Abstract
Interaction between the chemokine receptor XCR1 and its ligand is closely related to the immune function in animals; however, there are only a few reports on role of XCR1 in the immune system of fish. We aimed to analyze the expression of XCR1 in various organs or tissues of grass carp before and after Grass Carp Reovirus (GCRV) infection to better understand the function of XCR1 in resistance to GCRV infection. We cloned and sequenced the cDNA of grass carp XCR1 and analyzed the molecular structure of XCR1 based its amino acid sequence. Further, we analyzed the relative expression levels of XCR1 in different organs or tissues of male parent grass carp with GCRV resistance (P1) and their first-generation offspring (F1) before and after GCRV infection. Our results show that the total length of cDNA of the grass carp XCR1 gene is 1659 bp and encodes 365 amino acids. XCR1 contains seven conserved transmembrane helical domains. The homologous tertiary structure of XCR1 is similar to its homologs in other species. After artificial GCRV infection, there were significant differences in the expression of the grass carp XCR1 gene in different tissues, at different time points, and between P1 and F1 fish. These results will contribute to our understanding of the role of XCR1 in fish immune responses and contribute to the development of GCRV-resistant grass carp.
1. Introduction
Grass carp (Ctenopharyngodon idellus) is widely cultured because of its low feed cost and fast growth; therefore, it has become the most common species produced in aquaculture worldwide [1]. In 2018, the global production of cultured grass carp was 5,704,000 tons, accounting for 10.5% of the total output of cultured fish worldwide [1]. However, this industry has been seriously hampered because of the development of severe hemorrhagic diseases in this species caused by the Grass Carp Reovirus (GCRV) [2,3]. GCRV infection causes a mortality rate of up to 85% during the summer months [4], resulting in annual economic losses of at least one billion US dollars in China [5]. Therefore, exploring the prevention of severe hemorrhagic diseases caused by GCRV is of great value in the development of the grass carp culture industry.
Analyzing the immune factors involved in grass carp resistance to GCRV infection and their effects provides basic data for the development of new ways to prevent severe hemorrhagic disease caused by GCRV. We previously screened immune factors closely related to grass carp hemorrhage in the two pathways of coagulation complement (coagulation factors II, VII, and VIII, and profibrinolysin, q subcomponent of the first complement component, complement factors II, III, and V, and coagulation factor D) and cell-cytokine interaction (transforming growth factor-β3, chemokine XC receptor 1 [XCR1], chemokine receptor 4, nerve growth factor receptor, and unknown factor X) through transcriptome sequencing [6].
A chemokine is a type of cytokine that induces chemotaxis in leukocytes. More than 50 chemokines have been identified to date [7]. According to their structure and the arrangement of the four conserved cysteine residues, chemokines are organized into five subfamilies: CXC (α), CC (β), XC (γ), CX3C (δ), and CX [8]. Chemokines play biological roles by binding to specific receptors on the cell surface. Chemokines and chemokine receptors are involved in morphogenetic processes and play important roles in the immune system [9]. Among the chemokine receptors, XCR1 is the only member of the XCR family. XCR1 belongs to the chemokine receptor G protein-coupled receptor superfamily and is expressed in a variety of organs and cells [10,11,12]. In the early stages of innate immunity, natural killer (NK) cells activate and secrete chemokine (C motif) ligand 1 (XCL1) and chemotactic XCR1+DCs aggregate to activate NK cells. Subsequently, in the adaptive immune stage, XCR1+DCs cross-present cell-related antigens to CD8+T cells through the MHC-I molecular pathway, and CD8+T cells are activated to secrete XCL1, which promotes CD8+T cells to differentiate to T cells [13]. Chemokine receptors have been detected in a variety of fish, such as grass carp [14,15], Japanese killifish (Oryzias latipes) [9], and groupers (Epinephelus coioides) [16].
Although we previously found that XCR1 is related to the resistance of grass carp to GCRV infection [6], there have been no further studies conducted to support this idea. Therefore, we analyzed the expression of XCR1 in various organs or tissues of grass carp before and after GCRV infection to provide basic data for further analysis of the function of XCR1 in the process of grass carp resistance to GCRV infection. We first cloned and sequenced the cDNA sequence of grass carp XCR1 gene and analyzed the molecular structure of XCR1 based on the sequence. Then, the relative levels of expression of XCR1 in different organs or tissues of male parent grass carp with GCRV resistance (P1) and their first-generation offspring (F1) before and after GCRV infection were analyzed through quantitative reverse transcription polymerase chain reaction (qRT-PCR). The results of this study will contribute to our understanding of the role of XCR1 in fish immune responses and provide reference data for the breeding of GCRV-resistant grass carp using molecular biology technology.
2. Materials and Methods
2.1. Fish and Virus Sources
All procedures used in this experiment were approved by the Hunan Agricultural University Institutional Animal Care and Use Committee (approval number 2013008) and were performed in accordance with approved protocols. P1 and F1 fish were obtained as previously described [2] and farmed at the Wulong Fishery Base, Liuyang, China (28.30° N, 113.44° E). A total of 150 of each of P1 and F1 fish with an average weight of 21.0 ± 5.2 g were transferred to the Aquaculture Base of Hunan Agricultural University for temporary culture two weeks before the experiment [3]. The GCRV918 strain was donated by Lingbing Zeng of the Yangtze River Fisheries Research Institute, Chinese Academy of Fishery Sciences.
2.2. Experimental Design and Sample Collection
To avoid the accidental termination of the experiment due to the death of experimental fish during the experiment, we set up two more aquariums for each P1 and F1 fish. Therefore, after temporary culture for two weeks, the P1 and F1 fish were divided into ten aquariums (60 × 50 × 40 cm; 30 fish per aquarium). GCRV918 strain with 50% tissue culture infective dose (TCID50) value of 1.0 × 107/mL was injected intraperitoneally at 0.2 μL per gram of fish weight. The temperature was maintained at 28 °C and oxygen was supplied intermittently during the experiment. Six tissues (the gill, liver, midgut, head kidney, spleen, and muscle) were collected at 0, 12, 24, 48, 72, 96, 120, 144, and 168 h after artificial GCRV infection and quickly frozen in liquid nitrogen. P1 and F1 fish were randomly collected from three aquariums at each sampling time, respectively. The tissues were stored at −80 °C until further analysis.
2.3. RNA Extraction and cDNA Synthesis
Total RNA from each sample (approximately 30 mg) was extracted using the RNApure rapid total RNA extraction kit (BioTeke, Beijing, China) and cDNA was synthesized using the ReverAidTM first strand cDNA synthesis kit (Promega, Beijing, China) as previously described [3].
2.4. Primer Design and Amplification and Sequencing of cDNA Fragment of Grass Carp XCR1 Gene
According to the unigene of grass carp XCR1 reported by us [6], the primers were designed using Primer Premier 5.0 (Table 1). Primers were synthesized by Sangon Biotech (Shanghai, China). The 5′-RACE and 3′-RACE cDNA fragments of the grass carp XCR1 gene were amplified via PCR using RACE-Ready cDNA of grass carp spleen tissues as a template according to our previously described method [2]. The amplified products were detected using 1.0% agarose gel electrophoresis. After purification using a gel recovery kit (Tiangen, Beijing, China), DNA was incorporated into the pUCm-T plasmid vector (Sangon Biotech) and introduced into E. coli DH5α cells. The cells were then cultured and the plasmid was isolated for sequencing of the inserted fragment. DNA sequencing was conducted using M13 universal primers from Shanghai Majorbio Bio-pharm Biotechnology Co., Ltd., Shanghai, China.

Table 1.
Primers for XCR1 gene amplification and their qRT-PCR analysis.
2.5. Sequence Analysis
DNAStar was used to identify the open reading frame of the XCR1 sequence. CLC-ProteinWork-bench 5.3 was used to predict the encoded amino acid sequence. SWISS-MODEL was used to model the homologous tertiary structure of grass carp XCR1 through the SWISS-MODEL website [17]. The reference protein used for modeling was C-C chemokine receptor type 5 (accession number: 6akx.1.A) and the E value was 0.00 × 10−1. The online software ProtParam [18] was used to deduce the physical and chemical properties and secondary structures of XCR1. The conserved domain database [19] was used to analyze the conserved domains of XCR1 by RPS-BLAST [20]. Transmembrane helices were predicted using TMHMM-2.0 [21]. The BLAST tool in the NCBI database was used to determine the homologous protein sequence with the highest similarity and the other 12 vertebrate XCR1 protein sequences were downloaded to construct a phylogenetic tree. The sequences were aligned using DNAMAN and a phylogenetic neighbor-joining tree was constructed using MEGA 4 version 5.0 [22] with 1000 bootstraps.
2.6. qRT-PCR
Three biological replicates of each tissue and three technical replicates for each biological replicate were analyzed by qRT-PCR on an iCycler Thermal Cycler 96-Well Thermal Sealing Ring (Bio-Rad, Richmond, VA, USA) as previously described [3]. The β-actin gene sequence was used as the internal control [2,23]. The relative gene expressions were calculated using the comparative Ct (2−ΔΔCt) method [2,23].
2.7. Sequence Availability
The XCR1 gene sequences were obtained from the NCBI GenBank database with accession number KF937391.
2.8. Data Analysis
Wilcoxon test and Kruskal-Wallis test with Dunn post hoc tests were conducted using R 4.0.3 software with FSA package to detect whether there was significant difference between two or more groups. Statistical significance was set at p < 0.05.
3. Results
3.1. cDNA Sequences of Grass Carp XCR1 Gene
To get whole cDNA sequences of grass carp XCR1 gene, we amplified and sequenced the cDNA fragments of grass carp XCR1 gene. The total length of the cDNA of the grass carp XCR1 gene was 1659 bp and its open reading frame was 1098 bp, which encoded 365 amino acids. The length of the 5′ non-coding region of the cDNA was 160 bp and the length of the 3′ non-coding region was 401 bp. The 3′ non-coding region of the cDNA sequence contained two “AATAAA” mRNA tail signals and two mRNA unstable “ATTTA” motifs (Figure 1).
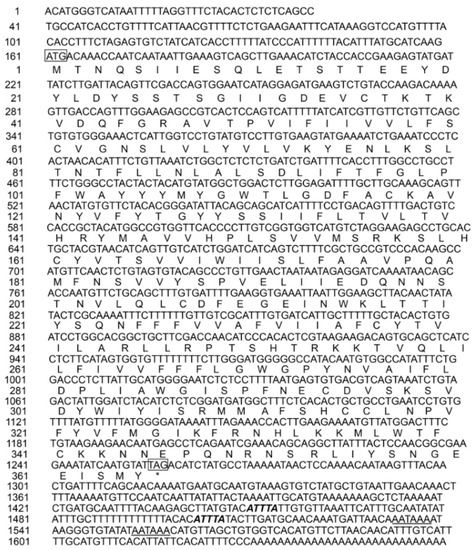
Figure 1.
The full-length cDNA sequence of Ctenopharynodon idellus XCR1. The initiation codon “ATG” and the termination codon “TAG” are labeled by square frame; the termination signal is labeled by “*”; the mRNA tailing signal is labeled by italic and bold; and the mRNA instability motif is labeled by underline.
3.2. Amino Acid Sequence of Grass Carp XCR1 Protein
To obtain the protein structure and function of grass carp XCR1 protein, we predicated the XCR1 protein structure by bioinformatics methods. The molecular formula of grass carp XCR1 is C1974H2972N460O524S20. The theoretical isoelectric point of grass carp XCR1 was 6.97 and its relative molecular weight was 42,173.2 Da. The protein instability coefficient was 38.54, which indicated that the protein was stable. The fat coefficient of the protein was 104.82 and the hydrophilicity was 0.455. Grass carp XCR1 contains seven conserved transmembrane helical domains (Figure 2A). The homologous tertiary structure of grass carp XCR1 is similar to that of other species (Figure 2B) [24,25,26], which further indicated that the function of grass carp XCR1 is likely to be the same as that of XCR1 found in other species. The amino acid sequence of grass carp XCR1 has high homology with the chemokine receptor members of other species (Table S1). Grass carp XCR1 and zebrafish XCR1 had the highest amino acid sequence homology (77%), followed by Astyanax mexicanus (63%). The amino acid sequence homology of grass carp XCR1 with mammalian XCR1 was approximately 50% (Table S1). The phylogenetic neighbor-joining tree showed that grass carp and zebrafish clustered first, followed by Astyanax mexicanus. Erinaceus europaeus, Microtus ochrogaster, Felis catus, Bos taurus, and Lipotes vexillifer clustered together (Figure 2C).
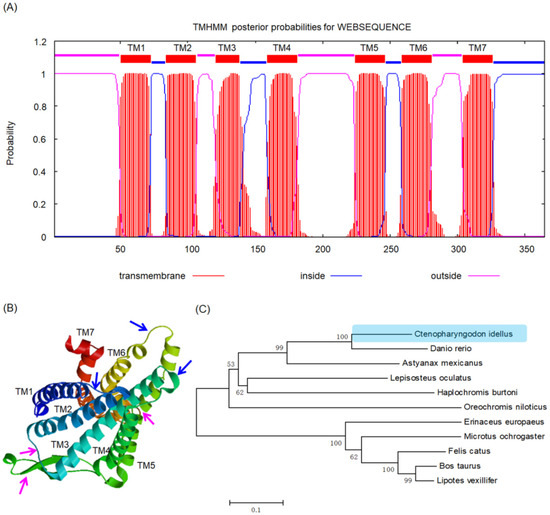
Figure 2.
Molecular structure and evolutionary relationship of grass carp XCR1 protein. (A) Prediction of conserved transmembrane helical domains; (B) homologous tertiary structure, blue and pink arrows indicate cell inside and outside regions, respectively; (C) phylogenetic neighbor-joining tree constructed based on amino acid sequences of XCR1 gene, light blue background indicates the evolutionary position of grass carp XCR1.
3.3. Expression of the XCR1 Gene in Various Grass Carp Tissues
To obtain the expressions of the XCR1 gene in different tissues of healthy grass carp, and compare differences of the XCR1 expression between P1 and F1 after the artificial GCRV infection, as the XCR1 expression between P1 and F1 after the artificial GCRV infection probably provide reference information for inferring the resistance of F1 to GCRV infection, we analyzed the expression of the XCR1 gene in various grass carp tissues. The XCR1 gene was found to be expressed in the gills, liver, midgut, head kidney, spleen, and muscle of grass carp, and there was a significant difference in expression among tissues (Kruskal-Wallis test, p < 0.05). The expression of the XCR1 gene was the highest in the gills of grass carp (Kruskal-Wallis test with Dunn post hoc tests, p < 0.01), followed by that in the spleen and head kidney (Kruskal-Wallis test with Dunn post hoc tests, p < 0.01, for each compared to the liver and midgut), and the lowest in muscle (Figure 3A).
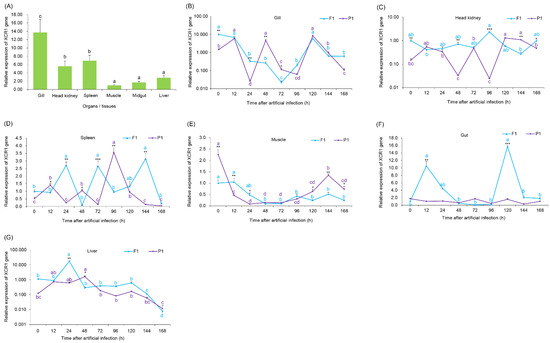
Figure 3.
Relative expression of XCR1 gene in different organs or tissues of grass carp. (A) The relative expression of the XCR1 gene in tissues or organs of grass carp, the different letters above the bars indicate that there were significant differences between the data; (B–F,G) The relative expression of the XCR1 gene in the indicated sites of grass carp at different times after artificial infection of GCRV. P1, male parent grass carp with GCRV resistance; F1, the first-generation offspring of P1. Asterisks indicate significant differences in XCR1 gene expression between P1 and F1. *, p < 0.05; **, p < 0.01. The different letters indicate that there were significant differences between different times after artificial infection of GCRV. * p < 0.05; ** p < 0.01; *** p < 0.001.
After artificial GCRV infection, the expression of XCR1 in the gills of F1 fish first decreased and then gradually recovered. It reached its lowest point at 72 h after infection and was significantly lower than that at other time points (p < 0.01; Figure 3B). Although the expression of this gene in the F1 head kidney fluctuated at different time points, it was maintained within an order of magnitude (Figure 3C). The expression of XCR1 in the F1 spleen fluctuated sharply at different time points (Figure 3D). The expression of XCR1 in the F1 muscle showed a downward trend within 48 h after infection and was maintained within a small range of fluctuation (Figure 3E). The expression of XCR1 in F1 gut peaked at two time points and then decreased rapidly (Figure 3F). The expression of XCR1 in the F1 liver increased first (peaking at 24 h after infection) and then decreased gradually (Figure 3G). However, the expression of this gene in the gills, head kidney, and spleen of P1 fish fluctuated sharply (Figure 3B–D). The expression of XCR1 in P1 muscle showed an initial sharp decline and then increased at later time points (Figure 3E). The expression of the gene in the P1 gut remained stable and was consistent with that in the absence of infection (Figure 3A,F). The expression of the gene in the P1 liver increased gradually at first, reaching a peak at 48 h after infection, and then decreased gradually (Figure 3G). It is worth noting that although the expression of XCR1 in both F1 and P1 spleens fluctuated sharply at each time point, their fluctuation directions were opposite (Figure 3D). The expression trend of this gene in the F1 and P1 livers with infection time was basically the same, although the peak time of expression in P1 was 12 h later than that in F1 (Figure 3G). The expression of this gene in the gills, head kidney, and spleen fluctuated greatly at different time points after infection. The expression in the P1 muscle decreased significantly faster than that in the F1 muscle within 48 h after infection (Figure 3E), whereas in the gut, the expression fluctuation after P1 infection was significantly less than that of F1 XCR1 (Figure 3F). Moreover, the expression of XCR1 in the organs were also significant differences at most times after artificial infection of GCRV between P1 and F1 (Wilcoxon test, p < 0.05; Figure 3B–G).
4. Discussion
Chemokine receptors are seven-transmembrane G-protein-coupled receptors, and their N-terminal and C-terminal regions are involved in ligand binding and triggering downstream signaling pathways [26]. XCR1 is the only member of the XC chemokine subfamily and plays an important role in abnormal physiological states such as chronic inflammation, allergic diseases, AIDS, autoimmune diseases, and tumors [26]. The XCR1 gene of grass carp was cloned in this study, which encodes a protein of 365 amino acids, similar to that of the orange-spotted grouper (Epinephelus coioides) (337–358 amino acids) [26]. Our results show that grass carp XCR1 contains seven conserved transmembrane helical domains (Figure 2), implying that grass carp XCR1 probably plays a function linking the chemokine and the corresponding ligand.
The head kidney is the main hematopoietic organ of fish, the spleen is the main immune organ of fish, the liver is the main organ involved in metabolism and immune, and the gut and gill are the main pathogenesis organs of grass carp hemorrhagic disease [2,3,27,28,29]. Therefore, we analyzed the expression of XCR1 in these organs in this study. XCR1 is expressed in various organs and cells of several organisms [26,30]. For instance, XCR1 is expressed in the mouse spleen, lungs, CD8+T cells, CD4+T cells, NK cells, B cells, neutrophils, and macrophages after stimulation by interferon-γ (IFN-γ) [10,31]. Our results show that XCR1 was expressed in the gills, head kidney, spleen, muscle, midgut, and liver of grass carp, and there were significant differences among tissues (p < 0.05). The expression of XCR1 was highest in the gill, followed by the spleen, head kidney, liver, and midgut, and the expression in the muscle was the lowest (p < 0.05; Figure 3A). These results implied that XCR1 may have a role in the immune response of grass carp.
The interaction between XCR1 and its ligand is closely related to immune function of the body [11,32]. The expression of the XCR1 ligand XCL1 in NK cells and CD8+T cells increased significantly during viral infection in humans and mice. Functionally, XCL1 can increase the number of antigen-specific CD8+T cells, enhance their ability to secrete IFN-γ, and induce cytotoxicity [11]. In Listeria monocytogenes infection in mice without XCL1, the CD8+T response was decreased and CD8α+DCs weakened the defense response mediated by CD8+T cells in the early stage of activation [30]. Compared with that in mammals, there are only a few reports on the immune role of XCR1 in fish. Ni et al. [26] reported that Cryptocaryon irritans infection upregulates the expression of XCR1 in the skin and spleen of E. coioides. In this study, the full-length cDNA sequence of grass carp XCR1 was cloned and the structure of XCR1 was predicted, which provides important reference information for the study of XCR1 involvement in the immune process of grass carp. Although our results show that there were significant differences in the expression of the grass carp XCR1 gene in different tissues and at different time points after artificial GCRV infection, as well as between P1 and F1, the interaction and regulation mechanism between XCR1 and other immune factors in grass carp after GCRV infection need to be further studied.
As our previously described [3], the P1 are the male parent grass carp with GCRV resistance, and F1 are their first-generation offspring. Comparing differences of the XCR1 expression between P1 and F1 probably provide reference information for inferring the resistance of F1 to GCRV infection. Therefore, we compared the XCR1 expressions in P1 and F1 organs. However, due to the lack of indicators of F1 resistance to GCRV infection, it was difficult for us to draw effective conclusions from present data. Establishing the relationship between mortality after GCRV infection and XCR1 expression in different populations in the future will provide technical support for judging the anti-infection ability of grass carp populations through XCR1 expression.
The dramatic inflections of XCR1 expressions after GCRV infection implied that GCRV infection was not the only factor affecting XCR1 expression. Additionally, considering the complex immune regulation processes of grass carp after GCRV infection [2,6], our results implied that more valuable information probably is obtained by analyzing all of the immune factors closely related to grass carp hemorrhage after GCRV infection simultaneously. The differences of XCR1 expression between P1 and F1, and between various organs indicated the complexity of XCR1 function. Other functions of grass carp XCR1 need further study.
Since the XCR1 expression in P1 and F1 organs fluctuated greatly after GCRV infection, the subsequent analysis of the XCR1 expression in uninfected grass carp organs at different time points during the experiment should be increased to determine whether the expression of XCR1 gene in uninfected grass carp organs is stable. Moreover, comparing the XCR1 expressions in grass carp organs at different time points after GCRV infection with those in uninfected grass carp organs can fully eliminate the impact of XCR1 expression fluctuation caused by changes of the external environment during the experiment. Therefore, it is necessary to add uninfected grass carp as a blank control in the follow-up study. Another noteworthy issue is gene expression does not necessarily translate to changes in protein expression. Examining the protein expression of XCR1 over time would enhance our understanding of the role of XCR1 during GCRV infection.
It should be noted that our results fluctuated greatly that was might due to the small sample size (three biological replicates and three technical replicates for each biological replicate) we analyzed at each sampling time point. Moreover, the expression patterns of the XCR1 gene of grass carps with different sizes or under different culture conditions might be different after GCRV infection. Therefore, further experimental verification of the expression patterns of the grass carp XCR1 gene is needed.
5. Conclusions
The grass carp XCR1 protein contains conserved domains that are consistent with those of other species. Its homology was closer to that of fish than other vertebrates, and its gene homology with other species is consistent with their evolutionary relationship. After artificial GCRV infection, there were significant differences in the expression of the grass carp XCR1 gene in different tissues and at different time points as well as in P1 and F1. The results of this study will contribute to our understanding of the role of XCR1 in fish immune responses and provide reference data for the breeding of GCRV-resistant grass carp using molecular biology technology.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/fishes7030130/s1, Table S1: The XCR1 sequence identities between grass carp and other species.
Author Contributions
Conceptualization, H.Y., H.W. and B.X.; methodology, H.Y. and H.W.; software, B.X.; validation, T.X.; formal analysis, H.Y., Y.L., T.X. and H.W.; investigation, H.Y. and Y.L.; resources, T.X.; data curation, T.X.; writing—original draft preparation, H.Y.; writing—review and editing, H.W. and B.X.; visualization, H.Y.; supervision, Y.L.; project administration, T.X.; funding acquisition, T.X. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Special Project for the Construction of Modern Agricultural Industrial Technology System, grant number CARS4548, and the National Key R & D Program of China, grant number 2018YFD0900302.
Institutional Review Board Statement
All procedures used in this experiment were approved by the Human Agricultural University Institutional Animal Care and Use Committee (approval number 2013008) and performed in accordance with approved protocols.
Data Availability Statement
The XCR1 gene sequences were obtained from the NCBI GenBank database with accession number KF937391.
Acknowledgments
The authors thank anonymous technicians at Guangdong Meilikang Bio-Science Ltd., China for assistance with data re-analysis.
Conflicts of Interest
The authors declare no conflict of interest.
References
- FAO. The State of World Fisheries and Aquaculture 2020. In Sustainability in Action; FAO: Rome, Italy, 2020. [Google Scholar] [CrossRef]
- Wang, H.; Ding, C.; Wang, J.; Zhao, X.; Jin, S.; Liang, J.; Luo, H.; Li, D.; Li, R.; Li, Y.; et al. Molecular cloning and expression analysis of coagulation factor VIII and plasminogen involved in immune response to GCRV, and immunity activity comparison of grass carp Ctenopharyngodon idella with different viral resistance. Fish Shellfish Immunol. 2019, 86, 794–804. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yang, H.; Li, R.; Li, Y.; Xiao, T. Different resistance potential to reovirus in grass carp (Ctenopharyngodon idella) populations and their immune characteristics. Aquac. Int. 2021, 29, 253–260. [Google Scholar] [CrossRef]
- Liang, H.-R.; Li, Y.-G.; Zeng, W.-W.; Wang, Y.-Y.; Wang, Q.; Wu, S.-Q. Pathogenicity and tissue distribution of grass carp reovirus after intraperitoneal administration. Virol. J. 2014, 11, 178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, W.; Wang, Y.; Liang, H.; Liu, C.; Song, X.; Shi, C.; Wu, S.; Wang, Q. A one-step duplex rRT-PCR assay for the simultaneous detection of grass carp reovirus genotypes I and II. J. Virol. Methods 2014, 210, 32–35. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Zhong, L.; Liu, Q.; Xiao, T.; Su, J.; Chen, K.; Wang, H.; Dai, Y.; Chen, J. Characterization of grass carp spleen transcriptome during GCRV infection. Genet. Mol. Res. 2016, 15, gnr.15026650. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Lu, Y.; Xiao, X. Progress in the research on XCR I and its ligand. Int. J. Pathol. Clin. Med. 2013, 33, 57–61. [Google Scholar] [CrossRef]
- Nomiyama, H.; Hieshima, K.; Osada, N.; Kato-Unoki, Y.; Otsuka-Ono, K.; Takegawa, S.; Izawa, T.; Yoshizawa, A.; Kikuchi, Y.; Tanase, S.; et al. Extensive expansion and diversification of the chemokine gene family in zebrafish: Identification of a novel chemokine subfamily CX. BMC Genom. 2008, 9, 222. [Google Scholar] [CrossRef] [Green Version]
- Aghaallaei, N.; Bajoghli, B.; Schwarz, H.; Schorpp, M.; Boehm, T. Characterization of mononuclear phagocytic cells in medaka fish transgenic for a cxcr3a:gfp reporter. Proc. Natl. Acad. Sci. USA 2010, 107, 18079–18084. [Google Scholar] [CrossRef] [Green Version]
- Dorner, B.G.; Scheffold, A.; Rolph, M.S.; Hüser, M.B.; Kaufmann, S.H.E.; Radbruch, A.; Flesch, I.E.A.; Kroczek, R.A. MIP-1alpha, MIP-1beta, RANTES and ATAC/lymphotactin function together with IFN-gamma as Type 1 cytokines. Proc. Natl. Acad. Sci. USA 2002, 99, 6181–6186. [Google Scholar] [CrossRef] [Green Version]
- Dorner, B.G.; Dorner, M.B.; Zhou, X.; Opitz, C.; Mora, A.; Güttler, S.; Hutloff, A.; Mages, H.W.; Ranke, K.; Schaefer, M.; et al. Selective Expression of the Chemokine Receptor XCR1 on Cross-presenting Dendritic Cells Determines Cooperation with CD8+ T Cells. Immunity 2009, 31, 823–833. [Google Scholar] [CrossRef] [Green Version]
- Khurram, S.A.; Whawell, S.A.; Bingle, L.; Murdoch, C.; McCabe, B.M.; Farthing, P.M. Functional expression of the chemokine receptor XCR1 on oral epithelial cells. J. Pathol. 2010, 221, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Kroczek, R.A.; Henn, V. The Role of XCR1 and its Ligand XCL1 in Antigen Cross-Presentation by Murine and Human Dendritic Cells. Front. Immunol. 2012, 3, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, M.; Sun, B.; Nie, P. The first non-mammalian CXCR3 in a teleost fish: Gene and expression in blood cells and central nervous system in the grass carp (Ctenopharyngodon idella). Mol. Immunol. 2007, 44, 1123–1134. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.Q.; Chang, M.X.; Sun, R.H.; Xiao, F.S.; Nie, P. The first non-mammalian CXCR5 in a teleost fish: Molecular cloning and expression analysis in grass carp (Ctenopharyngodon idella). BMC Immunol. 2010, 11, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Yang, M.; Li, C.; Wang, S.; Wang, Y.; Lin, F.; Zheng, L.; Yu, Y.; Qin, Q. Functional analysis of the CXCR1a gene response to SGIV viral infection in grouper. Fish Shellfish Immunol. 2019, 88, 217–224. [Google Scholar] [CrossRef] [PubMed]
- SWISS-MODEL Website. Available online: https://swissmodel.expasy.org/ (accessed on 1 January 2022).
- ProtParam Website. Available online: http://web.expasy.org/protparam/ (accessed on 1 January 2022).
- Conserved Domain Database. Available online: https://www.ncbi.nlm.nih.gov/cdd (accessed on 1 January 2022).
- Marchler-Bauer, A.; Lu, S.; Anderson, J.B.; Chitsaz, F.; Derbyshire, M.K.; DeWeese-Scott, C.; Fong, J.H.; Geer, L.; Geer, R.C.; Gonzales, N.R.; et al. CDD: A Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011, 39, D225–D229. [Google Scholar] [CrossRef] [Green Version]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S.; Notes, A. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Weis, W.I.; Kobilka, B.K. Structural insights into G-protein-coupled receptor activation. Curr. Opin. Struct. Biol. 2008, 18, 734–740. [Google Scholar] [CrossRef] [Green Version]
- Kobilka, B.K. Structural insights into adrenergic receptor function and pharmacology. Trends Pharmacol. Sci. 2011, 32, 213–218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ni, L.-Y.; Zhou, L.; Wang, H.-Q.; Luo, X.-C.; Dan, X.-M.; Li, Y.-W. Identification and expression analysis of three XCR1-like receptors from Epinephelus coioides after Cryptocaryon irritans infection. Fish Shellfish Immunol. 2017, 67, 95–102. [Google Scholar] [CrossRef] [PubMed]
- He, P.; Jiang, W.-D.; Liu, X.-A.; Feng, L.; Wu, P.; Liu, Y.; Jiang, J.; Tan, B.-P.; Yang, Q.-H.; Kuang, S.-Y.; et al. Dietary biotin deficiency decreased growth performance and impaired the immune function of the head kidney, spleen and skin in on-growing grass carp (Ctenopharyngodon idella). Fish Shellfish Immunol. 2020, 97, 216–234. [Google Scholar] [CrossRef] [PubMed]
- MacKenzie, S.; Balasch, J.C.; Novoa, B.; Ribas, L.; Roher, N.; Krasnov, A.; Figueras, A. Comparative analysis of the acute response of the trout, O. mykiss, head kidney to in vivo challenge with virulent and attenuated infectious hematopoietic necrosis virus and LPS-induced inflammation. BMC Genom. 2008, 9, 141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, N.; Song, Y.-L.; Wang, B.; Zhang, X.-Y.; Zhang, X.-J.; Wang, Y.-L.; Cheng, Y.-Y.; Chen, D.-D.; Xia, X.-Q.; Lu, Y.-S.; et al. Fish gut-liver immunity during homeostasis or inflammation revealed by integrative transcriptome and proteome studies. Sci. Rep. 2016, 6, 36048. [Google Scholar] [CrossRef] [Green Version]
- Crozat, K.; Guiton, R.; Contreras, V.; Feuillet, V.; Dutertre, C.-A.; Ventre, E.; Manh, T.-P.V.; Baranek, T.; Storset, A.K.; Marvel, J.; et al. The XC chemokine receptor 1 is a conserved selective marker of mammalian cells homologous to mouse CD8α+ dendritic cells. J. Exp. Med. 2010, 207, 1283–1292. [Google Scholar] [CrossRef]
- Zhu, C.; Ou, Z. Progress in the research on lymphotactin (Ltn). Foreign Med. Sci. Sec. Pathophysiol. Clin. Med. 2002, 22, 320–322. [Google Scholar]
- Lei, Y.; Ripen, A.M.; Ishimaru, N.; Ohigashi, I.; Nagasawa, T.; Jeker, L.; Bösl, M.R.; Holländer, G.A.; Hayashi, Y.; Malefyt, R.D.W.; et al. Aire-dependent production of XCL1 mediates medullary accumulation of thymic dendritic cells and contributes to regulatory T cell development. J. Exp. Med. 2011, 208, 383–394. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).