Genetic Structure across Isolated Virginia Populations of the Endangered Candy Darter (Etheostoma osburni)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Site
2.2. Field Methods
2.3. Mitochondrial DNA
2.4. Microsatellite DNA
3. Results
3.1. Mitochondrial DNA
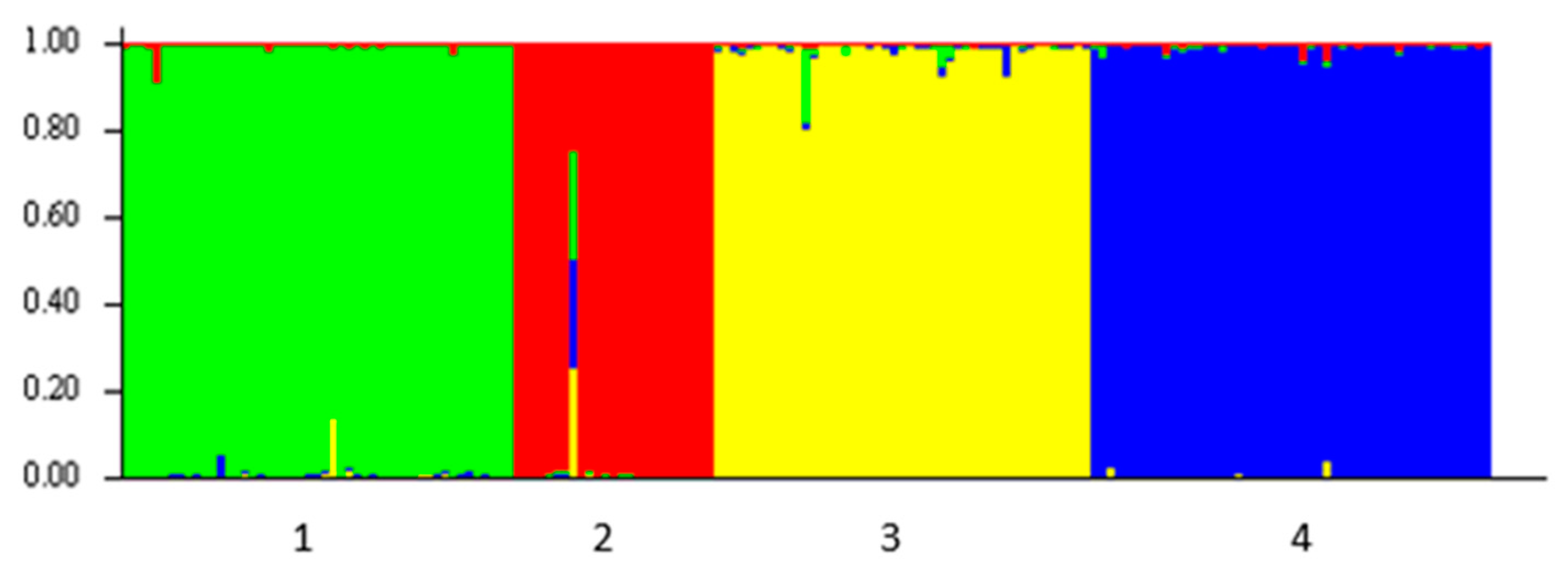
3.2. Microsatellite DNA
4. Discussion
4.1. Historical Demography and Genetics of Candy Darter in the Upper New River
4.2. Contemporary Population Genetics of Candy Darter in the Upper New River
4.3. Conservation Management Considerations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hallerman, E.M. (Ed.) Population viability analysis. In Population Genetics: Principles and Applications for Fisheries Scientists; American Fisheries Society: Bethesda, MD, USA, 2003; pp. 403–417. [Google Scholar]
- Whitlock, M.C.; Burger, R. Fixation of new mutations in small populations. In Evolutionary Conservation Biology; Ferriere, R., Dieckmann, U., Couvet, D., Eds.; Cambridge University Press: New York, NY, USA, 2004; pp. 155–170. [Google Scholar]
- Allendorf, F.W.; Luikart, G. Conservation and the Genetics of Populations; Blackwell Publishing: Malden, MA, USA, 2007. [Google Scholar]
- Whiteley, A.R.; Fitzpatrick, S.W.; Funk, W.C.; Tallmon, D.A. Genetic rescue to the rescue. Trends Ecol. Evol. 2015, 30, 42–49. [Google Scholar] [CrossRef]
- Jachowski, D.S.; Millspaugh, J.J.; Angermeier, P.L.; Slotow, R. (Eds.) Reintroduction of Fish and Wildlife Populations; University of California Press: Oakland, CA, USA, 2016. [Google Scholar]
- Jelks, H.L.; Walsh, S.J.; Burkhead, N.M.; Contreras-Balderas, S.; Diaz-Pardo, E.; Hendrickson, D.A.; Lyons, J.; Mandrak, N.E.; McCormick, F.; Nelson, J.S.; et al. Conservation status of imperiled North American freshwater and diadromous fishes. Fisheries 2008, 33, 372–407. [Google Scholar] [CrossRef]
- Angermeier, P.L. Ecological attributes of extinction-prone species: Loss of freshwater fishes of Virginia. Conserv. Biol. 1995, 9, 143–158. [Google Scholar] [CrossRef]
- Dunn, C.G. Comparison of habitat suitability among sites supporting strong, localized, and extirpated populations of candy darter Etheostoma osburni. In Final Report to Virginia Department of Game and Inland Fisheries; Virginia Department of Game and Inland Fisheries: Richmond, VA, USA, 2013. [Google Scholar]
- McBaine, K.M.; Hallerman, E.M. Demographic status and population genetic differentiation of candy darter populations in Virginia. In Final Report to Virginia Department of Game and Inland Fisheries; Virginia Department of Game and Inland Fisheries: Richmond, VA, USA, 2020; 95p. [Google Scholar]
- McBaine, K.E.; Hallerman, E.M.; Angermeier, P.L. Direct and molecular observation of movement and reproduction by candy darter, Etheostoma osburni, an endangered benthic stream fish. Fishes 2022, 7, 30. [Google Scholar] [CrossRef]
- U.S. Fish and Wildlife Service. Species Status Assessment (SSA) Report for the Candy Darter (Etheostoma osburni); USFWS Version 1.5; U.S. Fish and Wildlife Service: Hadley, MA, USA, 2018. [Google Scholar]
- Gibson, I.; Welsh, A.B.; Welsh, S.A.; Cincotta, D.A. Genetic swamping and possible species collapse: Tracking introgression between the native candy darter and introduced variegate darter. Conserv. Genet. 2019, 20, 287–298. [Google Scholar] [CrossRef]
- U.S. Fish and Wildlife Service. Endangered and threatened wildlife and plants; Endangered species status for the candy darter. Fed. Reg. 2018, 83, 58747–58754. [Google Scholar]
- U.S. Fish and Wildlife Service. Endangered and threatened wildlife and plants; Designation of critical habitat for candy darter. Fed. Reg. 2021, 86, 17956–17992. [Google Scholar]
- Helfrich, L.A.; Bye, M.B.; Dalton, D. Life history, status, and recovery of the candy darter Etheostoma osburni, in Virginia. In Final Report to the Virginia Department of Game and Inland Fisheries; Virginia Department of Game and Inland Fisheries: Richmond, VA, USA, 1996. [Google Scholar]
- Dunn, C.G.; Angermeier, P.L. Development of habitat suitability indices for the candy darter, with cross-scale validation across representative populations. Trans. Amer. Fish. Soc. 2016, 145, 1266–1281. [Google Scholar] [CrossRef]
- Near, T.J.; Bossu, C.M.; Bradburd, G.S.; Carlson, R.L.; Harrington, R.C.; Hollingsworth, P.R., Jr.; Keck, B.P.; Etnier, D.A. Phylogeny and temporal diversification of darters (Percidae: Etheostomatinae). Systemat. Biol. 2011, 60, 565–595. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. 2017. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Molec. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Clement, M.; Snell, Q.; Walker, P.; Posada, D.; Crandall, K. TCS: Estimating gene genealogies. In Parallel and Distributed Processing Symposium International; IEEE Computer Society: Los Alamitos, CA, USA, 2002; Volume 3, p. 184. [Google Scholar]
- Leigh, J.; Bryant, D.; Steel, M. PopART, ver. 1.7. 2015. Available online: http://www.popart.otago.ac.nz (accessed on 13 September 2023).
- Excoffier, L.; Lischer, H.E.L. Arlequin suite, version 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Molec. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Nylander, J.A. MrModeltest, version 2.2. 2004. Available online: http://www.abc.se/~nylander/ (accessed on 13 September 2023).
- Swofford, D.L. PAUP *: Phylogenetic Analysis Using Parsimony (* and Other Methods); Sinauer Associates: Sunderland, MA, USA, 2003. [Google Scholar]
- Hasegawa, M.; Kishino, H.; Yano, T. Dating the human-ape split by a molecular clock of mitochondrial DNA. J. Molec. Evol. 1985, 22, 160–174. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A. FigTree; ver.1. 4. Molecular Evolution, Phylogenetics and Epidemiology, University of Edinburgh, Institute of Evolutionary Biology: Edinburgh, UK, 2012; Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 13 September 2023).
- Switzer, J.F.; Welsh, S.A.; King, T.L. Microsatellite DNA primers for the candy darter, Etheostoma osburni and variegate darter, Etheostoma variatum, and cross-species amplification in other darters (Percidae). Molec. Ecol. Resour. 2008, 8, 335–338. [Google Scholar] [CrossRef] [PubMed]
- Van Oosterhout, C.; Hutchinson, W.F.; Willis, D.P.M.; Shipley, P. MICRO-CHECKER: Software for identifying and correcting genotyping errors in microsatellite data. Molec. Ecol. Notes 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Garza, J.C.; Williamson, E.G. Detection of reduction in population size using data from microsatellite loci. Molec. Ecol. 2001, 10, 305–318. [Google Scholar] [CrossRef]
- Hastings, A. Complex interactions between dispersal and dynamics: Lessons from coupled logistic equations. Ecology 1993, 74, 1362–1372. [Google Scholar] [CrossRef]
- Chybicki, I.J.; Burczyk, J. Simultaneous estimation of null alleles and inbreeding coefficients. J. Hered. 2009, 100, 106–113. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure from multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Waples, R.S.; Gaggiotti, O. What is a population? An empirical evaluation of some genetic methods for identifying the number of gene pools and their degree of connectivity. Molec. Ecol. 2006, 15, 1419–1439. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Molec. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Do, C.; Waples, R.S.; Peel, D.; Macbeth, G.M.; Tillett, B.J.; Ovenden, J.R. NeEstimator v2: Re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Molec. Ecol. Resour. 2014, 14, 209–214. [Google Scholar] [CrossRef] [PubMed]
- Waples, R.S.; Antao, T.; Luikart, G. Effects of overlapping generations on linkage disequilibrium estimates of effective population size. Genetics 2014, 197, 769–780. [Google Scholar] [CrossRef] [PubMed]
- Palsbøll, P.J.; Berube, M.; Allendorf, F.W. Identification of management units using population genetic data. Trends Ecol. Evol. 2007, 22, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, R.E.; Burkhead, N.M. Freshwater Fishes of Virginia; American Fisheries Society: Bethesda, MD, USA, 1994. [Google Scholar]
- Ray, J.M.; Lang, N.J.; Wood, R.M.; Mayden, R.L. History repeated: Recent and historical mitochondrial introgression between the current darter Etheostoma uniporum and rainbow darter Etheostoma caeruleum (Teleostei: Percidae). J. Fish Biol. 2008, 72, 418–434. [Google Scholar] [CrossRef]
- Bossu, C.M.; Near, T.J. Characterization of a contemporaneous hybrid zone between two darter species (Etheostoma bison and E. caeruleum) in the Buffalo River System. Genetica 2013, 141, 75–88. [Google Scholar]
- Castric, V.; Bonney, F.; Bernatchez, L. Landscape structure and hierarchical genetic diversity in the brook charr, Salvelinus fontinalis. Evolution 2001, 55, 1016–1028. [Google Scholar] [CrossRef]
- King, T.L.; Kalinowski, S.T.; Schill, W.B.; Spidle, A.P.; Lubinski, B.A. Population structure of Atlantic salmon (Salmo salar L.): A range-wide perspective from microsatellite DNA variation. Molec. Ecol. 2001, 10, 807–821. [Google Scholar] [CrossRef]
- Kazyak, D.C.; Lubinski, B.A.; Kulp, M.A.; Pregler, K.C.; Whiteley, A.R.; Hallerman, E.; Coombs, J.A.; Kanno, Y.; Rash, J.M.; Morgan, R.P.; et al. Population genetics of brook trout in the southern Appalachian Mountains. Trans. Amer. Fish. Soc. 2022, 151, 127–149. [Google Scholar] [CrossRef]
- Wright, S. The interpretation of population structure by F-statistics with special regard to systems of mating. Evolution 1965, 19, 395–420. [Google Scholar] [CrossRef]
- Waples, R.S.; Do, C. 2010. Linkage disequilibrium estimates of contemporary Ne using highly variable genetic markers: A largely untapped resource for applied conservation and evolution. Evol. Applic. 2010, 3, 244–262. [Google Scholar] [CrossRef]
- Frankham, R.; Bradshaw, C.J.; Brook, B.W. Genetics in conservation management: Revised recommendations for the 50/500 rules, Red List criteria and population viability analyses. Biol. Conserv. 2014, 170, 56–63. [Google Scholar] [CrossRef]
- Wright, S. Isolation by distance. Genetics 1943, 28, 114–138. [Google Scholar] [CrossRef]
- Blanton, R.E.; Cashner, M.F.; Thomas, M.R.; Brandt, S.L.; Floyd, M.A. Increased habitat fragmentation leads to isolation among and low genetic diversity within populations of the imperiled Kentucky arrow darter (Etheostoma sagitta spilotum). Conserv. Genet. 2019, 20, 1009–1022. [Google Scholar] [CrossRef]
- Argentina, J.E.; Angermeier, P.L.; Hallerman, E.M.; Welsh, S.A. Spatial extent of analysis influences observed patterns of population genetic structure in a widespread darter species (Percidae). Freshwat. Biol. 2018, 63, 1185–1198. [Google Scholar] [CrossRef]
- Roberts, J.H.; Angermeier, P.L.; Hallerman, E.M. Extensive dispersal of Roanoke logperch (Percina rex) inferred from genetic marker data. Ecol. Freshwat. Fish 2016, 25, 1–16. [Google Scholar] [CrossRef]
- Frankham, R.; Ballou, J.D.; Eldridge, M.D.B.; Lacy, R.C.; Ralls, K.; Dudash, M.R.; Fenster, C.B. Predicting the probability of outbreeding depression. Conserv. Biol. 2011, 25, 465–475. [Google Scholar] [CrossRef] [PubMed]
- Finger, A.J.; Parmenter, S.; May, B.P. Conservation of the Owens pupfish: Genetic effects of multiple translocations and extirpations. Trans. Amer. Fish. Soc. 2013, 142, 1430–1443. [Google Scholar] [CrossRef]
- Pavlova, A.; Beheregaray, L.B.; Coleman, R.; Gilligan, D.; Harrison, K.A.; Ingram, B.A.; Kearns, J.; Lamb, A.M.; Lintermans, M.; Lyon, J.; et al. Severe consequences of habitat fragmentation on genetic diversity of an endangered Australian freshwater fish: A call for assisted gene flow. Evol. Applic. 2017, 10, 531–550. [Google Scholar] [CrossRef] [PubMed]
- Ryman, N.; Laikre, L. Effects of supportive breeding on the genetically effective population size. Conserv. Biol. 1991, 5, 325–329. [Google Scholar] [CrossRef]



| Variable Sites | Counts | ||||||||||||||||||
| 1 | 3 | 3 | 4 | 4 | 5 | 6 | 8 | 8 | 8 | 8 | 9 | 9 | 9 | ||||||
| 6 | 3 | 7 | 4 | 5 | 9 | 7 | 0 | 4 | 4 | 8 | 2 | 3 | 3 | Dismal Creek | Cripple Creek | Laurel Creek | Stony Creek | ||
| Haplotypes | N | 7 | 6 | 7 | 3 | 2 | 0 | 7 | 0 | 2 | 8 | 4 | 6 | 4 | 6 | ||||
| Hap_1 | 33 | T | T | G | T | G | A | C | T | A | T | T | C | T | A | 0 | 18 | 15 | 0 |
| Hap_2 | 47 | · | · | · | · | · | G | · | C | · | C | · | · | · | · | 0 | 0 | 0 | 47 |
| Hap_3 | 1 | · | · | · | · | · | G | · | C | · | C | · | · | A | · | 0 | 0 | 0 | 1 |
| Hap_4 | 14 | C | · | · | C | A | G | T | C | · | C | · | · | · | G | 0 | 14 | 0 | 0 |
| Hap_5 | 16 | C | · | · | C | A | G | T | C | · | C | · | · | · | · | 13 | 0 | 3 | 0 |
| Hap_6 | 11 | C | C | · | C | A | G | T | C | · | C | · | · | · | · | 11 | 0 | 0 | 0 |
| Hap_7 | 3 | C | · | · | C | A | G | T | C | G | C | · | · | · | · | 0 | 3 | 0 | 0 |
| Hap_8 | 18 | · | · | · | · | · | · | · | C | · | · | C | · | · | · | 0 | 0 | 18 | 0 |
| Hap_9 | 6 | · | · | · | · | · | · | · | C | · | C | · | T | · | · | 0 | 0 | 6 | 0 |
| Hap_10 | 1 | · | · | A | · | · | · | · | C | · | C | · | · | · | · | 0 | 1 | 0 | 0 |
| A | ||||
| Source of Variation | df | Sum of Squares | Variance Components | % of Variation |
| Among populations | 3 | 140.61 | 1.25 Va | 58.7 |
| Among individuals within populations | 146 | 128.15 | 0.88 Vb | 41.3 |
| Total | 149 | 268.76 | 2.13 | |
| B | ||||
| Source of Variation | df | Sum of Squares | Variance Components | % of Variation |
| Among populations | 3 | 308.00 | 1.17 Va | 24.0 |
| Among individuals within populations | 167 | 714.36 | 0.56 Vb | 11.5 |
| Within individuals | 171 | 539.50 | 3.15 Vc | 64.5 |
| Total | 341 | 1561.86 | 4.89 | |
| Stream | Cripple Creek | Dismal Creek | Laurel Creek | Stony Creek |
|---|---|---|---|---|
| Cripple Creek | 0 | 0.44 | 0.27 | 0.49 |
| Dismal Creek | 0.28 | 0 | 0.77 | 0.95 |
| Laurel Creek | 0.19 | 0.33 | 0 | 0.63 |
| Stony Creek | 0.17 | 0.31 | 0.25 | 0 |
| Stream | N | A | Ar | Ho | He | Allelic Range | M |
|---|---|---|---|---|---|---|---|
| Cripple Creek | 98 | 6.67 | 52.33 | 0.64 | 0.70 | 161.00 | 0.11 |
| Dismal Creek | 50 | 3.92 | 26.96 | 0.25 | 0.46 | 247.33 | 0.02 |
| Laurel Creek | 94 | 7.00 | 50.50 | 0.56 | 0.63 | 188.67 | 0.08 |
| Stony Creek | 94 | 7.00 | 55.19 | 0.56 | 0.63 | 188.67 | 0.08 |
| Stream | Arlequin | INEST | ||
|---|---|---|---|---|
| FIS | P (Random FIS ≥ Observed FIS) | Fi Mean | 95% CI | |
| Cripple Creek | 0.09 | <0.001 | 0.02 | 0.0–0.05 |
| Dismal Creek | 0.45 | <0.001 | 0.06 | 0.0–0.15 |
| Laurel Creek | 0.14 | <0.001 | 0.02 | 0.0–0.06 |
| Stony Creek | 0.12 | <0.001 | 0.03 | 0.01–0.06 |
| Stream | n | Ne | Parametric CI | Jackknife CI |
|---|---|---|---|---|
| Cripple Creek | 49 | 172.6 | 85.3–1682 | 63.5–∞ |
| Dismal Creek | 25 | 39 | 10.3–∞ | 7.9–∞ |
| Laurel Creek | 47 | 136.1 | 73.9–520.2 | 57.7–∞ |
| Stony Creek | 50 | 192.7 | 82.3–∞ | 54–∞ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
McBaine, K.E.; Angermeier, P.L.; Hallerman, E.M. Genetic Structure across Isolated Virginia Populations of the Endangered Candy Darter (Etheostoma osburni). Fishes 2023, 8, 490. https://doi.org/10.3390/fishes8100490
McBaine KE, Angermeier PL, Hallerman EM. Genetic Structure across Isolated Virginia Populations of the Endangered Candy Darter (Etheostoma osburni). Fishes. 2023; 8(10):490. https://doi.org/10.3390/fishes8100490
Chicago/Turabian StyleMcBaine, Kathryn E., Paul L. Angermeier, and Eric M. Hallerman. 2023. "Genetic Structure across Isolated Virginia Populations of the Endangered Candy Darter (Etheostoma osburni)" Fishes 8, no. 10: 490. https://doi.org/10.3390/fishes8100490
APA StyleMcBaine, K. E., Angermeier, P. L., & Hallerman, E. M. (2023). Genetic Structure across Isolated Virginia Populations of the Endangered Candy Darter (Etheostoma osburni). Fishes, 8(10), 490. https://doi.org/10.3390/fishes8100490








