Abstract
The Yangtze finless porpoise (Neophocaena asiaeorientalis asiaeorientalis, YFP) is a critically endangered small odontocete species, mainly distributed in the middle and lower reaches of the Yangtze River, Poyang Lake, and Dongting Lake. Under the influence of human activities, many factors are threatening the survival and reproduction of YFPs in their natural habitat. Ex situ conservation is of great significance to strengthen the rescuing conservation of YFPs by providing suitable alternative habitats and promoting the reproduction and growth of the ex situ population. To reveal the differences in gene expression of YFPs in natural and ex situ protected waters, and to investigate the effects of environmental factors on YFPs and their mechanisms, we performed transcriptome sequencing for blood tissues of YFPs collected from natural waters and ex situ protected waters. Using RNA-seq we identified 4613 differentially expressed genes (DEGs), of which 4485 were up-regulated and 128 were down-regulated in the natural population. GO analysis showed that DEGs were significantly enriched in entries related to binding, catalytic activity, and biological regulation; KEGG analysis showed that DEGs were enriched mainly in signal transduction, endocrine system, immune system, and sensory system-related pathways. Further analysis revealed that water pollution in natural waters may affect the hormone secretion of YFPs by altering the expression pattern of endocrine genes, thus interfering with normal endocrine activities; noise pollution may induce oxidative stress and inflammatory responses in YFPs, thus impairing the auditory function of YFPs. This study provides a new perspective for further research on the effect of habitat conditions on the YFPs and suggests that improving the habitat environment may help in the conservation of YFPs.
1. Introduction
Along with the accelerated transformation of the natural environment by humans, ex situ conservation has become one of the important means of rare wildlife conservation [1]. Ex situ conservation refers to the removal of species whose survival and reproduction are seriously threatened to an appropriate environment for special protection and management [2]. Ex situ conservation has been widely used as the most fundamental species protection measure because it is less restricted by natural conditions and can fully improve the living environment of endangered species and formulate scientific management plans according to the growth and development law of species [3,4].
The Yangtze finless porpoise (Neophocaena asiaeorientalis asiaeorientalis) is a small odontocete species endemic to China, mainly distributed in the middle and lower reaches of the Yangtze River, Dongting Lake and Poyang Lake. The species is the only cetacean left in the Yangtze River after the baiji (Lipotes vexillifer) was declared functionally extinct in 2007 [5]. In 2013, the Yangtze finless porpoise was listed as a critically endangered species by the International Union for Conservation of Nature’s Species Survival Commission (IUCN/SSC) [6], and in 2021, it was listed as a national first-class key protected wild animal. To prevent the Yangtze finless porpoise from repeating the functional extinction of the baiji, in recent years, porpoise conservationists have been paying more attention to ex situ conservation while strengthening in situ conservation of the Yangtze finless porpoise [7]. At present, five Yangtze finless porpoise ex situ populations have been established in China, which have made positive contributions to strengthening the rescuing conservation of Yangtze finless porpoise.
Although the conservation of Yangtze finless porpoise has been heavily promoted, the Yangtze finless porpoise living in the natural waters of the middle and lower reaches of the Yangtze River still faces severe survival pressure. Research shows that the factors threatening the survival and reproduction of Yangtze finless porpoise are mainly from the following four aspects. First, limited food resources: before the “10-year ban on fishing” policy was formally implemented, long-term overfishing had significantly reduced fish resources in the Yangtze River basin, making it difficult to meet the demand of Yangtze finless porpoise for bait [8]. Second, water conservancy and hydropower projects: a large number of water-related projects have occupied the survival spaces of Yangtze finless porpoise, cutting off the living passage of fish and shrimp and other bait resources [9]. Third, frequent water traffic: the noise generated by ships and cross-river channels directly interferes with the hearing system of Yangtze finless porpoise and affects its daily life activities such as communicating, locating, and feeding [10]. Fourth, water pollution: the discharge of industrial wastewater and domestic sewage along the river seriously damages the survival environment of Yangtze finless porpoise [11]. Compared with natural waters, ex situ protected waters not only retain the natural environment of the original habitat but also carry out scientific transformation on this basis and strictly eliminate all forms of human interference. Therefore, the Yangtze finless porpoise living there is almost not restricted by the above factors.
It has been reported that changes in the living environment can lead to changes in the gene expression profile of animals, such as differential gene expression patterns in killifish living in heavily polluted and non-polluted waters [12], and transcriptome sequencing can reflect the gene expression in real time [13]. There are two advantages of using blood for transcriptome study. First, it is convenient to obtain peripheral blood and less harmful to experimental animals [14]. Second, blood flows to all parts of the body along the circulation system, involving a wide range of systems that can be used in many aspects of research, such as immunity [14], endocrine [15], stress response [16], and so on.
Studies have shown that there are significant differences in blood hormone levels [17], stress, and immune parameters [18] of the Yangtze finless porpoise between different habitats, suggesting that the natural habitat of the Yangtze finless porpoise is deteriorating. In addition, the researchers monitored underwater noise in the Yangtze Basin and found that noise levels detected at multiple points were sufficient to damage the auditory function of the Yangtze finless porpoise [19]. The above studies indicate that water pollution and noise pollution in natural waters are serious threats to the survival and reproduction of the Yangtze finless porpoise. However, due to the constraints of experimental subjects and conditions, these studies mostly focused on the population level or individual level and rarely involved the molecular level. The molecular mechanism of environmental pollution on the Yangtze finless porpoise remains unclear.
To understand the differences in gene expression of Yangtze finless porpoise in natural and ex situ protected waters, and to investigate the impact and molecular regulatory mechanism of habitat conditions on Yangtze finless porpoise, we used RNA-Seq to sequence and compare the transcriptomes of Yangtze finless porpoise blood tissues collected in natural and ex situ protected waters, and to discover the most significant functional pathways and key genes affected by environmental factors. The study will serve as a starting point for further investigation of the adaptive regulatory mechanisms of Yangtze finless porpoise to different environments and provides information to help plan conservation of the Yangtze finless porpoise.
2. Materials and Methods
2.1. Ethics Statement
We followed the national regulations and institutional policies for the care and use of animals in China, and the medical examinations and related experiments conducted were approved by the Anqing Fisheries Bureau of China. The research complies with the Chinese Aquatic Animal Protection Act promulgated in 1993 and amended in 2013.
2.2. Animals and Samples Collection
In November 2017, with the approval of the Yangtze River Basin Fisheries Administration Office of the Ministry of Agriculture and Rural Affairs of the People’s Republic of China, the Freshwater Fisheries Research Center of the Chinese Academy of Fishery Sciences organized and implemented the ex situ protection action of the Yangtze finless porpoise. During the mission, six healthy Yangtze finless porpoise were selected as experimental subjects, including three from Anqing Section of Yangtze River (CJ_1, CJ_2, CJ_3) and three from the Anqing Xijiang Yangtze finless porpoise ex situ protection base (XJ_1, XJ_2, XJ_3); see Table S1 for specific information. We captured the animals in batches using the traditional “sound chase and net capture” means [20]. To avoid stressing the Yangtze finless porpoise, soft spacious fishermen nets were used to form an enclosure, and the motorboat was turned off to make the animals calm down. The follow-up physical examination can be carried out only when the animals come out of the water gently, move freely, and have no obvious stress. Blood samples were extracted from the main tail vein using a 10 mL disposable syringe, and then transferred to a vacuum collecting vessel (PAXgeneTM, BD, USA), which was transported back to the laboratory and stored in a −80 °C ultra-low temperature refrigerator for future use.
2.3. Total RNA Extraction, mRNA Sequencing and Analysis
Total RNA was extracted from whole blood samples of six Yangtze finless porpoise according to the instructions of PAXgene Blood RNA Kit (QIAGEN, Hilden, Germany), and the quality of RNA was checked by Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA), and the qualified RNA was used for subsequent library construction. The cDNA library was constructed using MGIEasy RNA library preparation kit (Huada, Wuhan, China). After the library was successfully constructed, high-throughput sequencing was performed based on the MGI-2000 platform (Huada, Wuhan, China).
The raw data generated by sequencing were filtered using SOAPnuke (v1.4.0) software [21] to remove reads with sequencing adapter, ‘N’ base ratio over 5%, and low-quality base ratio over 20% to obtain clean reads. The obtained clean reads were analyzed by sequence alignment with the Yangtze finless porpoise reference genome using HISAT2 (v1.4.0) software [22], and then the gene expression level of each sample was calculated using RSEM software (v1.2.8) [23], and the gene expression level was expressed as FPKM (fragments per kilobase of transcript per million mapped fragments).
2.4. Differentially Expressed Gene Screening and Enrichment Analysis
DEseq2 (v1.4.5) [24] was used for the screening of differentially expressed genes (DEGs), with the screening criteria |log2foldchange| ≥ 1 and q ≤ 0.001 (adjusted p value). Functional classification and biological pathway classification of DEGs were performed based on the GO (Gene Ontology) and KEGG (Kyoto Encyclopedia of Genes and Genomes) annotation results, while enrichment analysis was performed using phyper function based on a hypergeometric test. p ≤ 0.05 was considered significantly enriched, p ≤ 0.001 was considered extremely significantly enriched.
2.5. Real-Time Quantitative PCR Validation
To verify the accuracy of transcriptome sequencing results, five differentially expressed genes (PTGS1, prostaglandin-endoperoxide synthase 1; FBXO11, F-box protein 11; SMAD4, SMAD family member 4; PRDX2, peroxiredoxin 2; GPX2, glutathione peroxidase 2) were randomly selected for real-time quantitative PCR. GAPDH (glyceraldehyde 3-phosphate dehydrogenase) was selected as the endogenous control gene. Primer sequences (Table S2) were designed using Primer Premier 5 (Permier Biosoft 5.0, USA). HiScript III RT SuperMix for qPCR kit (Vazyme, China) was used for reverse transcription of mRNA. All qRT-PCR was performed with ChamQ Universal SYBR qPCR Master Mix (Vazyme, China). The reaction was carried out with a total volume of 20 μL including 10 μL qPCR Mix, 0.8 μL of primers (10 μM), 2 μL of cDNA (500 ng), and 7.2 μL ddH2O. The following PCR program was applied in two steps: denaturation at 95 °C for 30 s followed by 40 cycles consisting of 95 °C for 10 s and 60 °C for 30 s. qRT-PCR was performed using a StepOne type fluorescent quantitative PCR instrument (ABI, Foster, CA, USA), and relative gene expression was calculated using the 2−ΔΔCT method.
2.6. Supporting Information
mRNA clean transcriptome data of XJ_1, XJ_2 and XJ_3 has been uploaded to the Sequence Read Archive (SRA) under accession number PRJNA789349. mRNA clean transcriptome data of CJ_1, CJ_2, and CJ_3 has been uploaded to SRA under accession number PRJNA699632 in another paper [25].
3. Results
3.1. Sequencing Data and Alignment Analysis
The transcriptome libraries of the sample of Yangtze finless porpoise were constructed using MGI-2000 platform, and a total of 455,560,000 raw reads were obtained. After removing low-quality reads, reads containing the sequencing adapter, and reads with excessive unknown base ‘N’ content, a total of 444,820,000 clean reads were obtained. The average size of each sample was 7.42 Gb and six samples had base quality values of Q20 ≥ 97% and Q30 ≥ 93%. The sequence alignment of the clean reads of each sample was compared with the reference genome, and the average alignment rate was 79.85%. The average alignment rate of uniquely mapped reads was 46.46% (Table 1). The transcriptome sequencing quality was satisfactory, meaning the constructed libraries could be used for subsequent gene expression analysis.

Table 1.
Sequencing and mapping statistics of blood samples for the Yangtze finless porpoise.
3.2. Analysis and Functional Annotation of Differentially Expressed Genes
Differential expression analysis of the above qualified transcriptome data showed that a total of 4613 DEGs were identified in the CJ and XJ groups, of which 4485 were significantly up-regulated in the CJ group and 128 were significantly down-regulated in the CJ group (Table S3). To further understand the physiological functions regulated by DEGs, functional annotation of DEGs was performed using GO and KEGG databases. The results showed that a total of 3259 DEGs were annotated to the GO database, including “biological process” (1875), “cellular component” (2024), and “molecular function” (2605). The function entry with the highest number of annotated genes was related to binding and belonged to “molecular function” (Figure S1). A total of 2176 DEGs were annotated to the KEGG database, including five functional categories: “environmental information processing” (808), “genetic information processing” (470), “cellular processes” (721), “organismal systems” (960), and “metabolism” (532). The functional pathway with the highest number of annotated genes is related to signal transduction, which belongs to “environmental information processing” (Figure S2).
3.3. Enrichment Analysis of Differentially Expressed Genes
GO enrichment analysis of DEGs showed that 4613 DEGs were significantly enriched to 345 GO terms and 58 GO terms were extremely significantly enriched (Figure S3), belonging to binding (24), catalytic activity (15), biological regulation (6), molecular function regulator (4), organelle part (4), metabolic process (2), protein-containing complex (1), cellular process (1), and cell (1). Figure 1A showed the top 10 GO terms with the most significant enrichment degree. The GO enrichment results indicate that DEGs are involved in a wide range of regulatory functions.
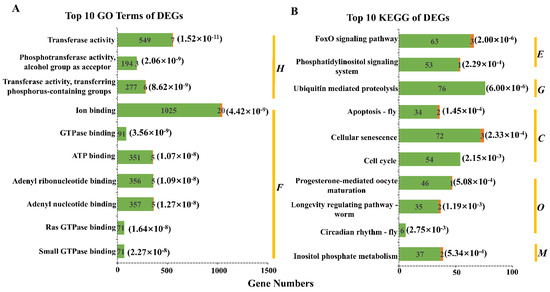
Figure 1.
Top 10 GO terms and KEGG pathways of DEGs. (A) Top 10 GO terms of DEGs. The numbers marked on green and orange part in each term were the number of downregulated and upregulated DEGs in the XJ group. The numbers in parentheses represent p value of each term. There are two categories: H, Catalytic Activity; F, Binding. (B) Top 10 KEGG pathways of DEGs. There are five categories: E, Environmental Information Processing; G, Genetic Information Processing; C, Cellular Processes; O, Organismal Systems; M, Metabolism. The meaning of the remaining parts can refer to (A).
KEGG enrichment analysis of DEGs showed that 4613 DEGs were significantly enriched to 43 KEGG pathways (Figure 2A), including 6 endocrine pathways, 5 immune pathways, 1 sensory pathway, etc. (Table 2), of which Figure 1B shows the top 10 KEGG pathways with the most significant enrichment degree. Considering the special environmental conditions of natural waters (water pollution, noise pollution), the changes of the above pathways and corresponding genes were most likely caused by environmental factors.
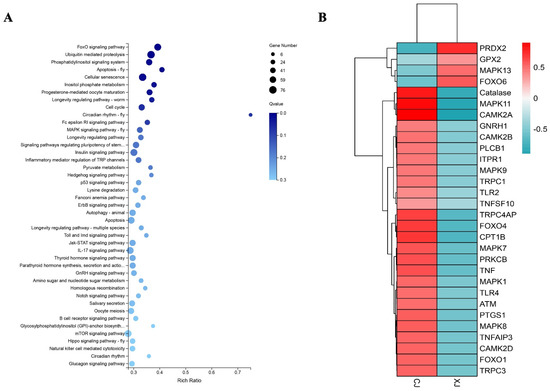
Figure 2.
KEGG enrichment analysis of DEGs and heatmap of key genes. (A) 43 KEGG pathways significantly enriched. (B) The heatmap of key genes. Red means up-regulating, blue means down-regulating, the shade of color means degree of regulating.

Table 2.
The enrichment results of several key KEGG pathways.
In addition, seven pathways were extremely significantly enriched, namely FOXO signaling pathway (ko04068), ubiquitin-mediated proteolysis (ko04120), apoptosis-fly (ko04214), phosphatidylinositol signaling system (ko04070), cellular senescence (ko04218), progesterone-mediated oocyte maturation (ko04914), and inositol phosphate metabolism (ko00562). These seven pathways are mainly involved in the regulation of important physiological processes such as intracellular signal transduction, endocrine system, substance metabolism, and cell growth and death.
3.4. Analysis of Key Genes
In this study, through enrichment analysis of DEGs and literature reading, candidate genes greatly affected by environmental factors were screened out, mainly genes related to endocrine system and noise-induced hearing loss. As shown in Figure 2B, most of these genes were up-regulated in the CJ group, suggesting that there may be stimulators inducing the expression of these genes in natural waters. The specific information of candidate genes is shown in Table 3.

Table 3.
Expression of key DEGs in the Yangtze finless porpoise.
3.5. Validation of RNA-Seq Results by qRT-PCR
The results of qRT-PCR validation of five randomly selected differentially expressed genes are shown in Figure 3. PTGS1, FBXO11, and SMAD4 genes were up-regulated in the CJ group, while PRDX2 and GPX2 were down-regulated in the CJ group compared with the XJ group. The differential gene expression verified by qRT-PCR was generally consistent with the transcriptome sequencing results, indicating that the transcriptome sequencing results were reliable in this study.
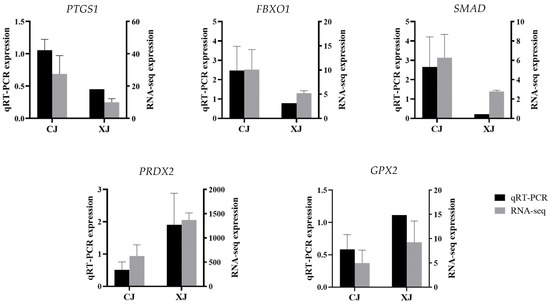
Figure 3.
Quantitative real-time PCR for validation of RNA-seq. qRT-PCR verification of five differentially expressed genes in CJ vs. XJ. The left vertical axis represents qRT-PCR relative expression. The right vertical axis represents RNA-seq expression. PTGS1, prostaglandin-endoperoxide synthase 1; FBXO11, F-box protein 11; SMAD4, SMAD family member 4; PRDX2, peroxiredoxin 2; GPX2, glutathione peroxidase 2.
4. Discussion
4.1. Water Pollution May Interfere with the Endocrine Activity of Yangtze Finless Porpoise
In mammals, the endocrine system is another important regulatory system besides the nervous system, mainly through the gonads, thyroid, and other endocrine glands to secrete hormones, thus regulating the metabolism, growth, and development of the organism, and maintaining the internal environmental homeostasis [26]. In the present study, DEGs were significantly enriched in endocrine-related KEGG pathways, such as GnRH signaling pathway (ko04912), insulin signaling pathway (ko04910), and thyroid hormone signaling pathway (ko04919), suggesting that the endocrine system of Yangtze finless porpoise is significantly different between natural and ex situ protected waters. Since the Yangtze finless porpoises in this study were of similar age and same sex, and none of them were in special periods such as pregnancy and lactation, the influence of their physiological factors on the endocrine system could be roughly excluded. In addition, we also calculated the body mass index (BMI) of the two groups of Yangtze finless porpoise, and using the BMI obesity standard for Yangtze finless porpoise established by Dai et al. [27] we found that all six Yangtze finless porpoise in this study were in the normal physiological range (Table S1) and there was no lean type or overweight, so the influence of nutritional status on the endocrine system was excluded.
Studies have shown that chemicals such as pesticides and fungicides, which are widely available in the environment, can seriously interfere with the endocrine activities of organisms; such substances are known as endocrine disruptors [28]. Typically, endocrine disruptors act on hormone receptors and their downstream signaling pathways to interfere with the synthesis, secretion, and action of hormones [29]. Huang et al. [30] found that azole fungicides could disrupt several endocrine-related signaling pathways, including progesterone-mediated oocyte maturation (ko04914) and insulin signaling pathway (ko04910). Ade et al. [31] inferred from acute toxicology experiments and transcriptomic studies that direct discharge of drugs such as fluoxetine into the water can lead to endocrine disorders in aquatic animals. Through 15 years of monitoring and analysis, Barra et al. [32] found that endocrine disruptors still exist in the wastewater discharged by pulp and paper mills into freshwater ecosystems after modern sewage treatment, and that fish affected by the pollution had a twofold increase in plasma vitellogenin content and a threefold increase in gonadosomatic indices.
The middle and lower reaches of the Yangtze River basin, one of the most economically developed regions in China, have been heavily polluted in recent years. The researchers measured levels of four types of organic micropollutants in the Yangtze River basin, including polycyclic aromatic hydrocarbons (PAHs), polychlorinated biphenyls (PCBs), organochlorine pesticides, medicines, and personal care products. They found that the main pollutants in the middle and lower reaches of the Yangtze River were PAHs and organochlorine pesticides, and the levels of the mixed pollutants were sufficient to cause great harm to aquatic organisms [33]. In addition, according to the historical data of heavy metal detection in the Yangtze River basin since the 1980s, the concentrations of Cu, Cd, Zn and Pb in the sediments of the middle and lower reaches of the Yangtze River have exceeded the critical effect value, and the concentrations of Cd and Hg have an increasing trend [34]. This suggests that the endocrine function of the Yangtze finless porpoise living in the natural waters of the middle and lower reaches of the Yangtze River is likely to change from the normal level due to frequent exposure to environmental pollutants such as pesticides and heavy metals.
Previous studies have proved that pollutants in the environment can induce up-regulated expression of important endocrine-related genes such as ITPR1 and GnRH, which is consistent with our findings. ITPR1 is an important blood glucose regulatory gene encoding the 1,4,5-inositol triphosphate receptor, which is mainly expressed in islet B cells. Wong et al. [35] found that Cd in the environment could induce overexpression of ITPR1 in the islets of mice, resulting in changes in blood glucose homeostasis (Figure 4). GNRH encodes gonadotropin releasing hormone, which can activate MAPK pathway by binding with receptors on the cell membrane and play a role in promoting gonadotropin secretion (Figure 4). Xi et al. [36] found that GNRH was up-regulated at the hypothalamic-pituitary level in both male and female pups exposed to bisphenol A (BPA) pollution, thus interfering with the synthesis and release of sex hormones.
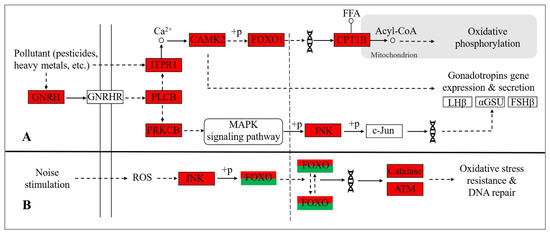
Figure 4.
Functional significance of differentially expressed key genes in two groups of Yangtze finless porpoise. Red represents upregulation in CJ group and green represents downregulation. Solid arrows indicate activation processes, dashed arrows indicate indirect activation processes. (A) The relationship between water pollution and endocrine gene expression. GNRH, gonadotropin releasing hormone; GNRHR, gonadotropin releasing hormone receptor; ITPR1, inositol 1,4,5-trisphosphate receptor type 1; CAMK2, calcium/calmodulin dependent protein kinase II; FOXO, forkhead box O; CPT1B, carnitine palmitoyltransferase 1B; PLCB, phospholipase C beta; PRKCB, protein kinase C beta; JNK (MAPK8/9), mitogen-activated protein kinase 8/9; c-Jun, transcription factor AP-1. (B) The relationship between noise stimulation and gene expression in FOXO pathway. Catalase (RASGEF1A), RasGEF domain family member 1A; ATM, ATM serine/threonine kinase.
4.2. Noise Pollution May Interfere with the Hearing Function of Yangtze Finless Porpoise
In this study, we found that several auditory-related genes, such as transient receptor potential (TRP) channel protein, were significantly differentially expressed between the natural water group and the ex situ protected water group. TRP channels are a kind of ion channel widely distributed in mammals, which are mainly responsible for sensory signal transmission and play an important role in the formation of hearing [37]. Some studies have suggested that this family of channel proteins can be expressed in immune cells and trigger cochlear inflammation when activated by stimulants such as noise [38], but this claim has not been proved.
In fact, for the Yangtze finless porpoise living in the natural waters of the middle and lower reaches of the Yangtze River, noise disturbance caused by human activities has seriously impaired its auditory function. Popov et al. [39] found that high-intensity, prolonged noise exposure caused temporary auditory threshold shift (TTS) in Yangtze finless porpoise, and the longer the noise exposure, the greater the increase in auditory threshold and the longer the recovery time to normal auditory ability. Tao et al. [40] found that riprap noise can be perceived by Yangtze finless porpoise and may affect its auditory function in a survey of riprap construction in the lower Yangtze River. Wang et al. [10] conducted noise monitoring at 25 sites in the middle and lower reaches of the Yangtze River and found that the noise detected at 8% of the sites caused direct permanent auditory threshold shift (PTS) damage to the Yangtze finless porpoise.
The damage caused by noise to hearing is largely caused by inflammation [41]. After noise exposure, a variety of inflammatory cells in the cochlea activate and secrete a large number of inflammatory factors, which on the one hand promote cell apoptosis and tissue damage, and on the other hand induce more inflammatory cells to mobilize and produce more inflammatory factors, aggravating tissue damage [42,43]. We found that the expression of inflammatory and immune-related genes, including TNF-α, TLR2, and TLR4, was significantly increased in the blood of Yangtze finless porpoise under environmental noise stimulation.
TNF-α is the most widely studied cellular inflammatory factor. Previous studies have shown that noise exposure increases TNF-α expression in the cochlea, while spiral artery vasoconstriction reduces blood and oxygen supply to the inner ear, leading to hair cell damage or death, which can be treated with TNF-α inhibitors [44]. TLR2 and TLR4 are both members of the toll-like receptor family. They bind to molecular signals released by damaged cells after noise exposure, activating the intrinsic cochlear immune system and inducing inflammatory cell infiltration and inflammatory factor synthesis, resulting in hearing damage [45,46].
In addition, strong noise or prolonged noise exposure can cause redox imbalance in the cochlea and generate large amounts of Reactive Oxygen Species (ROS) [47]. These oxygen radicals not only directly damage cellular DNA, proteins, and cell membranes, causing necrosis of cochlear hair cells and nerve cells [48], but also can act as intracellular messengers to activate a series of signal transduction pathways, indirectly leading to apoptosis of inner ear hair cells, hearing loss or even complete hearing loss [49]. Therefore, noise-stimulated Yangtze finless porpoise may enhance the expression of antioxidant genes under stress. We noted that in this study, several antioxidation-related genes, such as Catalase, ATM, GPX2, PRDX2, and PTGS1, were up-regulated in the natural water group of the Yangtze finless porpoise. Moreover, KEGG enrichment analysis showed that the FOXO signaling pathway (ko04068) was the most significant. This pathway is involved in biological regulation such as cellular anti-oxidative stress and promoting DNA repair and is of great significance to the stress response of cells [50]. In the FOXO pathway, the gene JNK is susceptible to ROS and other stimuli, leading to the phosphorylation of FOXO. The activated FOXO can further promote the expression of antioxidant genes in cells, thus reducing the level of ROS (Figure 4).
5. Conclusions
In this study, transcriptomic analysis of blood tissues collected from natural and ex situ protected populations of Yangtze finless porpoise was performed using RNA-Seq technology. 4613 DEGs were detected, which were mainly involved in signal transduction, endocrine system, immune system, and sensory system. Further analysis revealed that water pollution in natural waters may affect the hormone secretion of Yangtze finless porpoise by altering the expression pattern of endocrine genes, thus interfering with normal endocrine activities; noise pollution may induce oxidative stress and inflammatory responses in Yangtze finless porpoise, thus impairing the auditory function of Yangtze finless porpoise. This study reveals the differences in blood transcriptome expression profiles of Yangtze finless porpoise in different habitats and explains the multiple effects and mechanisms of environmental factors on Yangtze finless porpoise, providing an important theoretical reference for further strengthening the habitat conservation of Yangtze finless porpoise.
Particularly, because the species is highly endangered, the number of samples used to conduct the experiments is relatively limited, and the influence of chance factors other than habitat on the experimental results cannot be ruled out yet. However, the bias caused by chance factors was controlled within a reasonable range by improving the experimental and analytical protocols, such as shortening the time of physical examination as much as possible and strictly selecting the experimental animals to ensure that the influence of disturbing factors was minimized; the above conditions ensured that the experimental results obtained in this study were scientifically reasonable. However, since further validation experiments have not been conducted, a more in-depth and comprehensive study would therefore be valuable, if possible, in the future.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/fishes7030096/s1, Figure S1: GO classification of differentially expressed genes; Figure S2: KEGG pathway classification of differentially expressed genes; Figure S3: 58 GO terms extremely significantly enriched; Table S1: Morphological information of Yangtze finless porpoises for transcriptomic analysis; Table S2: Summary of qRT-PCR primer sequences for Yangtze finless porpoise; Table S3: 4613 differentially expressed genes in CJ vs. XJ comparison.
Author Contributions
Conceptualization, K.L. and D.Y.; methodology, W.L. and D.Y.; software, W.L., C.Y. and J.Z.; validation, K.L. and P.X.; formal analysis, W.L.; resources, Y.Y. and X.Z.; data curation, W.L., D.Y. and D.L.; writing—original draft preparation, W.L.; writing—review and editing, D.Y. and K.L.; visualization, W.L.; supervision, K.L.; project administration, K.L.; funding acquisition, K.L. and D.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Key R&D Program of China (2021YFD1200304), the Central Public-interest Scientific Institution Basal Research Fund, Freshwater Fisheries Research Center, CAFS (2017JBFM10, 2018JBFM04), Project of Yangtze Finless Porpoise Survey and Conservation (17200360), and the Project of Implementation of Yangtze Finless Porpoise Protection in the Middle and Lower Reaches of Yangtze River (2021).
Institutional Review Board Statement
The animal study protocol was approved by the Anqing Fisheries Bureau of China. The research complies with the Chinese Aquatic Animal Protection Act promulgated in 1993 and amended in 2013.
Data Availability Statement
mRNA clean transcriptome data were deposited in the NCBI Sequence Read Archive database with accession number PRJNA789349 and PRJNA699632.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Pimm, S.L.; Jenkins, C.N.; Abell, R.; Brooks, T.M.; Gittleman, J.L.; Joppa, L.N.; Raven, P.H.; Roberts, C.M.; Sexton, J.O. The biodiversity of species and their rates of extinction, distribution, and protection. Science 2014, 344, 1246752. [Google Scholar] [CrossRef] [PubMed]
- McGowan, P.J.; Traylor-Holzer, K.; Leus, K. IUCN Guidelines for Determining When and How Ex Situ Management Should Be Used in Species Conservation. Conserv. Lett. 2016, 10, 361–366. [Google Scholar] [CrossRef]
- Kleinman-Ruiz, D.; Soriano, L.; Casas-Marce, M.; Szychta, C.; Sánchez, I.; Fernández, J.; Godoy, J.A. Genetic evaluation of the Iberian lynx ex situ conservation programme. Heredity 2019, 123, 647–661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Oliveira, M.J.; Aguiar-Silva, F.H.; de Moraes, W.; Sanaiotti, T.M.; Banhos, A.; Moreira, N. Ex situ population of the Harpy Eagle and its potential for integrated conservation. ZooKeys 2022, 1083, 109–128. [Google Scholar] [CrossRef] [PubMed]
- Turvey, S.T.; Pitman, R.L.; Taylor, B.L.; Barlow, J.; Akamatsu, T.; Barrett, L.A.; Zhao, X.; Reeves, R.R.; Stewart, B.S.; Wang, K.; et al. First human-caused extinction of a cetacean species? Biol. Lett. 2007, 3, 537–540. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Turvey, S.; Zhao, X.; Mei, Z. Neophocaena asiaeorientalis ssp. asiaeorientalis. In The IUCN Red List of Threatened Species; Version 3.1. 2013. Available online: http://www.iucnredlist.org (accessed on 20 February 2021).
- Mei, Z.; Chen, M.; Han, Y.; Hao, Y.; Zheng, J.; Wang, K.; Wang, D. Thresholds of population persistence for the Yangtze finless porpoise: Implications for conservation managements. Integr. Zool. 2021, 16, 538–547. [Google Scholar] [CrossRef]
- Zhang, X.; Yu, D.; Wang, H.; Wan, A.; Chen, M.; Tao, F.; Song, Z. Effects of fish community on occurrences of Yangtze finless porpoise in confluence of the Yangtze and Wanhe Rivers. Environ. Sci. Pollut. Res. 2015, 22, 9524–9533. [Google Scholar] [CrossRef]
- Han, H.; Li, H.; Zhang, K. Spatial-Temporal Coupling Analysis of the Coordination between Urbanization and Water Ecosystem in the Yangtze River Economic Belt. Int. J. Environ. Res. Public Health 2019, 16, 3757. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.-T.; Akamatsu, T.; Duan, P.-X.; Zhou, L.; Yuan, J.; Li, J.; Lei, P.-Y.; Chen, Y.-W.; Yang, Y.-N.; Wang, K.-X.; et al. Underwater noise pollution in China’s Yangtze River critically endangers Yangtze finless porpoises (Neophocaena asiaeorientalis asiaeorientalis). Environ. Pollut. 2020, 262, 114310. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Qian, Z.; Ruan, Y.; Hao, Y.; Dong, W.; Li, K.; Mei, Z.; Wang, K.; Wu, C.; Wu, J.; et al. First evaluation of legacy persistent organic pollutant contamination status of stranded Yangtze finless porpoises along the Yangtze River Basin, China. Sci. Total Environ. 2020, 710, 136446. [Google Scholar] [CrossRef]
- Whitehead, A.; Triant, D.A.; Champlin, D.; Nacci, D. Comparative transcriptomics implicates mechanisms of evolved pollution tolerance in a killifish population. Mol. Ecol. 2010, 19, 5186–5203. [Google Scholar] [CrossRef] [PubMed]
- Van Dijk, E.L.; Jaszczyszyn, Y.; Naquin, D.; Thermes, C. The Third Revolution in Sequencing Technology. Trends Genet. 2018, 34, 666–681. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Huang, Y.; Wu, H.; Li, C.; Ling, S.; Sun, J.; Shen, H.; Yue, B.; Zhang, X. Blood transcriptome analysis revealed the immune changes and immunological adaptation of wildness training giant pandas. Mol. Genet. Genom. 2022, 297, 227–239. [Google Scholar] [CrossRef] [PubMed]
- Remy, S.; Govarts, E.; Wens, B.; De Boever, P.; Hond, E.D.; Croes, K.; Sioen, I.; Baeyens, W.; Van Larebeke, N.; Koppe, J.; et al. Metabolic targets of endocrine disrupting chemicals assessed by cord blood transcriptome profiling. Reprod. Toxicol. 2016, 65, 307–320. [Google Scholar] [CrossRef]
- Wang, Y.; Guo, J.; Wang, L.; Tian, H.; Sui, J. Transcriptome analysis revealed potential mechanisms of differences in physiological stress responses between caged male and female magpies. BMC Genom. 2019, 20, 447. [Google Scholar] [CrossRef]
- Nabi, G.; Hao, Y.; Robeck, T.R.; Jinsong, Z.; Wang, D. Physiological consequences of biologic state and habitat dynamics on the critically endangered Yangtze finless porpoises (Neophocaena asiaeorientalis ssp. asiaeorientalis) dwelling in the wild and semi-natural environment. Conserv. Physiol. 2018, 6, coy072. [Google Scholar]
- Nabi, G.; Li, Y.; McLaughlin, R.W.; Mei, Z.; Wang, K.; Hao, Y.; Zheng, J.; Wang, D. Immune Responses of the Critically Endangered Yangtze Finless Porpoises (Neophocaena asiaeorientalis ssp. asiaeorientalis) to Escalating Anthropogenic Stressors in the Wild and Seminatural Environments. Front. Physiol. 2020, 10, 1594. [Google Scholar] [CrossRef]
- Wang, Z.-T.; Duan, P.-X.; Akamatsu, T.; Chen, Y.-W.; An, X.; Yuan, J.; Lei, P.-Y.; Li, J.; Zhou, L.; Liu, M.-C.; et al. Riverside underwater noise pollution threaten porpoises and fish along the middle and lower reaches of the Yangtze River, China. Ecotoxicol. Environ. Saf. 2021, 226, 112860. [Google Scholar] [CrossRef]
- Hao, Y.-J.; Zhao, Q.-Z.; Wu, H.-P.; Chen, D.-Q.; Gong, C.; Li, L.; Wang, D. Physiological responses to capture and handling of free-ranging male Yangtze finless porpoises (Neophocaena phocaenoides asiaeorientalis). Mar. Freshw. Behav. Physiol. 2009, 42, 315–327. [Google Scholar] [CrossRef]
- Li, R.; Li, Y.; Kristiansen, K.; Wang, J. SOAP: Short oligonucleotide alignment program. Bioinformatis 2008, 24, 713–714. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, D.; Lin, D.; Guo, H.; Gu, H.; Ying, C.; Zhang, Y.; Zhang, J.; Liu, K.; Tang, W. Integrated analysis of blood mRNAs and microRNAs reveals immune changes with age in the Yangtze finless porpoise (Neophocaena asiaeorientalis). Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2021, 256, 110635. [Google Scholar] [CrossRef] [PubMed]
- Van den Berg, H.A. On a general theoretical foundation for endocrinology. Sci. Prog. 2019, 102, 43–60. [Google Scholar] [CrossRef] [Green Version]
- Dai, C.; Tang, B.; Hao, Y.; Wang, K.; Gong, C.; Yuan, J. Study on the health evaluation system of Yangtze finless porpoise (Neophocaena asiaeorientalis asiaeorientalis). J. Anhui Agric. Univ. 2021, 48, 403. [Google Scholar]
- Sokal, A.; Jarmakiewicz-Czaja, S.; Tabarkiewicz, J.; Filip, R. Dietary Intake of Endocrine Disrupting Substances Presents in Environment and Their Impact on Thyroid Function. Nutrients 2021, 13, 867. [Google Scholar] [CrossRef]
- Graceli, J.B.; Dettogni, R.S.; Merlo, E.; Niño, O.; da Costa, C.S.; Zanol, J.F.; Rios-Morris, E.A.; Miranda-Alves, L.; Denicol, A.C. The impact of endocrine-disrupting chemical exposure in the mammalian hypothalamic-pituitary axis. Mol. Cell Endocrinol. 2020, 518, 110997. [Google Scholar] [CrossRef]
- Huang, T.; Zhao, Y.; He, J.; Cheng, H.; Martyniuk, C.J. Endocrine disruption by azole fungicides in fish: A review of the evidence. Sci. Total Environ. 2022, 822, 153412. [Google Scholar] [CrossRef]
- Yamindago, A.; Lee, N.; Lee, N.; Jo, Y.; Woo, S.; Yum, S. Fluoxetine in the environment may interfere with the neurotransmission or endocrine systems of aquatic animals. Ecotoxicol. Environ. Saf. 2021, 227, 112931. [Google Scholar] [CrossRef]
- Barra, R.O.; Chiang, G.; Saavedra, M.F.; Orrego, R.; Servos, M.R.; Hewitt, L.M.; McMaster, M.E.; Bahamonde, P.; Tucca, F.; Munkittrick, K.R. Endocrine Disruptor Impacts on Fish from Chile: The Influence of Wastewaters. Front. Endocrinol. 2021, 12, 208. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Chen, Z.; Zhang, J.; Wu, S.; Yang, L.; Chen, L.; Shao, Y. The challenge of micropollutants in surface water of the Yangtze River. Sci. Total Environ. 2021, 780, 146537. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Tang, X.; Guo, W.; Lin, L.; Zhao, L.; Hu, Y.; Liu, M. Spatiotemporal distribution dynamics of heavy metals in water, sediment, and zoobenthos in mainstream sections of the middle and lower Changjiang River. Sci. Total Environ. 2020, 714, 136779. [Google Scholar] [CrossRef] [PubMed]
- Wong, W.P.; Wang, J.C.; Schipma, M.J.; Zhang, X.; Edwards, J.R.; El Muayed, M. Cadmium-mediated pancreatic islet transcriptome changes in mice and cultured mouse islets. Toxicol. Appl. Pharmacol. 2021, 433, 115756. [Google Scholar] [CrossRef]
- Xi, W.; Lee, C.K.F.; Yeung, W.S.B.; Giesy, J.P.; Wong, M.H.; Zhang, X.; Hecker, M.; Wong, C.K.C. Effect of perinatal and postnatal bisphenol A exposure to the regulatory circuits at the hypothalamus–pituitary–gonadal axis of CD-1 mice. Reprod. Toxicol. 2011, 31, 409–417. [Google Scholar] [CrossRef] [PubMed]
- Ramkumar, V.; Sheth, S.; Dhukhwa, A.; Al Aameri, R.; Rybak, L.P.; Mukherjea, D. Transient Receptor Potential (TRP) Channels and Auditory Functions. Antioxid. Redox Signal. 2021. [Google Scholar] [CrossRef] [PubMed]
- Tadros, S.F.; Kim, Y.; Phan, P.A.B.; Birnbaumer, L.; Housley, G.D. TRPC3 ion channel subunit immunolocalization in the cochlea. Histochem. Cell Biol. 2010, 133, 137–147. [Google Scholar] [CrossRef]
- Popov, V.V.; Supin, A.Y.; Wang, D.; Wang, K.; Dong, L.; Wang, S. Noise-induced temporary threshold shift and recovery in Yangtze finless porpoises Neophocaena phocaenoides asiaeorientalis. J. Acoust. Soc. Am. 2011, 130, 574–584. [Google Scholar] [CrossRef]
- Ju, T.; Zhang, T.C.; Wang, Z.T.; Xie, Y.; Zheng, C.H.; Wang, K.X.; Wang, D. Characteristics of riprapping underwater noise and its possible impacts on the Yangtze finless porpoise. Tech. Acoust. 2017, 36, 580–588. [Google Scholar]
- Frye, M.D.; Ryan, A.F.; Kurabi, A. Inflammation associated with noise-induced hearing loss. J. Acoust. Soc. Am. 2019, 146, 4020–4032. [Google Scholar] [CrossRef] [Green Version]
- Fuentes-Santamaría, V.; Alvarado, J.C.; Melgar-Rojas, P.; Gabaldón-Ull, M.C.; Miller, J.M.; Juiz, J.M. The Role of Glia in the Peripheral and Central Auditory System Following Noise Overexposure: Contribution of TNF-α and IL-1β to the Pathogenesis of Hearing Loss. Front. Neuroanat. 2017, 11, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abi-Hachem, R.N.; Zine, A.; Van De Water, T.R. The injured cochlea as a target for inflammatory processes, initiation of cell death pathways and application of related otoprotectives strategies. Recent Pat. CNS Drug Discov. 2010, 5, 147–163. [Google Scholar] [CrossRef] [PubMed]
- Arpornchayanon, W.; Canis, M.; Ihler, F.; Settevendemie, C.; Strieth, S. TNF-α inhibition using etanercept prevents noise-induced hearing loss by improvement of cochlear blood flow in vivo. Int. J. Audiol. 2013, 52, 545–552. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Pyykkö, I.; Zou, J. Involvement of Ubiquitin-Editing Protein A20 in Modulating Inflammation in Rat Cochlea Associated with Silver Nanoparticle-Induced CD68 Upregulation and TLR4 Activation. Nanoscale Res. Lett. 2016, 11, 240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moon, S.K.; Woo, J.I.; Lee, H.Y.; Park, R.; Shimada, J.; Pan, H.; Gellibolian, R.; Lim, D.J. Toll-like receptor 2-dependent NF-κB activation is involved in nontypeable Haemophilus influenzae-induced monocyte chemotactic protein 1 up-regulation in the spiral ligament fibrocytes of the inner ear. Infect. Immun. 2007, 75, 3361–3372. [Google Scholar] [CrossRef] [Green Version]
- Waqas, M.; Gao, S.; Salam, I.U.; Ali, M.K.; Ma, Y.; Li, W. Inner Ear Hair Cell Protection in Mammals against the Noise-Induced Cochlear Damage. Neural Plast. 2018, 2018, 3170801. [Google Scholar] [CrossRef] [Green Version]
- Slepecky, N. Overview of mechanical damage to the inner ear: Noise as a tool to probe cochlear function. Hear. Res. 1986, 22, 307–321. [Google Scholar] [CrossRef]
- Fetoni, A.R.; De Bartolo, P.; Eramo, S.L.M.; Rolesi, R.; Paciello, F.; Bergamini, C.; Fato, R.; Paludetti, G.; Petrosini, L.; Troiani, D. Noise-Induced Hearing Loss (NIHL) as a Target of Oxidative Stress-Mediated Damage: Cochlear and Cortical Responses after an Increase in Antioxidant Defense. J. Neurosci. Off. J. Soc. Neurosci. 2013, 33, 4011–4023. [Google Scholar] [CrossRef] [Green Version]
- Vurusaner, B.; Poli, G.; Basaga, H. Tumor suppressor genes and ROS: Complex networks of interactions. Free Radic. Biol. Med. 2012, 52, 7–18. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).