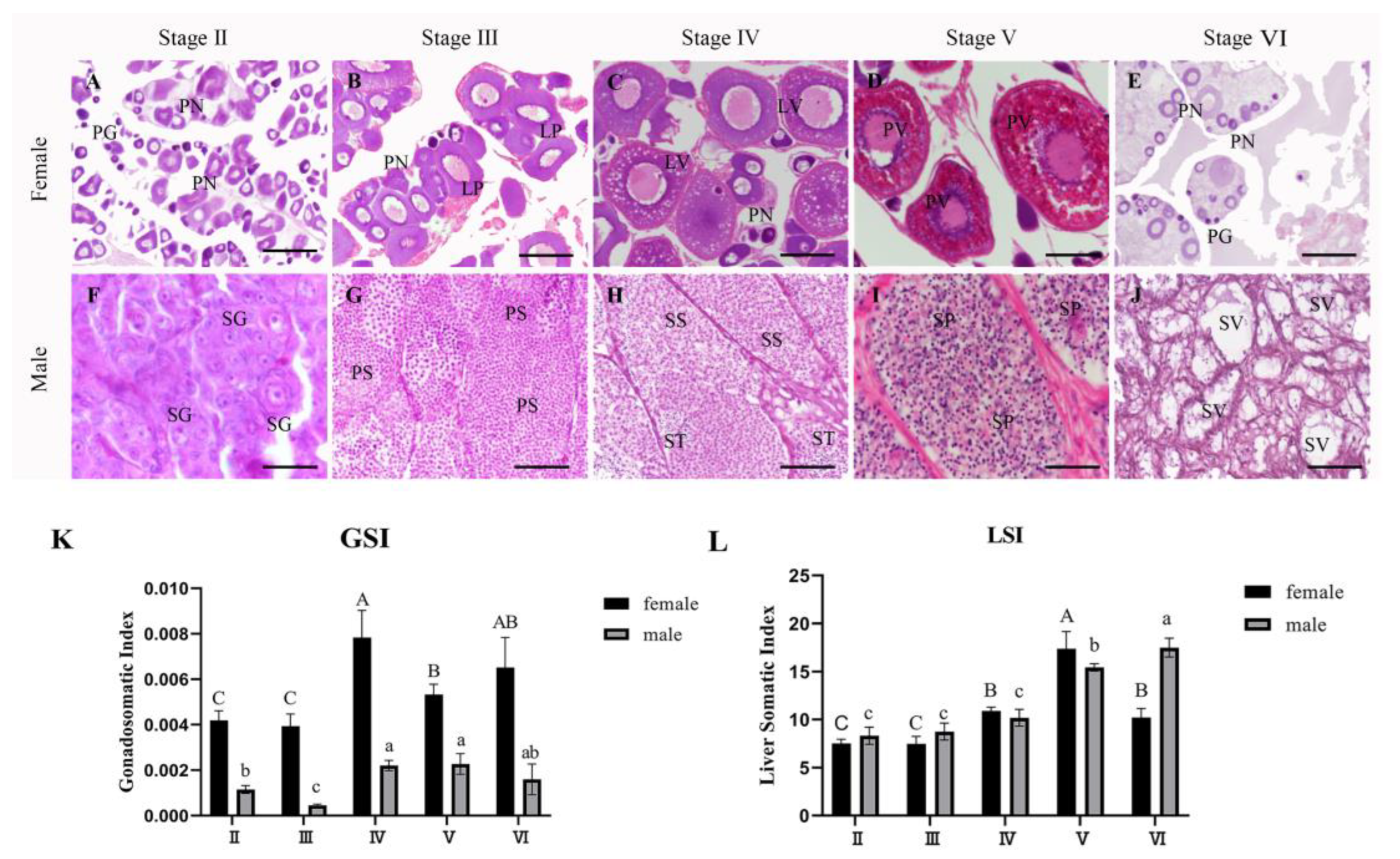
3.1. Gonadal Histology and Reproductive Stages Assessment
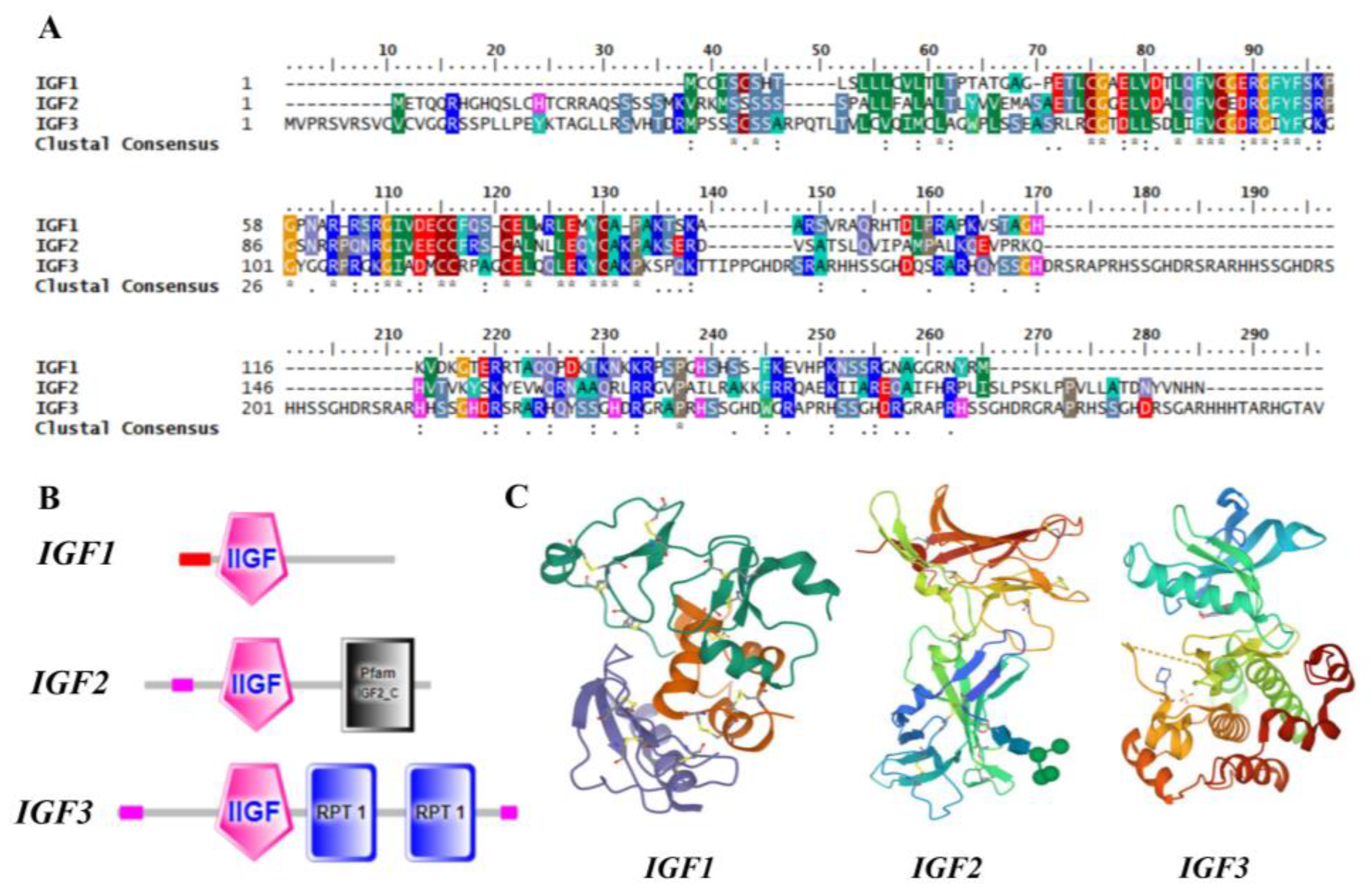
3.2. Molecular Cloning of Turbot igf3
3.3. Cross-Species Changes in igfs Members and Copy Numbers
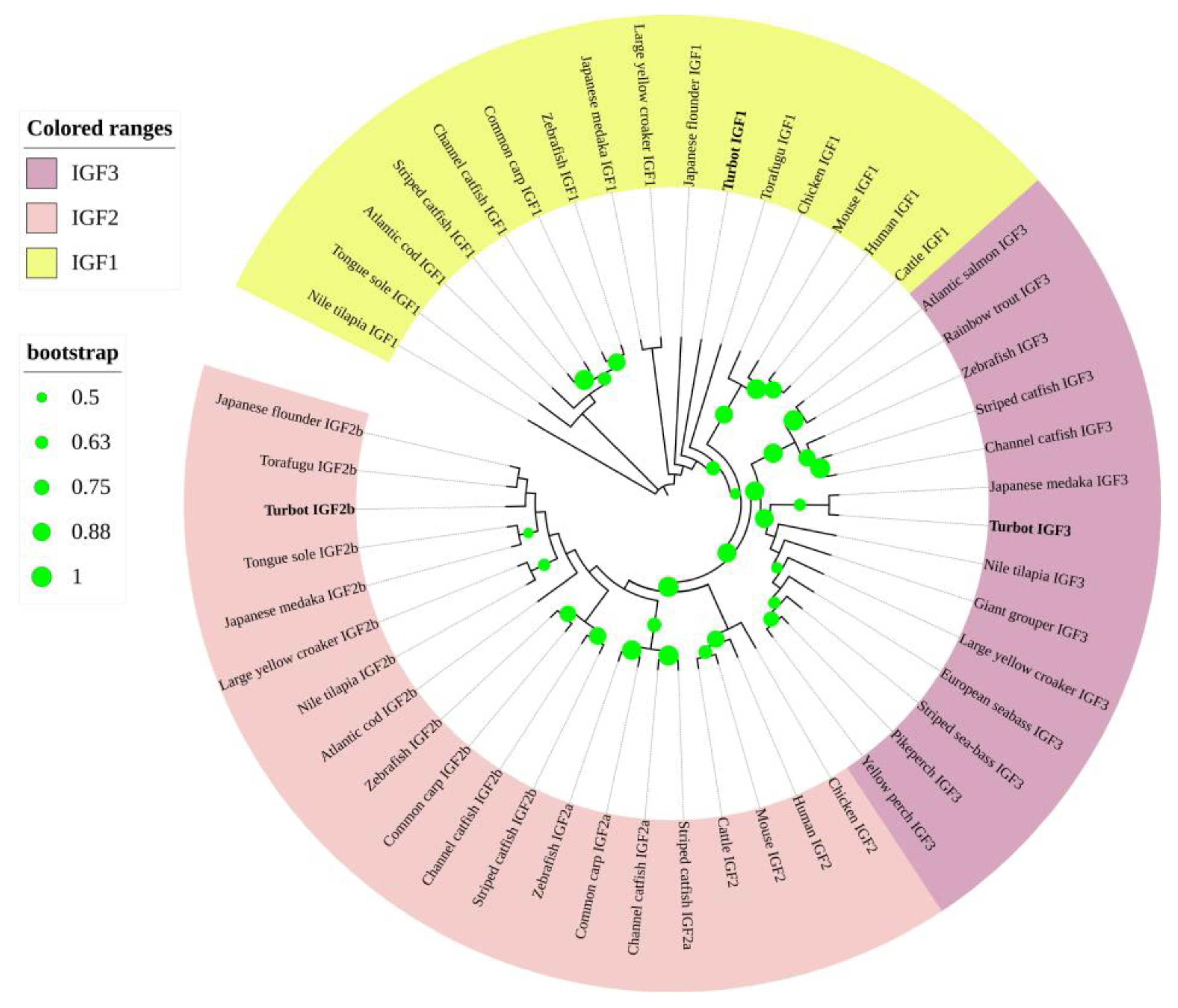
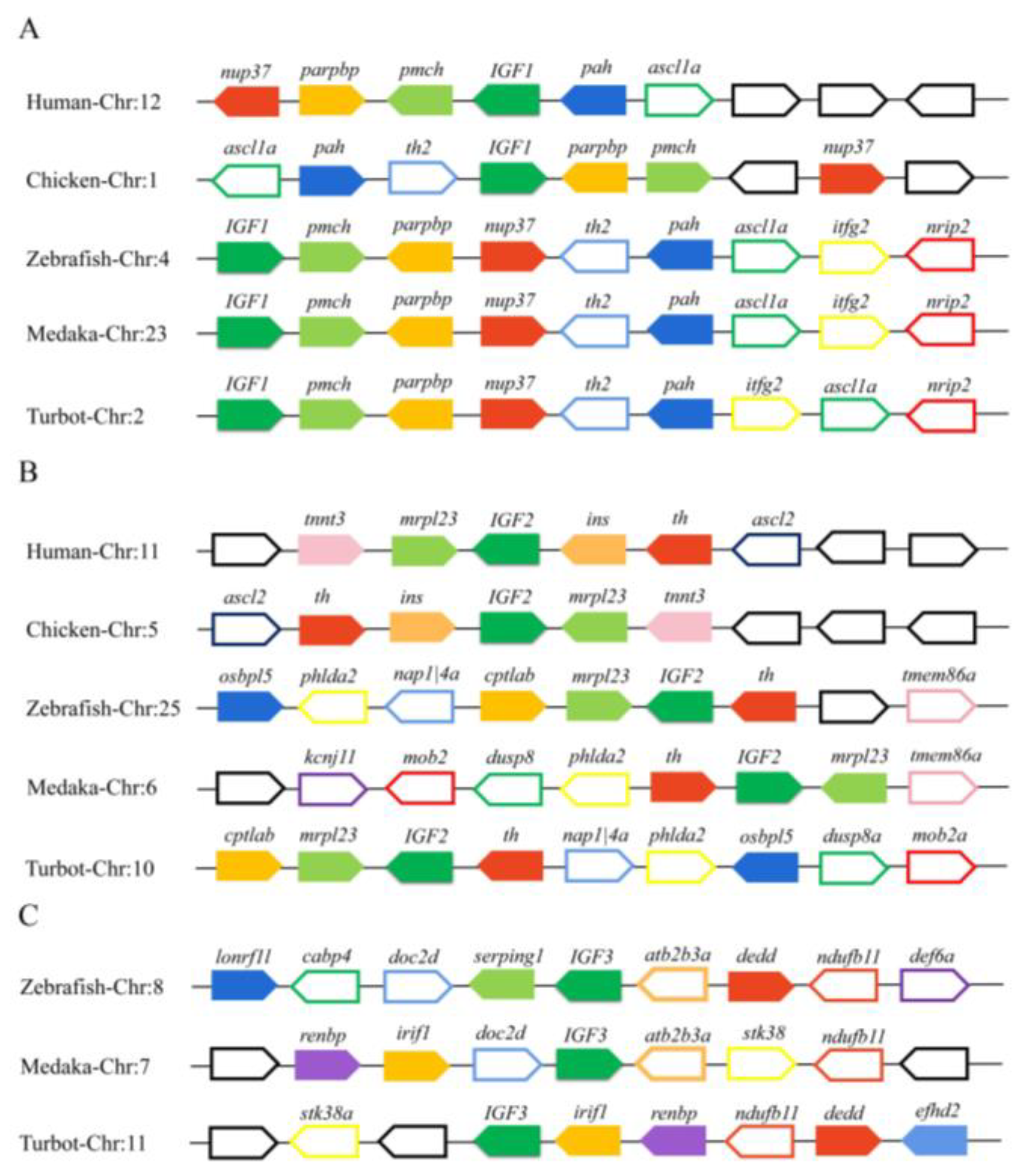
3.4. Phylogenetic and Synteny Analysis
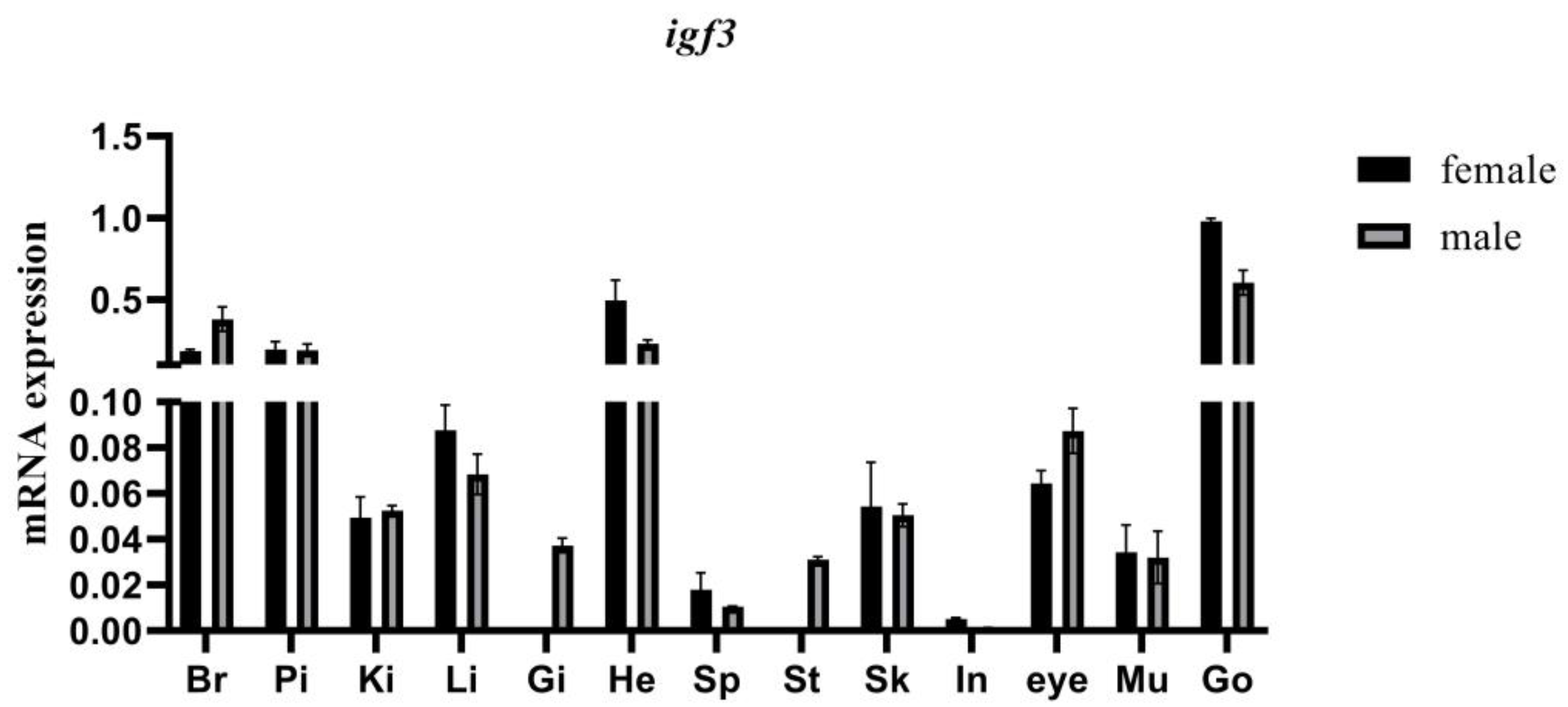
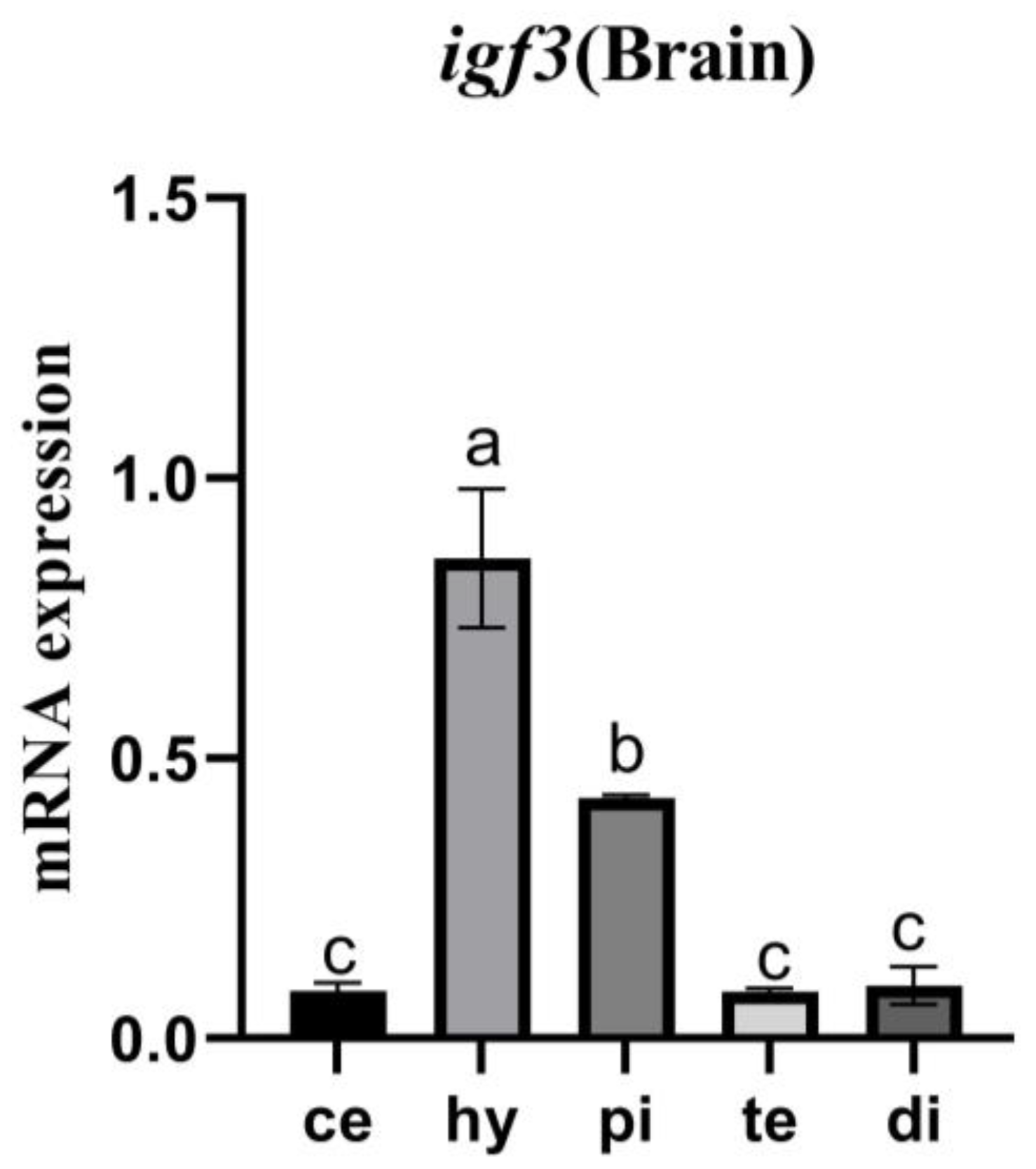
3.5. Tissue Expression Pattern of igf3 mRNA
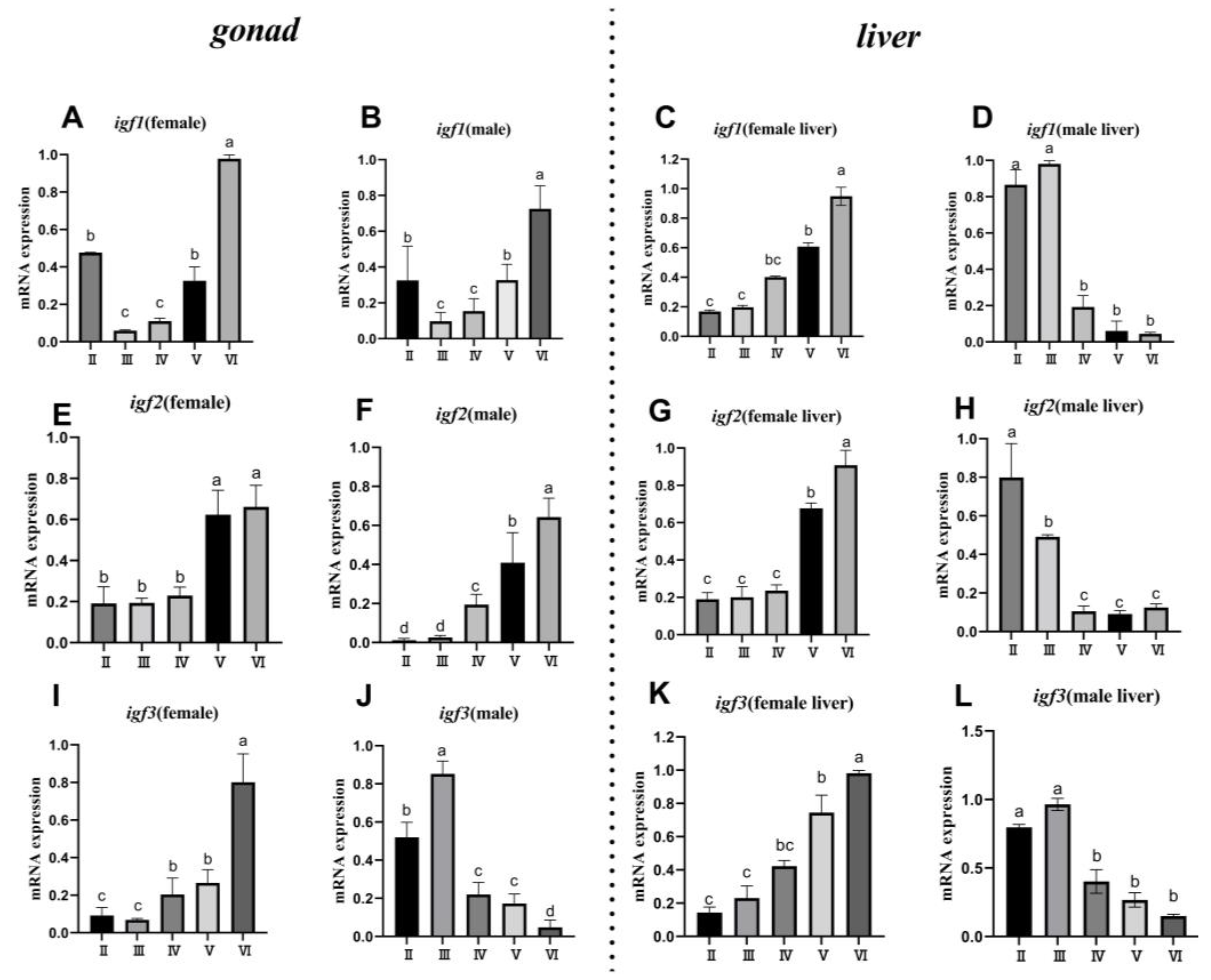
3.6. Expression Patterns igfs in Gonad and Liver during Gonadal Development
3.6.1. Gonad
3.6.2. Liver
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Biran, J.; Levavi-Sivan, B. Endocrine Control of Reproduction Fish. Encycl. Reprod. 2018, 3, 362–368. [Google Scholar] [CrossRef]
- Xue, R.; Wang, X.; Xu, S.; Liu, Y.; Feng, C.; Zhao, C.; Liu, Q.; Li, J. Expression profile and localization of vitellogenin mRNA and protein during ovarian development in turbot (Scophthalmus maximus). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2018, 226, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Della Torre, S.; Benedusi, V.; Fontana, R.; Maggi, A. Energy metabolism and fertility: A balance preserved for female health. Nat. Rev. Endocrinol. 2014, 10, 13–23. [Google Scholar] [CrossRef]
- Kwintkiewicz, J.; Giudice, L.C. The interplay of insulin-like growth factors, gonadotropins, and endocrine disruptors in ovarian follicular development and function. Semin. Reprod. Med. 2009, 27, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Shi, B.; Shao, P.; Shao, C.; Wang, C.; Li, J.; Liu, X.; Ma, X.; Zhao, X. The identification of miRNAs that regulate ovarian maturation in Cynoglossus semilaevis. Aquaculture 2022, 555, 738250. [Google Scholar] [CrossRef]
- Mahardini, A.; Yamauchi, C.; Takeuchi, Y.; Rizky, D.; Takekata, H.; Takemura, A. Changes in mRNA abundance of insulin-like growth factors in the brain and liver of a tropical damselfish, Chrysiptera cyanea, in relation to seasonal and food-manipulated reproduction. Gen. Comp. Endocrinol. 2018, 269, 112–121. [Google Scholar] [CrossRef] [PubMed]
- Reinecke, M. Insulin-Like Growth Factors and Fish Reproduction. Biol. Reprod. 2010, 82, 656–661. [Google Scholar] [CrossRef]
- Wen, H.; Qi, Q.; Hu, J.; Si, Y.; He, F.; Li, J. Expression analysis of the insulin-like growth factors I and II during embryonic and early larval development of turbot (Scophthalmus maximus). J. Ocean Univ. China 2015, 14, 309–316. [Google Scholar] [CrossRef]
- Nelson, S.N.; Van Der Kraak, G. Characterization and regulation of the insulin-like growth factor (IGF) system in the zebrafish (Danio rerio) ovary. Gen. Comp. Endocrinol. 2010, 168, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Kwok, H.F.; So, W.K.; Wang, Y.; Ge, W. Zebrafish gonadotropins and their receptors: I. Cloning and characterization of zebrafish follicle-stimulating hormone and luteinizing hormone receptors—Evidence for their distinct functions in follicle development. Biol. Reprod. 2005, 72, 1370–1381. [Google Scholar] [CrossRef]
- Li, J.; Liu, Z.; Wang, D.; Cheng, C.H. Insulin-like growth factor 3 is involved in oocyte maturation in zebrafish. Biol. Reprod. 2011, 4, 476–486. [Google Scholar] [CrossRef] [PubMed]
- Berishvili, G.; Baroiller, J.F.; Eppler, E.; Reinecke, M. Insulin-like growth factor-3 (IGF-3) in male and female gonads of the tilapia: Development and regulation of gene expression by growth hormone (GH) and 17alpha-ethinylestradiol (EE2). Gen. Comp. Endocrinol. 2010, 167, 128–134. [Google Scholar] [CrossRef]
- Wang, D.S.; Jiao, B.; Hu, C.; Huang, X.; Liu, Z.; Cheng, C.H. Discovery of a gonad-specific IGF subtype in teleost. Biochem. Biophys. Res. Commun. 2008, 367, 336–341. [Google Scholar] [CrossRef]
- Yang, H.; Chen, H.; Zhao, H.; Liu, L.; Xie, Z.; Xiao, L.; Li, S.; Zhang, Y.; Lin, H. Molecular cloning of the insulin-like growth factor 3 and difference in the expression of igf genes in orange-spotted grouper (Epinephelus coioides). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2015, 186, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Jing, Q.; Gao, Y.; Huang, B. Involvement and expression of growth hormone/insulin-like growth factor member mRNAs in the ovarian development of turbot (Scophthalmus maximus). Fish Physiol. Biochem. 2019, 45, 955–964. [Google Scholar] [CrossRef]
- Zhao, C.; Liu, Q.; Xu, S.; Xiao, Y.; Wang, W.; Yang, J.; Yang, Y.; Wang, Y.; Song, Z.; Li, J. Identification of type A spermatogonia in turbot (Scophthalmus maximus) using a new cell-surface marker of Lymphocyte antigen 75 (ly75/CD205). Theriogenology 2018, 113, 137–145. [Google Scholar] [CrossRef]
- Song, F.; Wang, L.; Zhu, W.; Fu, J.; Dong, J.; Dong, Z. A Novel igf3 Gene in Common Carp (Cyprinus carpio): Evidence for Its Role in Regulating Gonadal Development. PLoS ONE 2016, 11, e0168874. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Zhong, Y.; Zhao, Y.; Xie, W.; Guo, J.; Gui, L.; Li, M. Characterization and expression analysis of gonad specific igf3 in the medaka ovary. Aquac. Fish. 2020, 369, 13–18. [Google Scholar] [CrossRef]
- Gu, W.; Yang, Y.; Ning, C.; Wang, Y.; Hu, J.; Zhang, M.; Kuang, S.; Sun, Y.; Li, Y.; Zhang, Y.; et al. Identification and characteristics of insulin-like growth factor system in the brain, liver, and gonad during development of a seasonal breeding teleost, Pampus argenteus. Gen. Comp. Endocrinol. 2021, 300, 113645. [Google Scholar] [CrossRef]
- Meng, Z.; Hu, P.; Lei, J.; Jia, Y. Expression of insulin-like growth factors at mRNA levels during the metamorphic development of turbot (Scophthalmus maximus). Gen. Comp. Endocrinol. 2016, 235, 11–17. [Google Scholar] [CrossRef]
- Schulz, B.; Altendorf-Hofmann, A.; Kirchner, T.; Katenkamp, D.; Petersen, I.; Knosel, T. Loss of CD34 and high IGF2 are associated with malignant transformation in solitary fibrous tumors. Pathol. Res. Pract. 2014, 210, 92–97. [Google Scholar] [CrossRef]
- Dahle, R.; Taranger, G.L.; Karlsen, Ø.; Kjesbu, O.S.; Norberg, B. Gonadal development and associated changes in liver size and sexual steroids during the reproductive cycle of captive male and female Atlantic cod (Gadus morhua L.). Comp. Biochem. Phys. A 2003, 136, 641–653. [Google Scholar] [CrossRef] [PubMed]
- Morais, R.; Crespo, D.; Nobrega, R.H.; Lemos, M.S.; van de Kant, H.J.G.; de Franca, L.R.; Male, R.; Bogerd, J.; Schulz, R.W. Antagonistic regulation of spermatogonial differentiation in zebrafish (Danio rerio) by Igf3 and Amh. Mol. Cell. Endocrinol. 2017, 454, 112–124. [Google Scholar] [CrossRef]
- Yang, G.; Liang, X.; Xu, S.; Cai, H.; Ma, L.; Chang, X.; Zhang, Y.; Yang, L.; Meng, X. Molecular identification of Igf3 and its roles in grass carp (Ctenopharyngodon idella). Aquaculture 2022, 548, 737581. [Google Scholar] [CrossRef]
- Nobrega, R.H.; Morais, R.D.; Crespo, D.; de Waal, P.P.; de Franca, L.R.; Schulz, R.W.; Bogerd, J. Fsh Stimulates Spermatogonial Proliferation and Differentiation in Zebrafish via Igf3. Endocrinology 2015, 156, 3804–3817. [Google Scholar] [CrossRef] [PubMed]
- Jiao, F.; Fu, B.; Yang, Y.; Xue, H.; Wu, Y.; Zhao, H.; Wang, Q.; Yang, H. The Expression Pattern of Insulin-Like Growth Factor Subtype 3 (igf3) in the Orange-Spotted Grouper (Epinephelus coioides) and Its Function on Ovary Maturation. Int. J. Mol. Sci. 2023, 24, 2868. [Google Scholar] [CrossRef]
- Yan, R.; Ding, J.; Yang, Q.; Zhang, X.; Han, J.; Jin, T.; An, Y. Lead acetate induces cartilage defects and bone loss in zebrafish embryos by disrupting the GH/IGF-1 axis. Ecotoxicol. Environ. Saf. 2023, 253, 114666. [Google Scholar] [CrossRef]
- Si, Y.; He, F.; Wen, H.; Li, S.; He, H. Effects of low salinity on epigenetic changes of growth hormone and growth hormone receptor in half smooth tongue sole (Cynoglossus semilaevis). Reprod. Breed. 2021, 1, 11–21. [Google Scholar] [CrossRef]
- Shu, T.; Zhai, G.; Pradhan, A.; Olsson, P.E.; Yin, Z. Zebrafish cyp17a1 knockout reveals that androgen-mediated signaling is important for male brain sex differentiation. Gen. Comp. Endocrinol. 2020, 295, 113–490. [Google Scholar] [CrossRef]
- Gao, Y.; Lu, S.; Wu, M.; Yao, W.; Jin, Z.; Wu, X. Effects of dietary protein levels on growth, feed utilization and expression of growth-related genes of juvenile giant grouper (Epinephelus lanceolatus). Aquaculture 2019, 504, 369–374. [Google Scholar] [CrossRef]
- Pellissier, T.; Al Nafea, H.; Good, S.V. Divergence of insulin superfamily ligands, receptors and Igf binding proteins in marine versus freshwater stickleback: Evidence of selection in known and novel genes. Comp. Biochem. Physiol. Part D Genom. Proteom. 2018, 25, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Shen, W.; Cao, M.; Li, J.; Zheng, B.; Lou, Z.; Qian, B.; Xue, L. Dynamic alterations in methylation of global DNA and growth-related genes in large yellow croaker (Larimichthys crocea) in response to starvation stress. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2019, 227, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Yusuf, A.; Huang, X.; Chen, N.; Li, S.; Apraku, A.; Wang, W.; David, M.A. Growth and metabolic responses of juvenile largemouth bass (Micropterus salmoides) to dietary vitamin c supplementation levels. Aquaculture 2021, 534, 736243. [Google Scholar] [CrossRef]
- Yuan, B.; Chen, K.; Zhu, Y.; Yuan, Y.; Li, M. Medaka igf1 identifies somatic cells and meiotic germ cells of both sexes. Gene 2018, 642, 423–429. [Google Scholar] [CrossRef] [PubMed]
- Albornoz-Abud, N.A.; Canul-Marin, G.F.; Chan-Cua, I.; Hernandez-Nunez, E.; Canizares-Martinez, M.A.; Valdes-Lozano, D.; Rodriguez-Canul, R.; Albores-Medina, A.; Colli-Dula, R.C. Gene expression analysis on growth, development and toxicity pathways of male Nile tilapia (Oreochromis niloticus), after acute and sub-chronic benzo (α) pyrene exposures. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2021, 250, 109160. [Google Scholar] [CrossRef]
- Lynn, S.G.; Wallat, G.K.; Malison, J.A.; Shepherd, B.S. Developmental expression and estrogen responses of endocrine genes in juvenile yellow perch (Perca flavescens). Gen. Comp. Endocrinol. 2011, 171, 151–159. [Google Scholar] [CrossRef]
- Tran, T.T.H.; Nguyen, H.T.; Le, B.T.N.; Tran, P.H.; Van Nguyen, S.; Kim, O.T.P. Characterization of single nucleotide polymorphism in IGF1 and IGF1R genes associated with growth traits in striped catfish (Pangasianodon hypophthalmus Sauvage, 1878). Aquaculture 2021, 538, 736542. [Google Scholar] [CrossRef]
- Khoa, T.N.D.; Waqalevu, V.; Honda, A.; Shiozaki, K.; Kotani, T. An integrative description of the digestive system morphology and function of Japanese flounder (Paralichthys olivaceus) during early ontogenetic development. Aquaculture 2021, 531, 735855. [Google Scholar] [CrossRef]
- Swirplies, F.; Wuertz, S.; Baßmann, B.; Orban, A.; Schäfer, N.; Brunner, R.M.; Hadlich, F.; Goldammer, T.; Rebl, A. Identification of molecular stress indicators in pikeperch Sander lucioperca correlating with rising water temperatures. Aquaculture 2019, 501, 260–271. [Google Scholar] [CrossRef]
- Wei, Y.; Liang, M.; Xu, H. Fish protein hydrolysate affected amino acid absorption and related gene expressions of IGF-1/AKT pathways in turbot (Scophthalmus maximus). Aquacult. Nutr. 2019, 26, 145–155. [Google Scholar] [CrossRef]
- Vikeså, V.; Nankervis, L.; Remø, S.C.; Waagbø, R.; Hevrøy, E.M. Pre and postprandial regulation of ghrelin, amino acids and IGF1 in Atlantic salmon (Salmo salar L.) at optimal and elevated seawater temperatures. Aquaculture 2015, 438, 159–169. [Google Scholar] [CrossRef]
- Carnevali, O.; Maradonna, F.; Gioacchini, G. Integrated control of fish metabolism, wellbeing and reproduction: The role of probiotic. Aquaculture 2017, 472, 144–155. [Google Scholar] [CrossRef]
- Tenugu, S.; Pranoty, A.; Mamta, S.-K.; Senthilkumaran, B. Development and organisation of gonadal steroidogenesis in bony fishes—A review. Aquac. Fish. 2021, 6, 223–246. [Google Scholar] [CrossRef]
- Lubzens, E.; Bobe, J.; Young, G.; Sullivan, C.V. Maternal investment in fish oocytes and eggs: The molecular cargo and its contributions to fertility and early development. Aquaculture 2017, 472, 107–143. [Google Scholar] [CrossRef]
- Middleton, M.A.; Larsen, D.A.; Dickey, J.T.; Swanson, P. Evaluation of endocrine and transcriptomic markers of male maturation in winter-run Steelhead Trout (Oncorhynchus mykiss). Gen. Comp. Endocrinol. 2019, 81, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Yang, P.; Luo, F.; Wang, D.; Zhou, L. R-spondin1 signaling pathway is required for both the ovarian and testicular development in a teleosts, Nile tilapia (Oreochromis niloticus). Gen. Comp. Endocrinol. 2016, 230–231, 177–185. [Google Scholar] [CrossRef]
- Melo, M.C.; van Dijk, P.; Andersson, E.; Nilsen, T.O.; Fjelldal, P.G.; Male, R.; Nijenhuis, W.; Bogerd, J.; de Franca, L.R.; Taranger, G.L.; et al. Androgens directly stimulate spermatogonial differentiation in juvenile Atlantic salmon (Salmo salar). Gen. Comp. Endocrinol. 2015, 211, 52–61. [Google Scholar] [CrossRef] [PubMed]

| Aim | Primer | Primer Sequence (5′–3′) | Length |
|---|---|---|---|
| Partial cloning | igf3-1F | CTGTGCCAAACCAAAGAGCC | 504 bp |
| igf3-1R | CAAACTGCTGTGCCGTGTC | ||
| igf3-2F | CGGAGTGTGAGGAGTGTGTG | 411 bp | |
| igf3-2R | GGTGGTTTTCTGTGGGCTCT | ||
| Cloning 5′-end (RACE) | igf3-GSP5-1 | AGGAGGAATGGTGGTTTTCTGTG | |
| igf3-GSP5-nested | GGTGGTTTTCTGTGGGCTCT | ||
| Cloning 3′-end (RACE) | igf3-GSP3-1 | GAGAAATACTGTGCCAAACCAAAGAG | |
| igf3-GSP3-nested | CACAGAAAACCACCATTCCTCCT | ||
| expression analysis | igf1-F | CTGCTGAGGTTAAAGTGCGA | 165 bp |
| igf1-R | AAGCCTCTCTCTCCACACAC | ||
| igf2-F | GAGACGCTGTGCGGAGGAGA | 196 bp | |
| igf2-R | TTTCAGACTTGGCGGGTTT | ||
| igf3-3F | GTACGGATCTCCTCAGCGAC | 180 bp | |
| igf3-3R | GGCTCTTTGGTTTGGCACAG | ||
| β-actin-F | ATCGTGGGGCGCCCCAGGCACC | 543 bp | |
| β-actin-R | CTCCTTAATGTCACGCACGATTTC |
| Superorder | Species | Common Name | igf1 | igf2 (igf2a/igf2b) | igf3 |
|---|---|---|---|---|---|
| Numbers | |||||
| Percomorpha | S.maximus | Turbot | 1 | 1 | 1 |
| P.olivaceus | Japanese flounder | 1 | 1 | - | |
| C.semilaevis | Half-smooth tongue sole | 1 | 1 | - | |
| M.salmoides | Largemouth bass | 1 | 1 | - | |
| S.lucioperca | Pikeperch | 1 | 2 | 1 | |
| M.saxatilis | Striped sea-bass | 1 | 1 | 1 | |
| P.flavescens | Yellow perch | 1 | 1 | 1 | |
| E.lanceolatus | Giant grouper | 1 | 1 | 1 | |
| O.niloticus | Nile tilapia | 1 | 1 | 1 | |
| L.crocea | Large yellow croaker | 1 | 1 | 1 | |
| Atherinomorpha | O. latipes | Japanese medaka | 1 | 1 | 1 |
| X. maculatus | Southern platyfish | 1 | 1 | 1 | |
| Protacanthopterygii | S. salar | Atlantic salmon | 1 | 2 | 1 |
| O.mykiss | Rainbow trout | 1 | 1 | 1 | |
| Paracanthopterygii | G. morhua | Atlantic cod | 1 | 1 | - |
| Ostariophysi | D. rerio | Zebrafish | 1 | 2 | 1 |
| P.hypophthalmus | Striped catfish | 1 | 2 | 1 | |
| Clupeomorpha | C. harengus | Atlantic herring | 1 | 1 | 1 |
| Osteoglossomorpha | S. formosus | Asian arowana | 1 | - | 1 |
| Genes | Number of Species | Function | References |
|---|---|---|---|
| igf1 | 16 | Ovary: follicular cell proliferation follicle growth, oocyte maturation. Testis: stimulation of spermatogenesis. | C. semilaevis [29]; D. rerio [30]; E.lanceolatus [31]; G.aculeatus [32]; L.crocea [33]; M.salmoides [34]; O.latipes [35]; O.mykiss [28]; O.niloticus [36]; P.flavescens [37]; P.hypophthalmus [38]; P.olivaceus [39]; S.lucioperca [40]; S.maximus [41]; S. salar [42]; X. maculatus [43]; |
| igf2 (igf2a/igf2b) | 11 | Ovary: oocyte maturational competence, oocyte maturation. Testis: DNA synthesis of spermatogonia and spermatocytes. | C. semilaevis [29]; D. rerio [30]; E.lanceolatus [21]; G.aculeatus [32]; L.crocea [33]; M.salmoides [34]; O.mykiss [28]; O.niloticus [36]; P.flavescens [44]; P.hypophthalmus [38]; S.maximus [15]; X. maculatus [43]. |
| igf3 | 9 | Ovary: follicular cell survival, follicular cell development and ovulation. Testis: proliferation and division of type A spermatogonia and proliferation of Sertoli cells during spermatogenesis. | D. rerio [23]; E.lanceolatus [45]; M.salmoides [34]; O.latipes [18]; O.mykiss [45]; O.niloticus [46]; P.flavescens [44]; P.hypophthalmus [38]; S.salar [47]. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, C.; Zheng, S.; Dang, Y.; Wang, M.; Ren, Y. Identification of a New Insulin-like Growth Factor 3 (igf3) in Turbot (Scophthalmus maximus): Comparison and Expression Analysis of IGF System Genes during Gonadal Development. Fishes 2023, 8, 240. https://doi.org/10.3390/fishes8050240
Zhao C, Zheng S, Dang Y, Wang M, Ren Y. Identification of a New Insulin-like Growth Factor 3 (igf3) in Turbot (Scophthalmus maximus): Comparison and Expression Analysis of IGF System Genes during Gonadal Development. Fishes. 2023; 8(5):240. https://doi.org/10.3390/fishes8050240
Chicago/Turabian StyleZhao, Chunyan, Sujie Zheng, Yongji Dang, Mengshu Wang, and Yichao Ren. 2023. "Identification of a New Insulin-like Growth Factor 3 (igf3) in Turbot (Scophthalmus maximus): Comparison and Expression Analysis of IGF System Genes during Gonadal Development" Fishes 8, no. 5: 240. https://doi.org/10.3390/fishes8050240
APA StyleZhao, C., Zheng, S., Dang, Y., Wang, M., & Ren, Y. (2023). Identification of a New Insulin-like Growth Factor 3 (igf3) in Turbot (Scophthalmus maximus): Comparison and Expression Analysis of IGF System Genes during Gonadal Development. Fishes, 8(5), 240. https://doi.org/10.3390/fishes8050240








