First Evaluation of Associated Gut Microbiota in Wild Thick-Lipped Grey Mullets (Chelon labrosus, Risso 1827)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. DNA Extraction and Sequencing
2.3. Data Processing
2.4. Functional Analysis by PICRUSt
3. Results
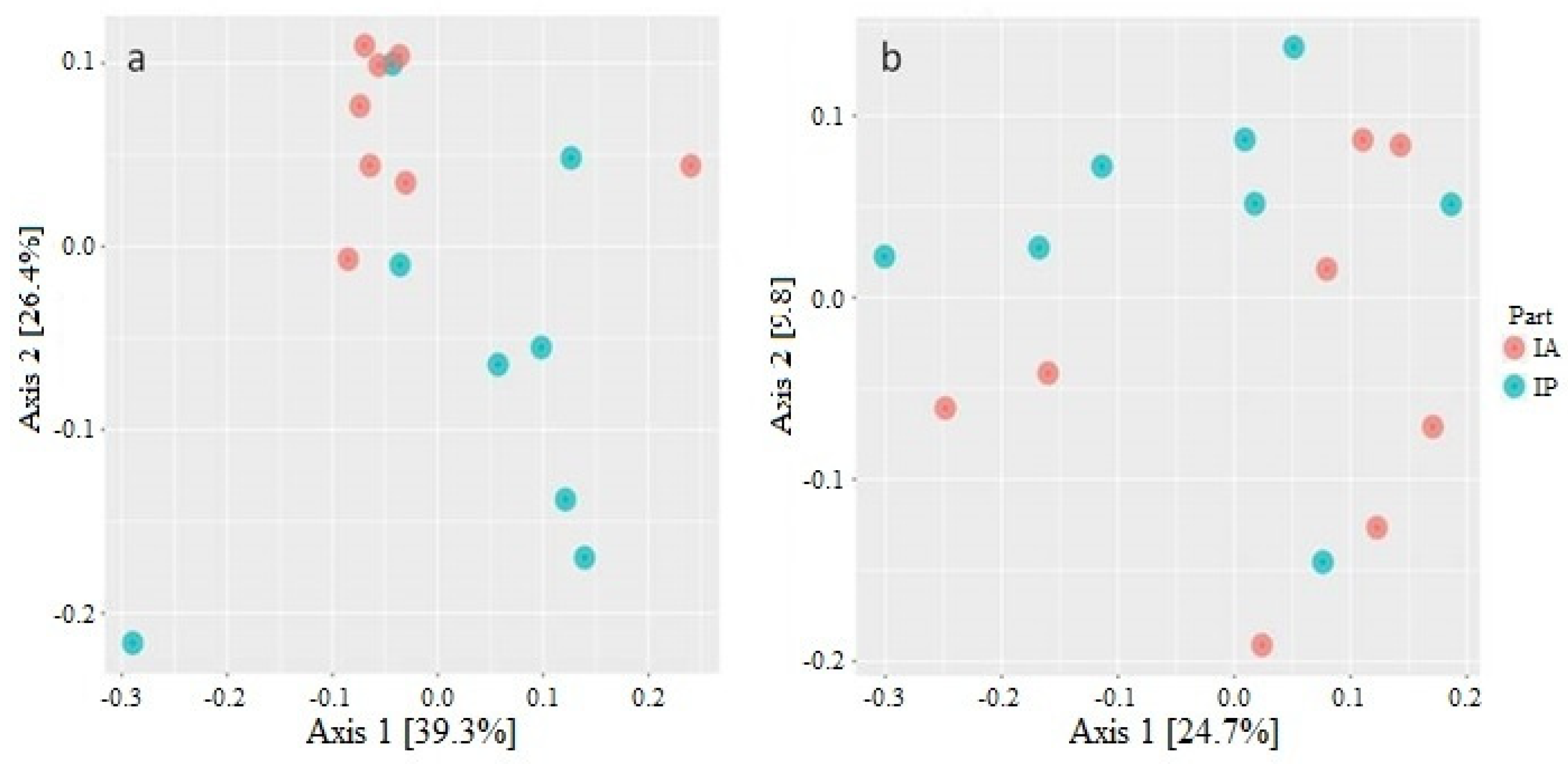
3.1. Diversity Analysis of Microbiota of Wild C. labrosus
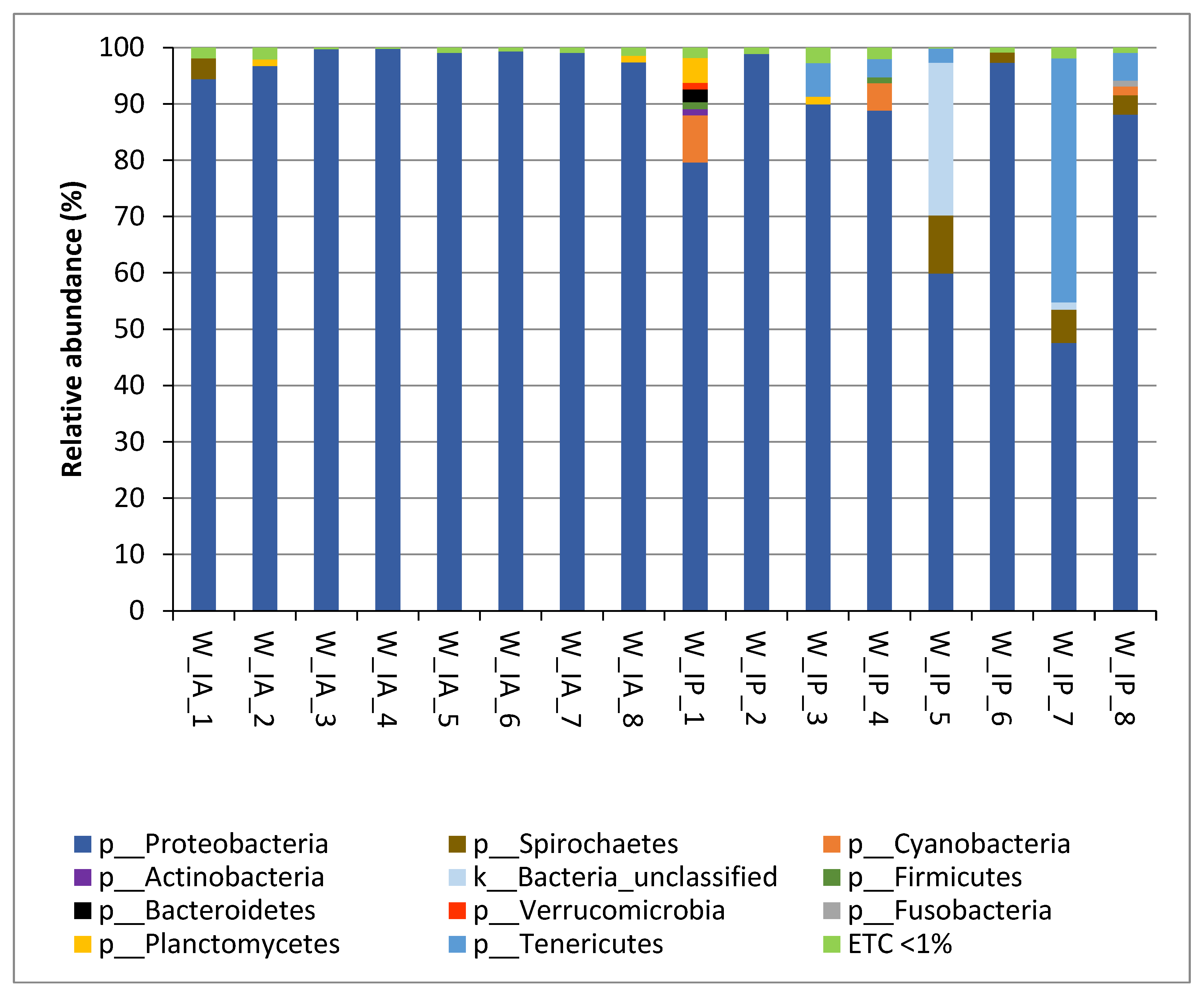
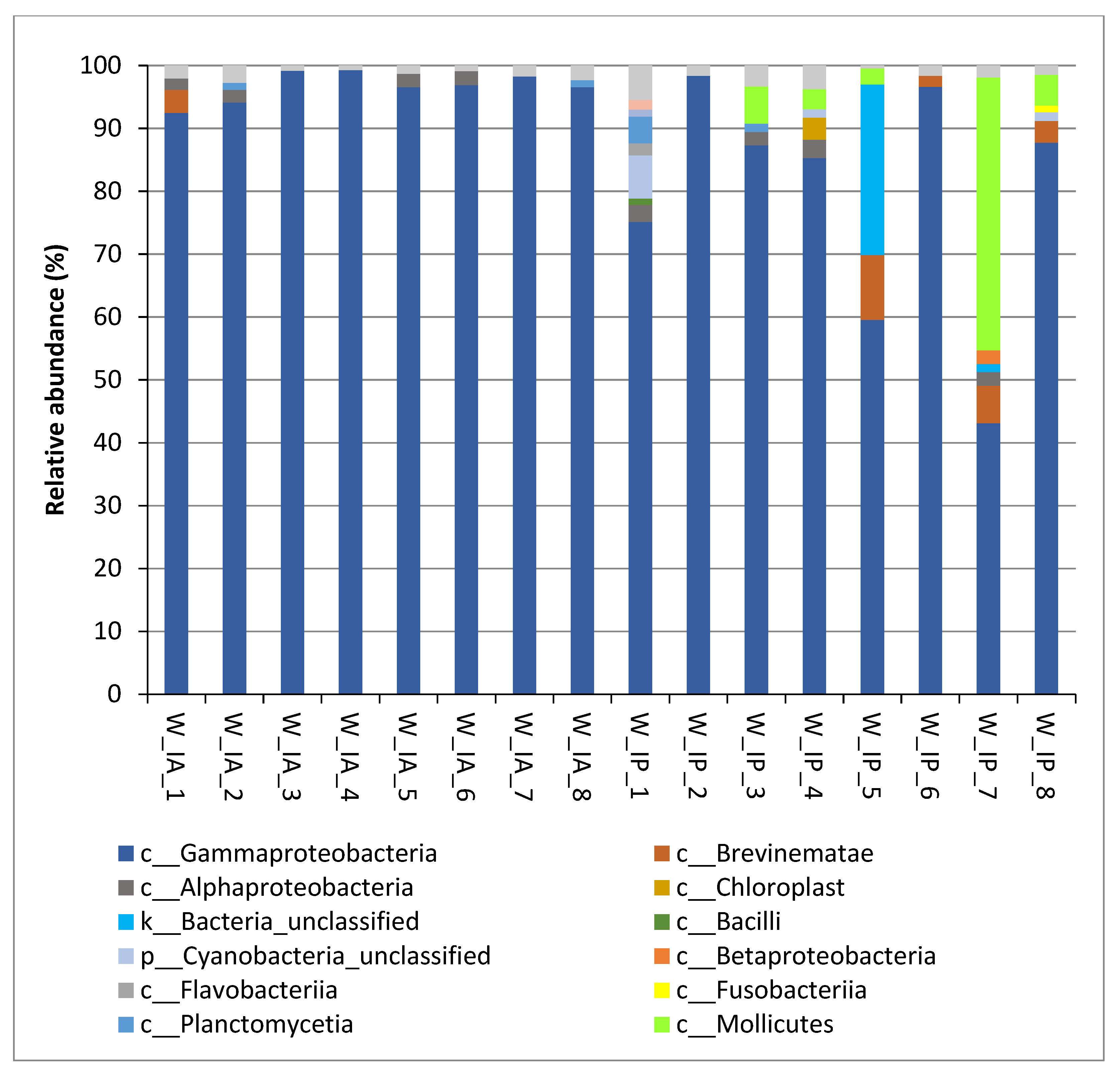
3.2. Composition of Intestinal Microbiota of Wild C. labrosus
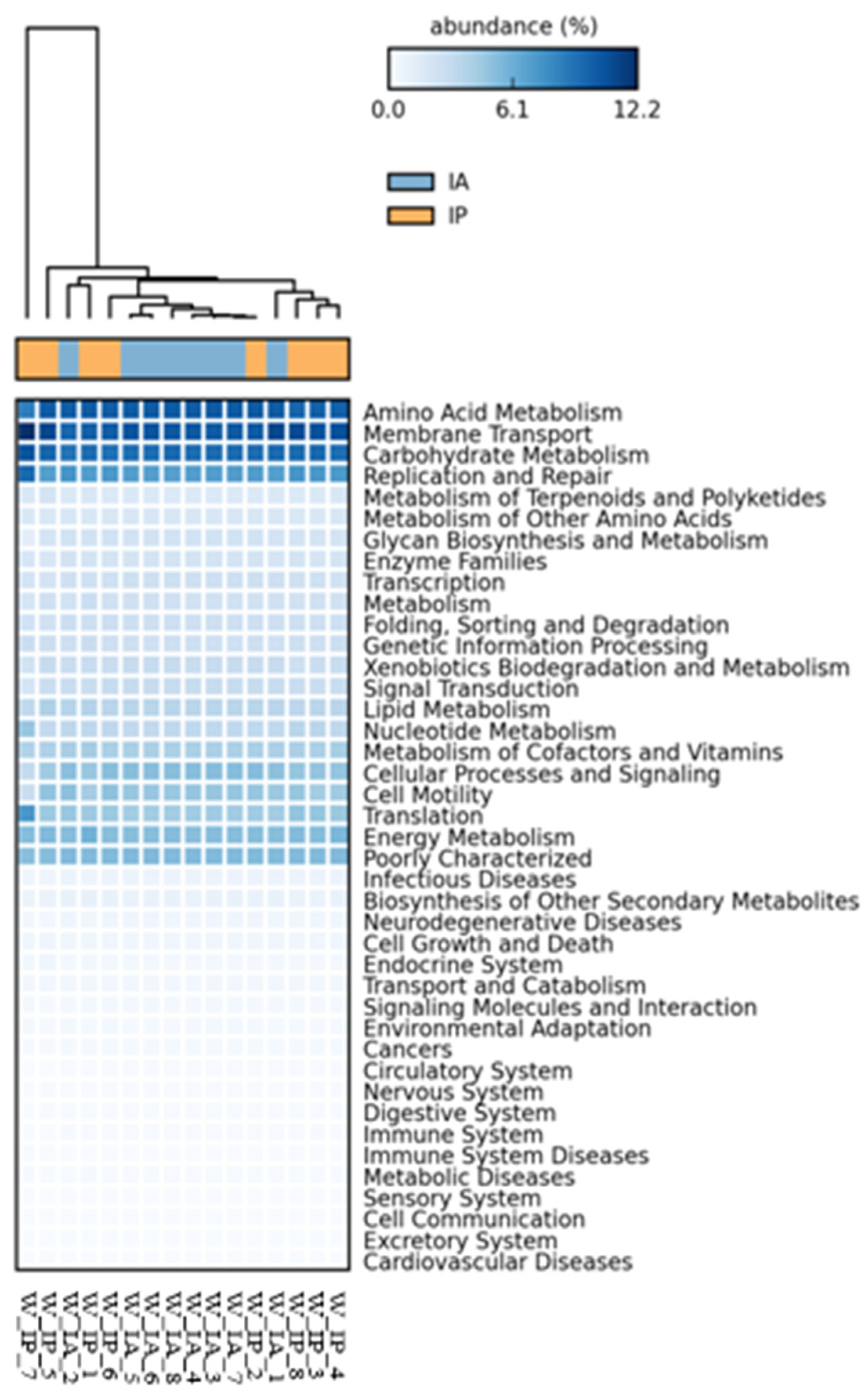
3.3. Functional Inference of the Gut Microbiota
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAO. The State of World Fisheries and Aquaculture 2020. In Sustainability in Action; FAO: Rome, Italy, 2020. [Google Scholar] [CrossRef]
- Naylor, R.L.; Hardy, R.W.; Buschmann, A.H.; Bush, S.R.; Cao, L.; Klinger, D.H.; Little, D.C.; Lubchenco, J.; Shumway, S.E.; Troell, M. A 20-year retrospective review of global aquaculture. Nature 2021, 591, 551–563. [Google Scholar] [CrossRef] [PubMed]
- Zander, K.; Risius, A.; Feucht, Y.; Janssen, M.; Hamm, U. Sustainable Aquaculture Products: Implications of Consumer Awareness and of Consumer Preferences for Promising Market Communication in Germany. J. Aquat. Food Prod. Technol. 2017, 27, 5–20. [Google Scholar] [CrossRef]
- Hua, K.; Cobcroft, J.M.; Cole, A.; Condon, K.; Jerry, D.R.; Mangott, A.; Praeger, C.; Vucko, M.J.; Zeng, C.; Zenger, K.; et al. The Future of Aquatic Protein: Implications for Protein Sources in Aquaculture Diets. One Earth 2019, 1, 316–329. [Google Scholar] [CrossRef]
- Boyd, C.E.; D’Abramo, L.R.; Glencross, B.D.; Huyben, D.C.; Juarez, L.M.; Lockwood, G.S.; McNevin, A.A.; Tacon, A.G.J.; Teletchea, F.; Tomasso, J.R., Jr.; et al. Achieving sustainable aquaculture: Historical and current perspectives and future needs and challenges. J. World Aquac. Soc. 2020, 51, 578–633. [Google Scholar] [CrossRef]
- Prabu, E.; Rajagopalsamy, C.; Ahilan, B.; Jeevagan, I.; Renuhadevi, M. Tilapia—An excellent candidate species for world aquaculture: A review. Annu. Res. Rev. Biol. 2019, 31, 1–14. [Google Scholar] [CrossRef]
- Koven, W.; Gisbert, E.; Meiri-Ashkenazi, I.; Nixon, O.; Israeli, D.; Tandler, A.; Soria, H.N.; Solovyev, M.; Rosenfeld, H. The effect of weaning diet type on grey mullet (Mugil cephalus) juvenile performance during the trophic shift from carnivory to omnivory. Aquaculture 2020, 518, 734848. [Google Scholar] [CrossRef]
- Whitfield, A.K. Ecological role of Mugilidae in the coastal zone. In Biology, Ecology and Culture of Grey Mullets (Mugilidae); CRC Press: Boca Raton, FL, USA, 2016; pp. 324–348. [Google Scholar]
- Crosetti, D.; Blaber, S.J.M. Biology, Ecology and Culture of Grey Mullets (Mugilidae); CRC Press: Boca Raton, FL, USA, 2015. [Google Scholar]
- Cardona, L. Habitat selection by grey mullets (Osteichthyes: Mugilidae) in Mediterranean estuaries: The role of salinity. Sci. Mar. 2006, 70, 443–455. [Google Scholar] [CrossRef]
- Whitfield, A.K.; Panfili, J.; Durand, J.-D. A global review of the cosmopolitan flathead mullet Mugil cephalus Linnaeus 1758 (Teleostei: Mugilidae), with emphasis on the biology, genetics, ecology and fisheries aspects of this apparent species complex. Rev. Fish Biol. Fish. 2012, 22, 641–681. [Google Scholar] [CrossRef]
- Alexander, K.; Potts, T.; Freeman, S.; Israel, D.; Johansen, J.; Kletou, D.; Meland, M.; Pecorino, D.; Rebours, C.; Shorten, M.; et al. The implications of aquaculture policy and regulation for the development of integrated multi-trophic aquaculture in Europe. Aquaculture 2015, 443, 16–23. [Google Scholar] [CrossRef]
- Abellan, E.; Arnal, I. Múgiles, mújoles o mugílidos. In Diversificación de Especies en la Piscicultura Marina Española; Ministerio de Agricultura, Alimentación y Medio Ambiente: Madrid, Spain, 2013; pp. 506–509. [Google Scholar]
- Heras, V.D.L.; Martos-Sitcha, J.; Yúfera, M.; Mancera, J.; Martínez-Rodríguez, G. Influence of stocking density on growth, metabolism and stress of thick- lipped grey mullet (Chelon labrosus) juveniles. Aquaculture 2015, 448, 29–37. [Google Scholar] [CrossRef]
- Pujante, I.M.; Moyano, F.J.; Martos-Sitcha, J.A.; Mancera, J.M.; Martínez-Rodríguez, G. Effect of different salinities on gene expression and activity of digestive enzymes in the thick-lipped grey mullet (Chelon labrosus). Fish Physiol. Biochem. 2018, 44, 349–373. [Google Scholar] [CrossRef] [PubMed]
- García-Márquez, J.; Galafat, A.; Alarcón, F.; Figueroa, F.; Martínez-Manzanares, E.; Arijo, S.; Abdala-Díaz, R. Cultivated and Wild Juvenile Thick-Lipped Grey Mullet, Chelon labrosus: A Comparison from a Nutritional Point of View. Animals 2021, 11, 2112. [Google Scholar] [CrossRef] [PubMed]
- García-Márquez, J.; Galafat, A.; Vizcaíno, A.J.; Barany, A.; Martos-Sitcha, J.A.; Mancera, J.M.; Acién, G.; Figueroa, F.L.; Alarcón, F.J.; Arijo, S.; et al. Dietary Use of the Microalga Chlorella fusca Improves Growth, Metabolism, and Digestive Functionality in Thick-Lipped Grey Mullet (Chelon labrosus, Risso 1827) Juveniles. Front. Mar. Sci. 2022, 9, 902203. [Google Scholar] [CrossRef]
- Egerton, S.; Culloty, S.; Whooley, J.; Stanton, C.; Ross, R.P. The Gut Microbiota of Marine Fish. Front. Microbiol. 2018, 9, 873. [Google Scholar] [CrossRef]
- Collins, S.L.; Patterson, A.D. The gut microbiome: An orchestrator of xenobiotic metabolism. Acta Pharm. Sin. B 2020, 10, 19–32. [Google Scholar] [CrossRef]
- Butt, R.L.; Volkoff, H. Gut Microbiota and Energy Homeostasis in Fish. Front. Endocrinol. 2019, 10, 9. [Google Scholar] [CrossRef]
- Yukgehnaish, K.; Kumar, P.; Sivachandran, P.; Marimuthu, K.; Arshad, A.; Paray, B.A.; Arockiaraj, J. Gut microbiota metagenomics in aquaculture: Factors influencing gut microbiome and its physiological role in fish. Rev. Aquac. 2020, 12, 1903–1927. [Google Scholar] [CrossRef]
- Floris, R.; Sanna, G.; Satta, C.; Piga, C.; Sanna, F.; Lugliè, A.; Fois, N. Intestinal Microbial Ecology and Fillet Metal Chemistry of Wild Grey Mullets Reflect the Variability of the Aquatic Environment in a Western Mediterranean Coastal Lagoon (Santa Giusta, Sardinia, Italy). Water 2021, 13, 879. [Google Scholar] [CrossRef]
- Le, M.H.; Wang, D. Structure and membership of gut microbial communities in multiple fish cryptic species under potential migratory effects. Sci. Rep. 2020, 10, 7547. [Google Scholar] [CrossRef]
- Tarnecki, A.M.; Patterson, W.F., III; Arias, C.R. Microbiota of wild-caught Red Snapper Lutjanus campechanus. BMC Microbiol. 2016, 16, 245. [Google Scholar] [CrossRef] [PubMed]
- Ou, W.; Yu, G.; Zhang, Y.; Mai, K. Recent progress in the understanding of the gut microbiota of marine fishes. Mar. Life Sci. Technol. 2021, 3, 434–448. [Google Scholar] [CrossRef]
- Martínez, G.; Shaw, E.M.; Carrillo, M.; Zanuy, S. Protein Salting-Out Method Applied to Genomic DNA Isolation from Fish Whole Blood. BioTechniques 1998, 24, 238–239. [Google Scholar] [CrossRef]
- Tapia-Paniagua, S.T.; Fumanal, M.; Anguis, V.; Fernandez-Diaz, C.; Alarcón, F.J.; Moriñigo, M.A.; Balebona, M.C. Modulation of Intestinal Microbiota in Solea senegalensis Fed Low Dietary Level of Ulva ohnoi. Front. Microbiol. 2019, 10, 171. [Google Scholar] [CrossRef] [PubMed]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of General 16S Ribosomal RNA Gene PCR Primers for Classical and Next-Generation Sequencing-Based Diversity Studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High throughput Sequence Data. 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 12 March 2021).
- Cerezo-Ortega, I.M.; Di Zeo-Sánchez, D.E.; García-Márquez, J.; Ruiz-Jarabo, I.; Sáez-Casado, M.I.; Balebona, M.C.; Moriñigo, M.A.; Tapia-Paniagua, S.T. Microbiota composition and intestinal integrity remain unaltered after the inclusion of hydrolysed Nannochloropsis gaditana in Sparus aurata diet. Sci. Rep. 2021, 11, 18779. [Google Scholar] [CrossRef]
- Cerezo, I.M.; Fumanal, M.; Tapia-Paniagua, S.T.; Bautista, R.; Anguís, V.; Fernández-Díaz, C.; Alarcón, F.J.; Moriñigo, M.A.; Balebona, M.C. Solea senegalensis Bacterial Intestinal Microbiota Is Affected by Low Dietary Inclusion of Ulva ohnoi. Front. Microbiol. 2022, 12, 801744. [Google Scholar] [CrossRef]
- Oksanen, J.; Kindt, R.; Legendre, P.; O’Hara, B.; Stevens, M.H.H.; Oksanen, M.J.; Suggests, M.A.S.S. The vegan package. Community Ecol. Package 2007, 10, 631–637. [Google Scholar]
- McMURDIE, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PloS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Pedrós-Alió, C. Marine microbial diversity: Can it be determined? Trends Microbiol. 2006, 14, 257–263. [Google Scholar] [CrossRef]
- Langille, M.G.I.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. Data, information, knowledge and principle: Back to metabolism in KEGG. Nucleic Acids Res. 2014, 42, D199–D205. [Google Scholar] [CrossRef] [PubMed]
- Dittmann, K.K.; Rasmussen, B.B.; Castex, M.; Gram, L.; Bentzon-Tilia, M. The aquaculture microbiome at the centre of business creation. Microb. Biotechnol. 2017, 10, 1279–1282. [Google Scholar] [CrossRef] [PubMed]
- Albertini-Berhaut, P.J. L’Intestin chez les Mugilidae (Poissons; Téléostéens) a différentes étapes de leur croissance I. Aspects morphologiques et histologiques. J. Appl. Ichthyol. 1987, 3, 1–12. [Google Scholar] [CrossRef]
- Ringø, E.; Olsen, R.E.; Mayhew, T.M.; Myklebust, R. Electron microscopy of the intestinal microflora of fish. Aquaculture 2003, 227, 395–415. [Google Scholar] [CrossRef]
- Amillano-Cisneros, J.M.; Hernández-Rosas, P.T.; Gomez-Gil, B.; Navarrete-Ramírez, P.; Ríos-Durán, M.G.; Martínez-Chávez, C.C.; Johnston-Monje, D.; Martínez-Palacios, C.A.; Raggi, L. Loss of gut microbial diversity in the cultured, agastric fish, Mexican pike silverside (Chirostoma estor: Atherinopsidae). PeerJ 2022, 10, e13052. [Google Scholar] [CrossRef] [PubMed]
- Lowrey, L.; Woodhams, D.C.; Tacchi, L.; Salinas, I. Topographical Mapping of the Rainbow Trout (Oncorhynchus mykiss) Microbiome Reveals a Diverse Bacterial Community with Antifungal Properties in the Skin. Appl. Environ. Microbiol. 2015, 81, 6915–6925. [Google Scholar] [CrossRef]
- Dhanasiri, A.K.S.; Brunvold, L.; Brinchmann, M.F.; Korsnes, K.; Bergh, Ø.; Kiron, V. Changes in the Intestinal Microbiota of Wild Atlantic cod Gadus morhua L. Upon Captive Rearing. Microb. Ecol. 2011, 61, 20–30. [Google Scholar] [CrossRef]
- Liu, H.; Guo, X.; Gooneratne, R.; Lai, R.; Zeng, C.; Zhan, F.; Wang, W. The gut microbiome and degradation enzyme activity of wild freshwater fishes influenced by their trophic levels. Sci. Rep. 2016, 6, 24340. [Google Scholar] [CrossRef]
- Ramírez, C.; Romero, J. Fine Flounder (Paralichthys adspersus) Microbiome Showed Important Differences between Wild and Reared Specimens. Front. Microbiol. 2017, 8, 271. [Google Scholar] [CrossRef]
- Ramírez, C.; Romero, J. The Microbiome of Seriola lalandi of Wild and Aquaculture Origin Reveals Differences in Composition and Potential Function. Front. Microbiol. 2017, 8, 1844. [Google Scholar] [CrossRef]
- Escalas, A.; Auguet, J.-C.; Avouac, A.; Seguin, R.; Gradel, A.; Borrossi, L.; Villéger, S. Ecological Specialization Within a Carnivorous Fish Family Is Supported by a Herbivorous Microbiome Shaped by a Combination of Gut Traits and Specific Diet. Front. Mar. Sci. 2021, 8, 91. [Google Scholar] [CrossRef]
- Llewellyn, M.S.; McGinnity, P.; Dionne, M.; Letourneau, J.; Thonier, F.; Carvalho, G.R.; Creer, S.; Derome, N. The biogeography of the atlantic salmon (Salmo salar) gut microbiome. ISME J. 2016, 10, 1280–1284. [Google Scholar] [CrossRef] [PubMed]
- Kashinskaya, E.N.; Simonov, E.P.; Izvekova, G.I.; Parshukov, A.N.; Andree, K.B.; Solovyev, M.M. Composition of the microbial communities in the gastrointestinal tract of perch (Perca fluviatilis L. 1758) and cestodes parasitizing the perch digestive tract. J. Fish Dis. 2020, 43, 23–38. [Google Scholar] [CrossRef] [PubMed]
- Daly, K.; Kelly, J.; Moran, A.W.; Bristow, R.; Young, I.S.; Cossins, A.R.; Bravo, D.; Shirazi-Beechey, S. Host selectively contributes to shaping intestinal microbiota of carnivorous and omnivorous fish. J. Gen. Appl. Microbiol. 2019, 65, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yan, Q.; Ringø, E.; Wu, X.; He, Y.; Yang, D. The influence of weight and gender on intestinal bacterial community of wild largemouth bronze gudgeon (Coreius guichenoti, 1874). BMC Microbiol. 2016, 16, 191. [Google Scholar] [CrossRef]
- Ei-Jakee, J.; Elshamy, S.; Hassan, A.-W.; Abdelsalam, M.; Younis, N.; El-Hady, M.A.; Eissa, A.E. Isolation and characterization of Mycoplasmas from some moribund Egyptian fishes. Aquac. Int. 2020, 28, 901–912. [Google Scholar] [CrossRef]
- Legrand, T.P.R.A.; Wynne, J.W.; Weyrich, L.S.; Oxley, A.P.A. Investigating Both Mucosal Immunity and Microbiota in Response to Gut Enteritis in Yellowtail Kingfish. Microorganisms 2020, 8, 1267. [Google Scholar] [CrossRef]
- Webster, T.M.U.; Rodriguez-Barreto, D.; Castaldo, G.; Gough, P.; Consuegra, S.; de Leaniz, C.G. Environmental plasticity and colonisation history in the Atlantic salmon microbiome: A translocation experiment. Mol. Ecol. 2020, 29, 886–898. [Google Scholar] [CrossRef]
- Domínguez-Maqueda, M.; Cerezo, I.M.; Tapia-Paniagua, S.T.; De La Banda, I.G.; Moreno-Ventas, X.; Moriñigo, M.Á.; Balebona, M.C. A tentative study of the effects of heat-inactivation of the probiotic strain Shewanella putrefaciens pdp11 on senegalese sole (Solea senegalensis) intestinal microbiota and immune response. Microorganisms 2021, 9, 808. [Google Scholar] [CrossRef]
- Brown, R.M.; Wiens, G.D.; Salinas, I. Analysis of the gut and gill microbiome of resistant and susceptible lines of rainbow trout (Oncorhynchus mykiss). Fish Shellfish Immunol. 2019, 86, 497–506. [Google Scholar] [CrossRef]
- Gupta, S.; Lokesh, J.; Abdelhafiz, Y.; Siriyappagouder, P.; Pierre, R.; Sørensen, M.; Fernandes, J.M.O.; Kiron, V. Macroalga-Derived Alginate Oligosaccharide Alters Intestinal Bacteria of Atlantic Salmon. Front. Microbiol. 2019, 10, 2037. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wang, J.; He, T.; Becker, S.; Zhang, G.; Li, D.; Ma, X. Butyrate: A Double-Edged Sword for Health? Adv. Nutr. 2018, 9, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Mielko, K.A.; Jabłoński, S.J.; Milczewska, J.; Sands, D.; Łukaszewicz, M.; Młynarz, P. Metabolomic studies of Pseudomonas aeruginosa. World J. Microbiol. Biotechnol. 2019, 35, 178. [Google Scholar] [CrossRef] [PubMed]
- Hoque, F.; Abraham, T.J.; Nagesh, T.S.; Kamilya, D. Pseudomonas aeruginosa FARP72 Offers Protection Against Aeromonas hydrophila Infection in Labeo rohita. Probiotics Antimicrob. Proteins 2019, 11, 973–980. [Google Scholar] [CrossRef]
- Giri, S.S.; Jun, J.W.; Yun, S.; Kim, H.J.; Kim, S.G.; Woo, K.J.; Han, S.J.; Oh, W.T.; Kwon, J.; Sukumaran, V.; et al. Effects of dietary heat-killed Pseudomonas aeruginosa strain VSG2 on immune functions, antioxidant efficacy, and disease resistance in Cyprinus carpio. Aquaculture 2020, 514, 734489. [Google Scholar] [CrossRef]
- Givens, C.; Ransom, B.; Bano, N.; Hollibaugh, J. Comparison of the gut microbiomes of 12 bony fish and 3 shark species. Mar. Ecol. Prog. Ser. 2015, 518, 209–223. [Google Scholar] [CrossRef]
- Cámara-Ruiz, M.; Cerezo, I.; Guardiola, F.; García-Beltrán, J.; Balebona, M.; Moriñigo, M.; Esteban, M. Alteration of the Immune Response and the Microbiota of the Skin during a Natural Infection by Vibrio harveyi in European Seabass (Dicentrarchus labrax). Microorganisms 2021, 9, 964. [Google Scholar] [CrossRef]
- Reyes-Becerril, M.; Alamillo, E.; Angulo, C. Probiotic and Immunomodulatory Activity of Marine Yeast Yarrowia lipolytica Strains and Response Against Vibrio parahaemolyticus in Fish. Probiotics Antimicrob. Proteins 2021, 13, 1292–1305. [Google Scholar] [CrossRef]
- Gatesoupe, F.-J.; Infante, J.-L.Z.; Cahu, C.; Quazuguel, P. Early weaning of seabass larvae, Dicentrarchus labrax: The effect on microbiota, with particular attention to iron supply and exoenzymes. Aquaculture 1997, 158, 117–127. [Google Scholar] [CrossRef]
- Sugita, H.; Ito, Y. Identification of intest\inal bacteria from Japanese flounder (Paralichthys olivaceus) and their ability to digest chitin. Lett. Appl. Microbiol. 2006, 43, 336–342. [Google Scholar] [CrossRef]
- MacDonald, N.; Stark, J.; Austin, B. Bacterial microflora in the gastro-intestinal tract of Dover sole (Solea solea L.), with emphasis on the possible role of bacteria in the nutrition of the host. FEMS Microbiol. Lett. 1986, 35, 107–111. [Google Scholar] [CrossRef]
- Pier, G.B. Pseudomonas aeruginosa lipopolysaccharide: A major virulence factor, initiator of inflammation and target for effective immunity. Int. J. Med Microbiol. 2007, 297, 277–295. [Google Scholar] [CrossRef] [PubMed]
- Green, E.R.; Mecsas, J. Bacterial Secretion Systems: An Overview. Microbiol. Spectr. 2016, 4, 1. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Cheng, C.; Xie, J.; Liu, S.; Ma, H.; Feng, J.; Su, Y.; Guo, Z. Coupled changes of bacterial community and function in the gut of mud crab (Scylla Paramamosain) in response to Baimang disease. AMB Express 2019, 9, 18. [Google Scholar] [CrossRef]
- DeWitt, R.C.; Kudsk, K.A. The gut’s role in metabolism, mucosal barrier function, and gut immunology. Infect. Dis. Clin. N. Am. 1999, 13, 465–481. [Google Scholar] [CrossRef]
- Li, T.; Long, M.; Li, H.; Gatesoupe, F.-J.; Zhang, X.; Zhang, Q.; Feng, D.; Li, A. Multi-Omics Analysis Reveals a Correlation between the Host Phylogeny, Gut Microbiota and Metabolite Profiles in Cyprinid Fishes. Front. Microbiol. 2017, 8, 454. [Google Scholar] [CrossRef]
- Agus, A.; Planchais, J.; Sokol, H. Gut Microbiota Regulation of Tryptophan Metabolism in Health and Disease. Cell Host Microbe 2018, 23, 716–724. [Google Scholar] [CrossRef]
- Kuz’Mina, V. Physiological Adaptations (By the Example of the Exotrophy Process in Fish). J. Evol. Biochem. Physiol. 2001, 37, 285–299. [Google Scholar] [CrossRef]
- Pujante, I.M.; Díaz-López, M.; Mancera, J.M.; Moyano, F.J. Characterization of digestive enzymes protease and alpha-amylase activities in the thick-lipped grey mullet (Chelon labrosus, Risso 1827). Aquac. Res. 2017, 48, 367–376. [Google Scholar] [CrossRef]
- Uscanga-Martínez, A.; Perales-García, N.; Alvarez-González, C.A.; Moyano, F.J.; Ramírez, D.T.; Gisbert, G.E.; Márquez-Couturier, G.; Contreras-Sánchez, W.M.; Arias-Rodriguez, L.; Indy, J.R. Changes in digestive enzyme activity during initial ontogeny of bay snook Petenia splendida. Fish Physiol. Biochem. 2011, 37, 667–680. [Google Scholar] [CrossRef]
- Hernández-Sámano, A.; Guzmán-García, X.; García-Barrientos, R.; Guerrero-Legarreta, I. Actividad enzimática de proteasas de Cyprinus carpio (Cypriniformes: Cyprinidae) extraídas de una laguna contaminada en México. Rev. Biol. Trop. 2017, 65, 589–597. [Google Scholar] [CrossRef]
- Staats, C.C.; Boldo, J.; Broetto, L.; Vainstein, M.; Schrank, A. Comparative genome analysis of proteases, oligopeptide uptake and secretion systems in Mycoplasma spp. Genet. Mol. Biol. 2007, 30, 225–229. [Google Scholar] [CrossRef]
- Ray, A.; Ghosh, K.; Ringø, E. Enzyme-producing bacteria isolated from fish gut: A review. Aquac. Nutr. 2012, 18, 465–492. [Google Scholar] [CrossRef]
- Villasante, A.; Ramírez, C.; Rodríguez, H.; Catalán, N.; Díaz, O.; Rojas, R.; Opazo, R.; Romero, J. In-depth analysis of swim bladder-associated microbiota in rainbow trout (Oncorhynchus mykiss). Sci. Rep. 2019, 9, 8974. [Google Scholar] [CrossRef]
- Wu, Z.; Wang, S.; Zhang, Q.; Hao, J.; Lin, Y.; Zhang, J.; Li, A. Assessing the intestinal bacterial community of farmed Nile tilapia (Oreochromis niloticus) by high-throughput absolute abundance quantification. Aquaculture 2020, 529, 735688. [Google Scholar] [CrossRef]
- Cui, W.; Ma, A.; Huang, Z.; Liu, Z.; Yang, K.; Zhang, W. myo-inositol facilitates salinity tolerance by modulating multiple physiological functions in the turbot Scophthalmus maximus. Aquaculture 2020, 527, 735451. [Google Scholar] [CrossRef]
- Li, P.; Mai, K.; Trushenski, J.; Wu, G. New developments in fish amino acid nutrition: Towards functional and environmentally oriented aquafeeds. Amino Acids 2009, 37, 43–53. [Google Scholar] [CrossRef]
- Hakomori, S.-I.; Igarashi, Y. Functional Role of Glycosphingolipids in Cell Recognition and Signaling. J. Biochem. 1995, 118, 1091–1103. [Google Scholar] [CrossRef]
- Jennemann, R.; Gröne, H.-J. Cell-specific in vivo functions of glycosphingolipids: Lessons from genetic deletions of enzymes involved in glycosphingolipid synthesis. Prog. Lipid Res. 2013, 52, 231–248. [Google Scholar] [CrossRef]
- Li, J.; Chi, Z. Siderophores from marine microorganisms and their applications. J. Ocean Univ. China 2004, 3, 40–47. [Google Scholar] [CrossRef]
- Qian, Y.; Li, X.-F.; Zhang, D.-D.; Cai, D.-S.; Tian, H.-Y.; Liu, W.-B. Effects of Dietary Pantothenic Acid on Growth, Intestinal Function, Anti-Oxidative Status and Fatty Acids Synthesis of Juvenile Blunt Snout Bream Megalobrama amblycephala. PLoS ONE 2015, 10, e0119518. [Google Scholar] [CrossRef] [PubMed]
- Solovyev, M.M.; Kashinskaya, E.N.; Bochkarev, N.A.; Andree, K.B.; Simonov, E. The effect of diet on the structure of gut bacterial community of sympatric pair of whitefishes (Coregonus lavaretus): One story more. PeerJ 2019, 7, e8005. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Jian, S.Q.; Cao, H.; Wen, C.; Hu, B.; Peng, M.; Peng, L.; Yuan, J.; Liang, L. Changes in microbiota along the intestine of grass carp (Ctenopharyngodon idella): Community, interspecific interactions, and functions. Aquaculture 2019, 498, 151–161. [Google Scholar] [CrossRef]
- Catron, T.R.; Gaballah, S.; Tal, T. Using Zebrafish to Investigate Interactions Between Xenobiotics and Microbiota. Curr. Pharmacol. Rep. 2019, 5, 468–480. [Google Scholar] [CrossRef]
- Petriello, M.C.; Hoffman, J.B.; Vsevolozhskaya, O.; Morris, A.J.; Hennig, B. Dioxin-like PCB 126 increases intestinal inflammation and disrupts gut microbiota and metabolic homeostasis. Environ. Pollut. 2018, 242, 1022–1032. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Tang, L.; Liu, Y.; Hu, C.; Zhou, B.; Lam, P.K.; Lam, J.C.; Chen, L. Activation of aryl hydrocarbon receptor by dioxin directly shifts gut microbiota in zebrafish. Environ. Pollut. 2019, 255, 113357. [Google Scholar] [CrossRef]
- DeBofsky, A.; Xie, Y.; Grimard, C.; Alcaraz, A.J.; Brinkmann, M.; Hecker, M.; Giesy, J.P. Differential responses of gut microbiota of male and female fathead minnow (Pimephales promelas) to a short-term environmentally-relevant, aqueous exposure to benzo[a]pyrene. Chemosphere 2020, 252, 126461. [Google Scholar] [CrossRef]
- Vitali, F.; Mandalakis, M.; Chatzinikolaou, E.; Dailianis, T.; Senatore, G.; Casalone, E.; Mastromei, G.; Sergi, S.; Lussu, R.; Arvanitidis, C.; et al. Benthic Prokaryotic Community Response to Polycyclic Aromatic Hydrocarbon Chronic Exposure: Importance of Emission Sources in Mediterranean Ports. Front. Mar. Sci. 2019, 6, 590. [Google Scholar] [CrossRef]

| IA | IP | p | |
|---|---|---|---|
| Chao1 | 124.15 ± 44.26 | 158.61 ± 64.87 | 0.235 |
| Shannon | 1.40 ± 0.23 | 1.91 ± 0.42 * | 0.009 |
| Simpson | 0.66 ± 0.04 | 0.76 ± 0.07 * | 0.002 |
| KEGG Pathways | IA | IP | p |
|---|---|---|---|
| Lipid metabolism | |||
| Biosynthesis of unsaturated fatty acids | 0.355 ± 0.006 | 0.308 ± 0.062 | 0.080 |
| Fatty acid biosynthesis | 0.496 ± 0.016 | 0.466 ± 0.084 | 0.360 |
| Lipid biosynthesis proteins | 0.750 ± 0.017 | 0.744 ± 0.073 | 0.827 |
| Carbohydrate metabolism | |||
| Inositol phosphate metabolism | 0.228 ± 0.008 * | 0.203 ± 0.002 | 0.032 |
| Amino acid metabolism | |||
| Lysine biosynthesis | 0.603 ± 0.014 | 0.560 ± 0.096 | 0.252 |
| Lysine degradation | 0.377 ± 0.009 | 0.384 ± 0.060 | 0.736 |
| Phenylalanine metabolism | 0.388 ± 0.020 * | 0.345 ± 0.016 | 0.013 |
| Phenylalanine, tyrosine and tryptophan biosynthesis | 0.713 ± 0.017 | 0.670 ± 0.104 | 0.280 |
| Valine, leucine and isoleucine biosynthesis | 0.663 ± 0.020 | 0.658 ± 0.025 | 0.653 |
| Valine, leucine and isoleucine degradation | 0.758 ± 0.027 | 0.734 ± 0.096 | 0.512 |
| Metabolism of cofactors and vitamin | |||
| Folate biosynthesis | 0.476 ± 0.015 | 0.442 ± 0.056 | 0.137 |
| Pantothenate and CoA biosynthesis | 0.479 ± 0.012 | 0.507 ± 0.017 * | 0.023 |
| Ubiquinone and other terpenoid-quinone biosynthesis | 0.398 ± 0.007 | 0.368 ± 0.069 | 0.255 |
| Energy metabolism | |||
| Carbon fixation in photosynthetic organisms | 0.380 ± 0.007 | 0.463 ± 0.108 | 0.069 |
| Carbon fixation pathways in prokaryotes | 0.992 ± 0.014 | 0.986 ± 0.126 | 0.896 |
| Photosynthesis | 0.274 ± 0.008 | 0.361 ± 0.093 | 0.063 |
| Metabolism of terpenoids and polyketides | |||
| Biosynthesis of ansamycins | 0.040 ± 0.002 | 0.052 ± 0.015 | 0.054 |
| Biosynthesis of siderophore group non ribosomal peptides | 0.092 ± 0.004 * | 0.077 ± 0.013 | 0.049 |
| Biosynthesis of vancomicyn group antibiotics | 0.047 ± 0.003 | 0.047 ± 0.010 | 0.936 |
| Geraniol degradation | 0.301 ± 0.010 | 0.299 ± 0.081 | 0.948 |
| Limonene and pinene degradation | 0.252 ± 0.009 | 0.267 ± 0.036 | 0.280 |
| Polyketide sugar unit biosynthesis | 0.133 ± 0.004 | 0.126 ± 0.023 | 0.463 |
| Terpenoid backbone biosynthesis | 0.399 ± 0.008 | 0.456 ± 0.072 | 0.058 |
| Tetracycline biosynthesis | 0.143 ± 0.005 | 0.133 ± 0.023 | 0.280 |
| Zeatin biosynthesis | 0.029 ± 0.001 | 0.036 ± 0.012 | 0.139 |
| Glycan biosynthesis and metabolism | |||
| Glycosaminoglycan degradation | 0.038 ± 0.004 | 0.037 ± 0.014 | 0.827 |
| Glycosphingolipid biosynthesis–ganglio series | 0.035 ± 0.001 | 0.028 ± 0.008 | 0.069 |
| Glycosphingolipid biosynthesis–globo series | 0.082 ± 0.007 * | 0.066 ± 0.011 | 0.028 |
| Lipopolysaccharide biosynthesis | 0.392 ± 0.010 | 0.357 ± 0.068 | 0.190 |
| Lipopolysaccharide biosynthesis proteins | 0.543 ± 0.013 | 0.487 ± 0.083 | 0.097 |
| N-Glycan biosynthesis | 0.027 ± 0.003 | 0.029 ± 0.011 | 0.697 |
| Other glycan degradation | 0.103 ± 0.003 | 0.096 ± 0.012 | 0.167 |
| Peptidoglycan biosynthesis | 0.622 ± 0.012 | 0.576 ± 0.098 | 0.229 |
| Biosynthesis of other secondary metabolites | |||
| beta-Lactam resistance | 0.045 ± 0.003 | 0.037 ± 0.011 | 0.105 |
| Butirosin and neomycin biosynthesis | 0.046 ± 0.004 | 0.043 ± 0.009 | 0.434 |
| Isoquinoline alkaloid biosynthesis | 0.045 ± 0.002 | 0.053 ± 0.012 | 0.110 |
| Novobiocin biosynthesis | 0.133 ± 0.002 | 0.129 ± 0.006 | 0.104 |
| Penicillin and cephalosporin biosynthesis | 0.071 ± 0.004 | 0.062 ± 0.014 | 0.146 |
| Phenylpropanoid biosynthesis | 0.108 ± 0.006 | 0.109 ± 0.017 | 0.965 |
| Streptomycin biosynthesis | 0.235 ± 0.004 | 0.223 ± 0.039 | 0.384 |
| Tropane, piperidine and pyridine alkaloid biosynthesis | 0.127 ± 0.001 | 0.125 ± 0.003 | 0.063 |
| Xenobiotics biodegradation and metabolism | |||
| Aminobenzoate degradation | 0.323 ± 0.005 | 0.307 ± 0.063 | 0.507 |
| Atrazine degradation | 0.032 ± 0.009 | 0.029 ± 0.008 | 0.528 |
| Benzoate degradation | 0.408 ± 0.043 | 0.384 ± 0.090 | 0.509 |
| Bisphenol degradation | 0.045 ± 0.005 | 0.056 ± 0.019 | 0.163 |
| Caprolactam degradation | 0.192 ± 0.006 | 0.192 ± 0.047 | 0.983 |
| Chloroalkane and chloroalkene degradation | 0.284 ± 0.009 | 0.282 ± 0.020 | 0.737 |
| Chlorocyclohexane and chlorobenzene degradation | 0.094 ± 0.006 | 0.083 ± 0.020 | 0.174 |
| Dioxin degradation | 0.043 ± 0.011 | 0.035 ± 0.008 | 0.104 |
| Drug metabolism–cytochrome P450 | 0.220 ± 0.013 | 0.192 ± 0.040 | 0.099 |
| Ethylbenzene degradation | 0.076 ± 0.003 | 0.069 ± 0.012 | 0.130 |
| Fluorobenzoate degradation | 0.057 ± 0.001 * | 0.046 ± 0.001 | 0.037 |
| Metabolism of xenobiotics by cytochrome P450 | 0.218 ± 0.013 | 0.189 ± 0.041 | 0.089 |
| Naphthalene degradation | 0.215 ± 0.007 | 0.202 ± 0.035 | 0.337 |
| Nitrotoluene degradation | 0.059 ± 0.004 | 0.064 ± 0.021 | 0.563 |
| Polycyclic aromatic hydrocarbon degradation | 0.025 ± 0.006 | 0.036 ± 0.017 | 0.113 |
| Styrene degradation | 0.104 ± 0.005 * | 0.088 ± 0.012 | 0.017 |
| Toluene degradation | 0.198 ± 0.003 | 0.172 ± 0.033 | 0.061 |
| Xylene degradation | 0.042 ± 0.011 | 0.034 ± 0.008 | 0.083 |
| Membrane transport | |||
| ABC transporters | 2.520 ± 0.199 | 2.654 ± 0.242 | 0.245 |
| Bacterial secretion system | 0.849 ± 0.034 | 0.821 ± 0.041 | 0.153 |
| Secretion system | 2.423 ± 0.070 | 2.213 ± 0.293 | 0.083 |
| Transporters | 4.286 ± 0.280 | 4.660 ± 0.625 | 0.144 |
| Cell motility | |||
| Bacterial chemotaxis | 0.895 ± 0.025 | 0.855 ± 0.139 | 0.451 |
| Bacterial motility proteins | 2.668 ± 0.082 | 2.422 ± 0.431 | 0.156 |
| Flagellar assembly | 0.980 ± 0.025 | 0.893 ± 0.164 | 0.179 |
| Other | |||
| Bacterial toxins | 0.124 ± 0.007 | 0.115 ± 0.013 | 0.147 |
| Cell cycle–Caulobacter | 0.381 ± 0.008 | 0.402 ± 0.023 | 0.088 |
| Two-component system | 2.787 ± 0.032 | 2.569 ± 0.295 | 0.076 |
| Pertussis | 0.062 ± 0.004 | 0.054 ± 0.008 | 0.072 |
| Tuberculosis | 0.138 ± 0.002 | 0.151 ± 0.013 | 0.091 |
| Vibrio cholerae infection | 0.031 ± 0.006 | 0.027 ± 0.007 | 0.267 |
| Vibrio cholerae pathogenic cycle | 0.335 ± 0.020 | 0.285 ± 0.061 | 0.060 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
García-Márquez, J.; Cerezo, I.M.; Figueroa, F.L.; Abdala-Díaz, R.T.; Arijo, S. First Evaluation of Associated Gut Microbiota in Wild Thick-Lipped Grey Mullets (Chelon labrosus, Risso 1827). Fishes 2022, 7, 209. https://doi.org/10.3390/fishes7040209
García-Márquez J, Cerezo IM, Figueroa FL, Abdala-Díaz RT, Arijo S. First Evaluation of Associated Gut Microbiota in Wild Thick-Lipped Grey Mullets (Chelon labrosus, Risso 1827). Fishes. 2022; 7(4):209. https://doi.org/10.3390/fishes7040209
Chicago/Turabian StyleGarcía-Márquez, Jorge, Isabel M. Cerezo, Félix L. Figueroa, Roberto Teófilo Abdala-Díaz, and Salvador Arijo. 2022. "First Evaluation of Associated Gut Microbiota in Wild Thick-Lipped Grey Mullets (Chelon labrosus, Risso 1827)" Fishes 7, no. 4: 209. https://doi.org/10.3390/fishes7040209
APA StyleGarcía-Márquez, J., Cerezo, I. M., Figueroa, F. L., Abdala-Díaz, R. T., & Arijo, S. (2022). First Evaluation of Associated Gut Microbiota in Wild Thick-Lipped Grey Mullets (Chelon labrosus, Risso 1827). Fishes, 7(4), 209. https://doi.org/10.3390/fishes7040209






