Genetic Diversity and Population Structure of the Chinese Mitten Crab (Eriocheir sinensis) from Six Different Lakes Using Microsatellites
Abstract
1. Introduction
2. Materials and Methods
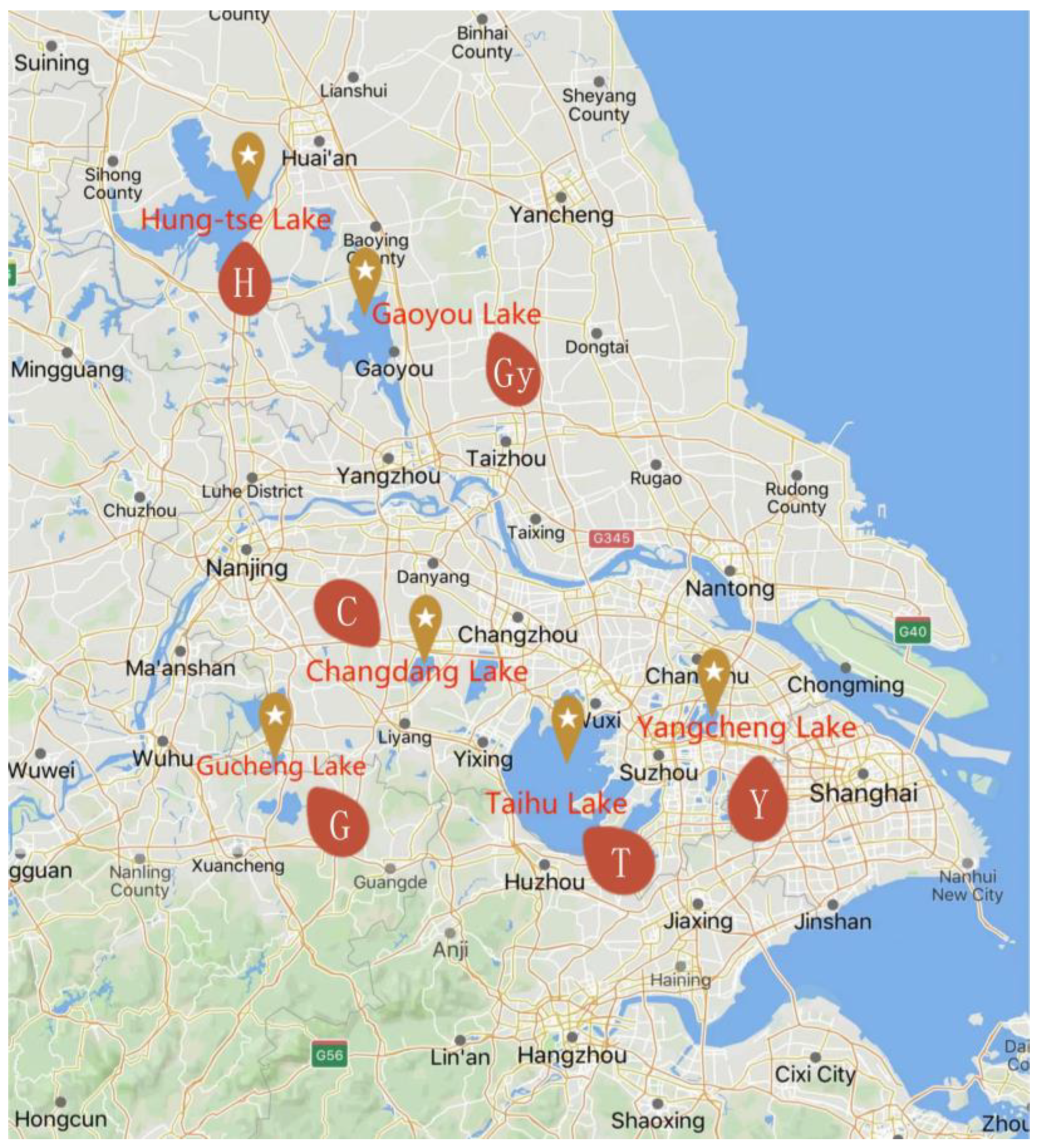
2.1. Study Area and Sample Collection
2.2. DNA Extraction
2.3. Microsatellite Primer Amplification
2.4. Genetic Diversity
2.5. Population Structure and Genetic Differentiation
3. Results
3.1. Microsatellite
3.2. Genetic Diversity
3.3. Hardy–Weinberg Equilibrium and Mutation–Drift Equilibrium Significant Test Using IAM, TPM, and SMM Model
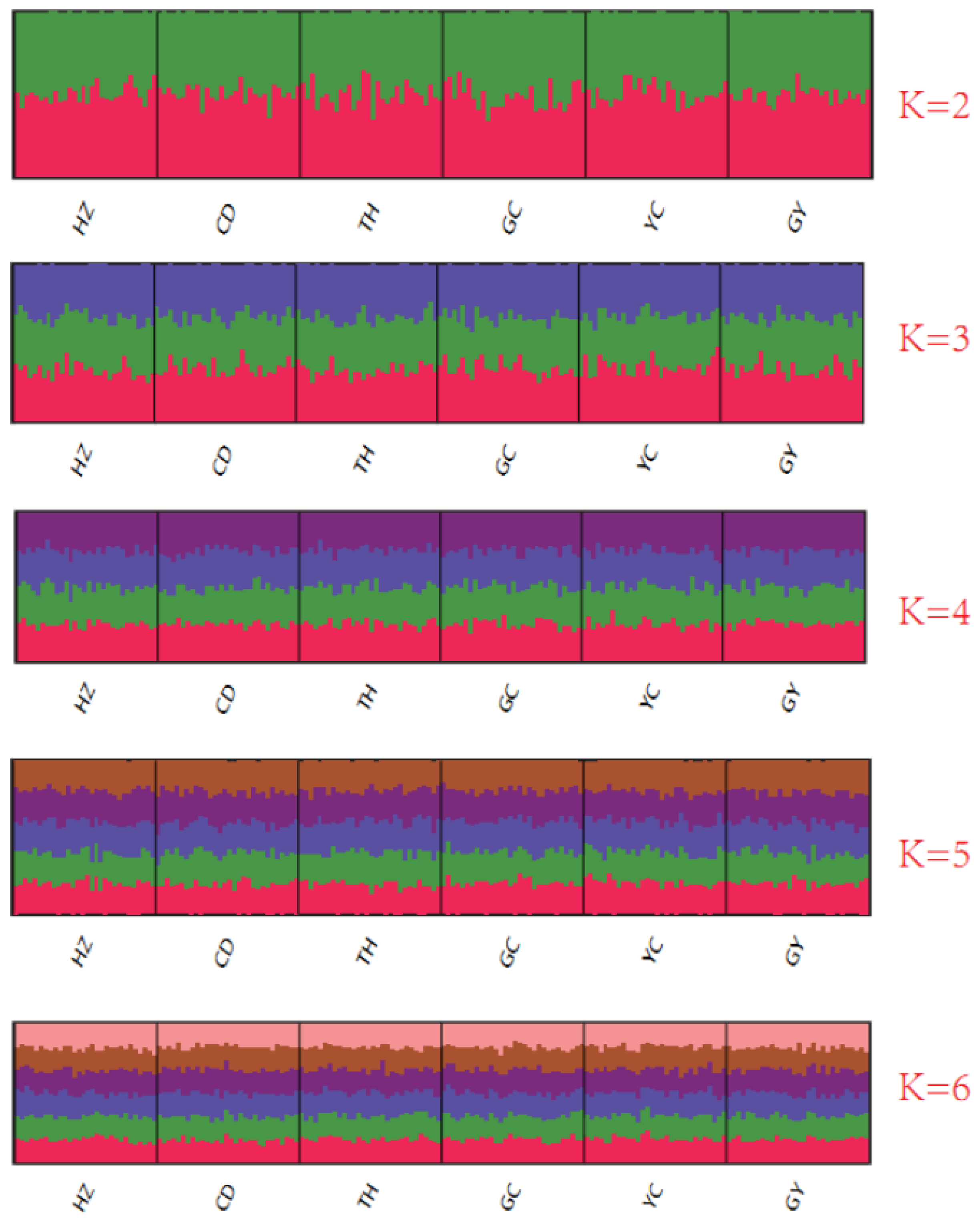
3.4. Population Structure of Six Crab Populations
4. Discussion
4.1. Chinese Mittens Crab Diversity
4.2. Mutation–Drift Equilibrium Significant Test Using IAM, TPM, and SMM Model
4.3. Genetic Distance of Chinese Mitten Crabs Population
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fishery Bureau. China Fishery Statistical Yearbook 2016; China Agriculture Press: Beijing, China, 2016.
- Dong, J.; Li, X.; Zhang, R.; Zhao, Y.; Wu, G.; Liu, J.; Zhu, X.; Li, L. Comparative analysis of the intestinal bacterial community and expression of gut immunity genes in the Chinese Mitten Crab (Eriocheir sinensis). AMB Express 2018, 8, 192. [Google Scholar] [CrossRef] [PubMed]
- Reverter, M.; Tapissier-Bontemps, N.; Sasal, P.; Saulnier, D. Use of Medicinal Plants in Aquaculture. Diagn. Control. Dis. Fish Shellfish. 2017, 9, 223–261. [Google Scholar]
- Sui, L.; Wille, M.; Cheng, Y.; Wu, X.; Sorgeloos, P. Larviculture techniques of Chinese mitten crab Eriocheir sinensis. Aquaculture 2011, 315, 16–19. [Google Scholar] [CrossRef]
- Zhang, T.; Li, Z.; Cui, Y. Survival, growth, sex ratio, and maturity of the Chinese mitten crab (Eriocheir sinensis) reared in a Chinese pond. J. Freshw. Ecol. 2001, 16, 633–640. [Google Scholar] [CrossRef]
- Crozier, L.G.; Hutchings, J. Plastic and evolutionary responses to climate change in fish. Evol. Appl. 2014, 7, 68–87. [Google Scholar] [CrossRef]
- Levitus, S.; Antonov, J.I.; Boyer, T.P.; Baranova, O.K.; Garcia, H.E.; Locarnini, R.A.; Mishonov, A.V.; Reagan, J.R.; Seidov, D.; Yarosh, E.S.; et al. World ocean heat content and thermosteric sea level change (0–2000 m), 1955–2010. Geophys. Res. Lett. 2012, 39, 1–5. [Google Scholar] [CrossRef]
- O’Reilly, C.M.; Sharma, S.; Gray, D.K.; Hampton, S.E.; Read, J.S.; Rowley, R.J.; Schneider, P.; Lenters, J.D.; McIntyre, P.B.; Kraemer, B.M.; et al. Rapid and highly variable warming of lake surface waters around the globe. Geophys. Res. Lett. 2015, 42, 10773–10781. [Google Scholar] [CrossRef]
- Miao, H.; An, C. Challenges for Protected Areas Management in China. Sustainability 2020, 12, 5879. [Google Scholar]
- Zaya, D.N.; Molano-Flores, B.; Feist, M.A.; Koontz, J.A.; Coons, J. Assessing genetic diversity for the USA endemic carnivorous plant Pinguicula ionantha R.K. Godfrey (Lentibulariaceae). Conserv. Genet. 2016, 18, 171–180. [Google Scholar] [CrossRef]
- Frankham, R. Genetics and extinction. Biol. Conserv. 2005, 126, 131–140. [Google Scholar] [CrossRef]
- Sean, H.; Bruford, M.W.; Chris, F.W.; Peter, G.; Patrick, G.M.; Grueber, C.E.; Myriam, H.; Hunter, M.E.; Christina, H.; Kalamujic, S.B. Global Commitments to Conserving and Monitoring Genetic Diversity Are Now Necessary and Feasible. Bioscience 2021, 71, 964–976. [Google Scholar]
- Monika, S.; Gabor, S.; Katarzyna, W.O.; Jakub, S.; Dusan, G. Genetic Diversity and Population Structure of the Rare and Endangered Plant Species Pulsatilla patens (L.) Mill in East Central Europe. PLoS ONE 2016, 11, e0151730. [Google Scholar]
- Páez-Triana, L.; Muoz, M.; Herrera, G.; Moreno-Pérez, D.A.; Rmírez, J.D. Genetic diversity and population structure of Rhipicephalus sanguineus sensu lato across different regions of Colombia. Parasites Vectors 2021, 14, 424. [Google Scholar] [CrossRef] [PubMed]
- Sui, L.; Zhang, F.; Wang, X.; Bossier, P.; Sorgeloos, P.; Hänfling, B. Genetic diversity and population structure of the Chinese mitten crab Eriocheir sinensis in its native range. Mar. Biol. 2009, 156, 1573–1583. [Google Scholar] [CrossRef]
- Zhang, C.; Li, Q.; Wu, X.; Liu, Q.; Cheng, Y. Genetic diversity and genetic structure of farmed and wild Chinese mitten crab (Eriocheir sinensis) populations from three major basins by mitochondrial DNA COI and Cyt b gene sequences. Mitochondrial DNA A DNA. Mapp. Seq. Anal. 2017, 29, 1081–1089. [Google Scholar]
- Cunningham, G.M.; Roman, M.G.; Flores, L.C.; Hubbard, G.B.; Salmon, A.B.; Zhang, Y.; Gelfond, J.; Ikeno, Y. The paradoxical role of thioredoxin on oxidative stress and aging. Arch. Biochem. Biophys. 2015, 576, 32–38. [Google Scholar] [CrossRef]
- Cowman, P.F.; Bellwood, D.R. The historical biogeography of coral reef fishes: Global patterns of origination and dispersal. J. Biogeogr. 2013, 40, 209–224. [Google Scholar] [CrossRef]
- Harrison, H.B.; Williamson, D.H.; Evans, R.D.; Almany, G.R.; Thorrold, S.R.; Russ, G.R.; Feldheim, K.A.; van Herwerden, L.; Planes, S.; Srinivasan, M.; et al. Larval Export from Marine Reserves and the Recruitment Benefit for Fish and Fisheries. Curr. Biol. 2012, 22, 1023–1028. [Google Scholar] [CrossRef]
- Yeh, F.C.; Yang, R.-C.; Boyle, T.; Ye, Z.; Xiyan, J. POPGENE 32, Microsoft Windows-Based Freeware for Population Genetic Analysis; University of Alberta: Edmonton, AB, Canada, 2000. [Google Scholar]
- Rousset, F. Equilibrium Values of Measures of Population Subdivision for Stepwise Mutation Processes. Genetics 1996, 142, 1357–1362. [Google Scholar] [CrossRef]
- Excoffier, L.; Smouse, P.E.; Quattro, J.M. Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics 1992, 131, 479–491. [Google Scholar] [CrossRef]
- Wilson, G.A.; Rannala, B. Bayesian Inference of Recent Migration Rates Using Multilocus Genotypes. Genetics 2003, 163, 1177–1191. [Google Scholar] [CrossRef] [PubMed]
- Evanno, G.S.; Regnaut, S.J.; Goudet, J. Detecting the number of clusters of individuals using the software structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Austin, Z.; Sutton, J. Qualitative research: Getting started. Can. J. Hosp. Pharm. 2014, 67, 436. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.J.; Donnelly, P.J. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Simental-Rodríguez, S.L.; Quiñones-Pérez, C.Z.; Moya, D.; Hernández-Tecles, E.; López-Sánchez, C.A.; Wehenkel, C. The Relationship between Species Diversity and Genetic Structure in the Rare Picea chihuahuana Tree Species Community, Mexico. PLoS ONE 2014, 9, e111623. [Google Scholar] [CrossRef]
- Stange, M.; Barrett, R.D.H.; Hendry, A.P. The importance of genomic variation for biodiversity, ecosystems and people. Nat. Rev. Genet. 2020, 22, 89–105. [Google Scholar] [CrossRef]
- Amos, W.; Harwood, J. Factors affecting levels of genetic diversity in natural populations. Philos. Trans. R. Soc. B Biol. Sci. 1998, 353, 177–186. [Google Scholar] [CrossRef]
- Coates, D.J.; Byrne, M.; Moritz, C. Genetic Diversity and Conservation Units: Dealing with the Species-Population Continuum in the Age of Genomics. Front. Ecol. Evol. 2018, 6, 165. [Google Scholar] [CrossRef]
- Gaggiotti, O.E.; Chao, A.; Peres-Neto, P.; Chiu, C.H.; Edwards, C.; Fortin, M.J.; Jost, L.; Richards, C.M.; Selkoe, K.A. Diversity from genes to ecosystems: A unifying framework to study variation across biological metrics and scales. Evol. Appl. 2018, 11, 1176–1193. [Google Scholar] [CrossRef]
- Wang, C.; Chenhong, L.I.; Sifa, L.I. Mitochondrial DNA-inferred population structure and demographic history of the mitten crab (Eriocheir sensu stricto) found along the coast of mainland China. Mol. Ecol. 2008, 17, 3515–3527. [Google Scholar] [CrossRef]
- Jun, W.; Lei, H.; Qixuan, C.; Guoqing, L.; Chenghui, W. Complete mitochondrial genomes of three mitten crabs, Eriocheir sinensis, E. hepuensis, and E. japonica. Mitochondrial DNA Part A 2016, 27, 1175–1176. [Google Scholar]
- Wang, H.; Feng, G.; Zhang, Y. Studies on genetic diversity of Chinese mitten crab Eriocheir sinensis of Yangtze River system based on mitochondrial DNA control region. J. Phys. Conf. Ser. 2020, 1549, 032010. [Google Scholar] [CrossRef]
- Estoup, A.L.; Garnery, L.; Solignac, M.; Cornuet, J.M. Microsatellite variation in honey bee (Apis mellifera L.) populations: Hierarchical genetic structure and test of the infinite allele and stepwise mutation models. Genetics 1995, 140, 679–695. [Google Scholar] [CrossRef] [PubMed]
- Cornuet, J.M.; Luikart, G. Description and Power Analysis of Two Tests for Detecting Recent Population Bottlenecks from Allele Frequency Data. Genetics 1996, 144, 2001–2014. [Google Scholar] [CrossRef]
- Wu, Z.; Yu, D.; Wang, Z.; Li, X.; Xu, X. Great influence of geographic isolation on the genetic differentiation of Myriophyllum spicatum under a steep environmental gradient. Sci. Rep. 2015, 5, 15618. [Google Scholar] [CrossRef]
- Prentice, M.B.; Bowman, J.; Khidas, K.; Koen, E.L.; Row, J.R.; Murray, D.L.; Wilson, P.J. Selection and drift influence genetic differentiation of insular Canada lynx (Lynx canadensis) on Newfoundland and Cape Breton Island. Ecol. Evol. 2017, 7, 3281–3294. [Google Scholar] [CrossRef]
- Wu, Z.; Xu, X.; Zhang, J.; Wiegleb, G.; Hou, H. Influence of environmental factors on the genetic variation of the aquatic macrophyte Ranunculus subrigidus on the Qinghai-Tibetan Plateau. BMC Evol. Biol. 2019, 19, 237. [Google Scholar] [CrossRef]
- Cheng, Y.; Wu, X.; Yang, X.; Hines, A.H. Current Trends in Hatchery Techniques and Stock Enhancement for Chinese Mitten Crab. Eriocheir. Jpn. Sin. Rev. Fish. Sci. 2008, 16, 377–384. [Google Scholar] [CrossRef]
- Huang, W.; Zhao, X.; Zhao, X.; Li, Y.; Lian, J. Effects of environmental factors on genetic diversity of Caragana microphylla in Horqin Sandy Land, northeast China. Ecol. Evol. 2016, 6, 8256–8266. [Google Scholar] [CrossRef]
| Crab1 | Crab20 | Crab2 | Crab11 | Crab18 | Crab15 | Crab6 | Crab17 | Crab8 | Crab12 | Esin42 | HLJEsa42 | Crab21 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AO | |||||||||||||
| GC | 2.00 | 5.00 | 1.00 | 6.00 | 7.00 | 8.00 | 5.00 | 6.00 | 9.00 | 10.00 | 14.00 | 10.00 | 5.00 |
| HZ | 2.00 | 6.00 | 2.00 | 6.00 | 6.00 | 7.00 | 3.00 | 5.00 | 8.00 | 9.00 | 17.00 | 9.00 | 6.00 |
| YC | 5.00 | 5.00 | 2.00 | 7.00 | 8.00 | 7.00 | 4.00 | 5.00 | 11.00 | 9.00 | 15.00 | 11.00 | 8.00 |
| CD | 2.00 | 5.00 | 1.00 | 8.00 | 7.00 | 7.00 | 4.00 | 5.00 | 8.00 | 6.00 | 13.00 | 15.00 | 6.00 |
| GY | 3.00 | 5.00 | 1.00 | 6.00 | 7.00 | 7.00 | 4.00 | 5.00 | 10.00 | 9.00 | 13.00 | 12.00 | 4.00 |
| TH | 2.00 | 7.00 | 1.00 | 5.00 | 7.00 | 5.00 | 3.00 | 4.00 | 10.00 | 7.00 | 15.00 | 14.00 | 6.00 |
| All | 5.00 | 9.00 | 2.00 | 10.00 | 13.00 | 8.00 | 5.00 | 6.00 | 15.00 | 13.00 | 19.00 | 19.00 | 8.00 |
| AE | |||||||||||||
| GC | 1.98 | 3.25 | 1.00 | 2.03 | 3.86 | 4.05 | 1.70 | 3.32 | 3.75 | 6.14 | 11.11 | 5.59 | 2.85 |
| HZ | 1.86 | 2.89 | 1.06 | 2.98 | 3.44 | 2.31 | 1.18 | 2.86 | 4.09 | 5.80 | 10.84 | 6.33 | 3.57 |
| YC | 2.13 | 2.65 | 1.03 | 2.40 | 4.35 | 2.44 | 1.45 | 2.47 | 4.87 | 6.16 | 9.62 | 8.03 | 3.46 |
| CD | 1.98 | 2.14 | 1.00 | 2.51 | 2.62 | 3.07 | 1.46 | 2.45 | 3.94 | 4.54 | 9.72 | 8.45 | 3.23 |
| GY | 1.82 | 2.25 | 1.00 | 2.57 | 3.07 | 2.72 | 1.18 | 2.61 | 4.74 | 5.73 | 9.83 | 8.78 | 2.82 |
| TH | 1.89 | 3.61 | 1.00 | 3.19 | 3.60 | 1.92 | 1.22 | 2.31 | 4.23 | 5.57 | 10.05 | 9.00 | 3.44 |
| All | 1.96 | 2.88 | 1.01 | 2.63 | 3.71 | 2.75 | 1.36 | 2.73 | 4.38 | 6.14 | 11.95 | 8.47 | 3.25 |
| HO | |||||||||||||
| GC | 0.90 | 0.46 | 0.00 | 0.53 | 0.40 | 0.33 | 0.33 | 0.60 | 0.76 | 0.76 | 0.83 | 0.40 | 0.56 |
| HZ | 0.73 | 0.43 | 0.06 | 0.83 | 0.40 | 0.36 | 0.16 | 0.53 | 0.70 | 0.76 | 0.83 | 0.53 | 0.80 |
| YC | 0.86 | 0.33 | 0.03 | 0.63 | 0.40 | 0.46 | 0.36 | 0.46 | 0.80 | 0.83 | 0.83 | 0.53 | 0.86 |
| CD | 0.9 | 0.36 | 0.00 | 0.63 | 0.60 | 0.46 | 0.36 | 0.40 | 0.70 | 0.80 | 0.90 | 0.90 | 0.70 |
| GY | 0.63 | 0.40 | 0.00 | 0.56 | 0.63 | 0.60 | 0.16 | 0.23 | 0.73 | 0.93 | 0.76 | 0.93 | 0.53 |
| TH | 0.70 | 0.33 | 0.00 | 0.70 | 0.66 | 0.46 | 0.20 | 0.43 | 0.80 | 0.90 | 0.66 | 0.93 | 0.80 |
| All | 0.78 | 0.38 | 0.01 | 0.65 | 0.51 | 0.45 | 0.26 | 0.44 | 0.75 | 0.83 | 0.80 | 0.70 | 0.71 |
| HE | |||||||||||||
| GC | 0.50 | 0.70 | 0.00 | 0.51 | 0.75 | 0.76 | 0.42 | 0.71 | 0.74 | 0.85 | 0.92 | 0.83 | 0.66 |
| HZ | 0.47 | 0.66 | 0.06 | 0.67 | 0.72 | 0.57 | 0.15 | 0.66 | 0.76 | 0.84 | 0.92 | 0.85 | 0.73 |
| YC | 0.54 | 0.63 | 0.03 | 0.59 | 0.78 | 0.60 | 0.31 | 0.60 | 0.80 | 0.85 | 0.91 | 0.89 | 0.72 |
| CD | 0.50 | 0.54 | 0.00 | 0.61 | 0.62 | 0.68 | 0.32 | 0.60 | 0.75 | 0.79 | 0.91 | 0.89 | 0.70 |
| GY | 0.45 | 0.56 | 0.00 | 0.62 | 0.68 | 0.64 | 0.15 | 0.62 | 0.80 | 0.83 | 0.91 | 0.90 | 0.65 |
| TH | 0.48 | 0.73 | 0.00 | 0.69 | 0.73 | 0.48 | 0.18 | 0.57 | 0.77 | 0.83 | 0.91 | 0.90 | 0.72 |
| All | 0.49 | 0.65 | 0.01 | 0.62 | 0.73 | 0.63 | 0.26 | 0.63 | 0.77 | 0.83 | 0.91 | 0.88 | 0.69 |
| HWE | |||||||||||||
| GC | 0.00 | 0.04 | - | 0.92 | 0.00 | 0.00 | 0.68 | 0.19 | 0.90 | 0.58 | 0.77 | 0.00 | 0.09 |
| HZ | 0.00 | 0.05 | 0.85 | 0.16 | 0.00 | 0.05 | 0.94 | 0.44 | 0.63 | 0.43 | 0.99 | 0.01 | 0.65 |
| YC | 0.02 | 0.00 | 1.00 | 0.99 | 0.00 | 0.15 | 0.89 | 0.38 | 0.97 | 0.48 | 0.99 | 0.06 | 0.22 |
| CD | 0.00 | 0.17 | - | 0.91 | 0.87 | 0.11 | 0.89 | 0.45 | 0.63 | 0.44 | 0.82 | 0.99 | 0.40 |
| GY | 0.03 | 0.06 | - | 0.39 | 0.42 | 0.39 | 0.99 | 0.00 | 0.91 | 0.86 | 0.45 | 0.74 | 0.00 |
| TH | 0.00 | 0.00 | - | 0.90 | 0.88 | 0.67 | 0.90 | 0.50 | 0.96 | 0.54 | 0.95 | 0.99 | 0.49 |
| All | 0.00 | 0.00 | 0.89 | 0.99 | 0.00 | 0.00 | 0.32 | 0.00 | 0.20 | 0.54 | 0.20 | 0.03 | 0.01 |
| IAM | TPM | SMM | |
|---|---|---|---|
| Expected number of loci with excess of heterozygosity | 42.67 | 42.86 | 43.11 |
| Observed number of loci with excess of heterozygosity | 55.00 | 40.00 | 21.00 |
| Observed number of loci with deficient heterozygosity | 19.00 | 34.00 | 53.00 |
| All loci fit mutation–drift equilibrium | 1.30 | 2.09 | 0.37 |
| Pop | Crab1 | Crab2 | Crab6 | Crab8 | Crab11 | Crab12 | Crab15 | Crab17 | Crab18 | Crab21 | Esin42 | HLJEsa42 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Shannon’s Information index | GC | 0.68 | 0.00 | 0.85 | 1.62 | 1.09 | 1.99 | 1.66 | 1.36 | 1.58 | 1.26 | 2.51 | 1.94 |
| HZ | 0.65 | 0.14 | 0.34 | 1.62 | 1.32 | 1.88 | 1.25 | 1.20 | 1.45 | 1.44 | 2.56 | 2.00 | |
| YC | 0.89 | 0.08 | 0.61 | 1.88 | 1.20 | 1.95 | 1.25 | 1.05 | 1.64 | 1.51 | 2.42 | 2.21 | |
| CD | 0.68 | 0.00 | 0.65 | 1.61 | 1.33 | 1.63 | 1.44 | 1.07 | 1.33 | 1.33 | 2.39 | 2.38 | |
| GY | 0.73 | 0.00 | 0.37 | 1.81 | 1.26 | 1.88 | 1.34 | 1.12 | 1.42 | 1.16 | 2.41 | 2.29 | |
| TH | 0.66 | 0.00 | 0.37 | 1.74 | 1.36 | 1.81 | 0.96 | 0.98 | 1.52 | 1.38 | 2.47 | 2.39 | |
| All | 0.74 | 0.04 | 0.61 | 1.82 | 1.33 | 1.97 | 1.43 | 1.19 | 1.63 | 1.39 | 2.62 | 2.40 |
| Pop | Crab1 | Crab2 | Crab6 | Crab8 | Crab11 | Crab12 | Crab15 | Crab17 | Crab18 | Crab21 | Esin42 | HLJEsa42 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PIC | GC | 0.37 | 0.00 | 0.31 | 0.74 | 0.60 | 0.77 | 0.67 | 0.58 | 0.62 | 0.69 | 0.90 | 0.88 |
| HZ | 0.37 | 0.00 | 0.41 | 0.73 | 0.51 | 0.83 | 0.75 | 0.69 | 0.73 | 0.64 | 0.90 | 0.81 | |
| YC | 0.45 | 0.00 | 0.14 | 0.79 | 0.61 | 0.82 | 0.63 | 0.62 | 0.67 | 0.64 | 0.90 | 0.88 | |
| CD | 0.35 | 0.05 | 0.14 | 0.75 | 0.66 | 0.82 | 0.57 | 0.65 | 0.70 | 0.71 | 0.90 | 0.84 | |
| GY | 0.36 | 0.00 | 0.17 | 0.76 | 0.68 | 0.81 | 0.48 | 0.56 | 0.72 | 0.70 | 0.90 | 0.89 | |
| TH | 0.52 | 0.03 | 0.30 | 0.79 | 0.58 | 0.83 | 0.59 | 0.59 | 0.77 | 0.71 | 0.89 | 0.87 |
| Crab1 | Crab20 | Crab2 | Crab11 | Crab18 | Crab15 | Crab6 | Crab17 | Crab8 | Crab12 | Esin42 | HLJEsa42 | Crab21 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fis | −0.62 | 0.38 | −0.02 | −0.06 | 0.26 | 0.27 | −0.03 | 0.28 | 0.01 | −0.01 | 0.10 | 0.18 | −0.03 |
| Fit | −0.60 | 0.40 | −0.01 | −0.04 | 0.29 | 0.29 | 0.00 | 0.29 | 0.02 | 0.01 | 0.12 | 0.200 | −0.02 |
| Fst | 0.01 | 0.03 | 0.01 | 0.01 | 0.03 | 0.03 | 0.03 | 0.02 | 0.01 | 0.02 | 0.02 | 0.01 | 0.01 |
| Nm * | 20.78 | 7.04 | 12.50 | 13.64 | 7.08 | 7.84 | 6.77 | 11.04 | 24.85 | 13.06 | 15.33 | 13.42 | 35.27 |
| Locus | Obs. F | Min F | Max F | Mean | SE | L95 | U95 |
|---|---|---|---|---|---|---|---|
| crab1 | 0.50 | 0.20 | 0.97 | 0.57 | 0.03 | 0.29 | 0.93 |
| crab20 | 0.34 | 0.11 | 0.95 | 0.38 | 0.02 | 0.18 | 0.72 |
| crab2 | 0.98 | 0.50 | 0.99 | 0.84 | 0.02 | 0.50 | 0.99 |
| crab11 | 0.37 | 0.10 | 0.95 | 0.34 | 0.017 | 0.17 | 0.69 |
| crab18 | 0.26 | 0.07 | 0.93 | 0.27 | 0.01 | 0.14 | 0.53 |
| crab15 | 0.36 | 0.12 | 0.96 | 0.41 | 0.02 | 0.21 | 0.78 |
| crab6 | 0.73 | 0.20 | 0.97 | 0.56 | 0.03 | 0.28 | 0.92 |
| crab17 | 0.36 | 0.16 | 0.97 | 0.50 | 0.03 | 0.25 | 0.88 |
| crab8 | 0.22 | 0.06 | 0.92 | 0.24 | 0.00 | 0.12 | 0.51 |
| crab12 | 0.16 | 0.07 | 0.93 | 0.27 | 0.01 | 0.14 | 0.56 |
| Esin42 | 0.08 | 0.05 | 0.90 | 0.19 | 0.00 | 0.10 | 0.38 |
| HLJEsa42 | 0.11 | 0.05 | 0.90 | 0.18 | 0.00 | 0.10 | 0.38 |
| crab21 | 0.30 | 0.12 | 0.96 | 0.40 | 0.02 | 0.20 | 0.78 |
| GC | HZ | YC | CD | GY | TH | |
|---|---|---|---|---|---|---|
| GC | 0.95 | 0.95 | 0.95 | 0.94 | 0.94 | |
| HZ | 0.04 | 0.97 | 0.96 | 0.97 | 0.96 | |
| YC | 0.04 | 0.02 | 0.95 | 0.96 | 0.95 | |
| CD | 0.05 | 0.04 | 0.04 | 0.97 | 0.95 | |
| GY | 0.05 | 0.02 | 0.03 | 0.02 | 0.97 | |
| TH | 0.05 | 0.03 | 0.04 | 0.04 | 0.02 |
| Source of Variation | d.f | Sum of Squares | Variance Components | Percentage of Variation |
|---|---|---|---|---|
| Among populations | 5 | 22.91 | 0.013 | 0.36 |
| Among individuals Within populations | 174 | 662.62 | 0.20 | 5.56 |
| Within individuals | 180 | 613.00 | 3.41 | 94.08 |
| Total | 359 | 1298.53 | 3.62 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, S.; Nsekanabo, J.D.; Munganga, B.P.; He, X.; Li, J.; Yu, F.; Wang, M.; Tang, Y. Genetic Diversity and Population Structure of the Chinese Mitten Crab (Eriocheir sinensis) from Six Different Lakes Using Microsatellites. Fishes 2023, 8, 220. https://doi.org/10.3390/fishes8050220
Su S, Nsekanabo JD, Munganga BP, He X, Li J, Yu F, Wang M, Tang Y. Genetic Diversity and Population Structure of the Chinese Mitten Crab (Eriocheir sinensis) from Six Different Lakes Using Microsatellites. Fishes. 2023; 8(5):220. https://doi.org/10.3390/fishes8050220
Chicago/Turabian StyleSu, Shengyan, Jean Damascene Nsekanabo, Brian Pelekelo Munganga, Xinjin He, Jianlin Li, Fan Yu, Meiyao Wang, and Yongkai Tang. 2023. "Genetic Diversity and Population Structure of the Chinese Mitten Crab (Eriocheir sinensis) from Six Different Lakes Using Microsatellites" Fishes 8, no. 5: 220. https://doi.org/10.3390/fishes8050220
APA StyleSu, S., Nsekanabo, J. D., Munganga, B. P., He, X., Li, J., Yu, F., Wang, M., & Tang, Y. (2023). Genetic Diversity and Population Structure of the Chinese Mitten Crab (Eriocheir sinensis) from Six Different Lakes Using Microsatellites. Fishes, 8(5), 220. https://doi.org/10.3390/fishes8050220