Cu/Zn Superoxide Dismutase and Catalase of Yangtze Sturgeon, Acipenser dabryanus: Molecular Cloning, Tissue Distribution and Response to Fasting and Refeeding
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Design
2.2. Sample Collection
2.3. Molecular Cloning of Cu/Zn-SOD and CAT from Yangtze Sturgeon
2.4. Sequence Analysis and Phylogenetic Analysis
2.5. Quantitative Real-Time PCR (qPCR)
2.6. Statistical Analysis
3. Results
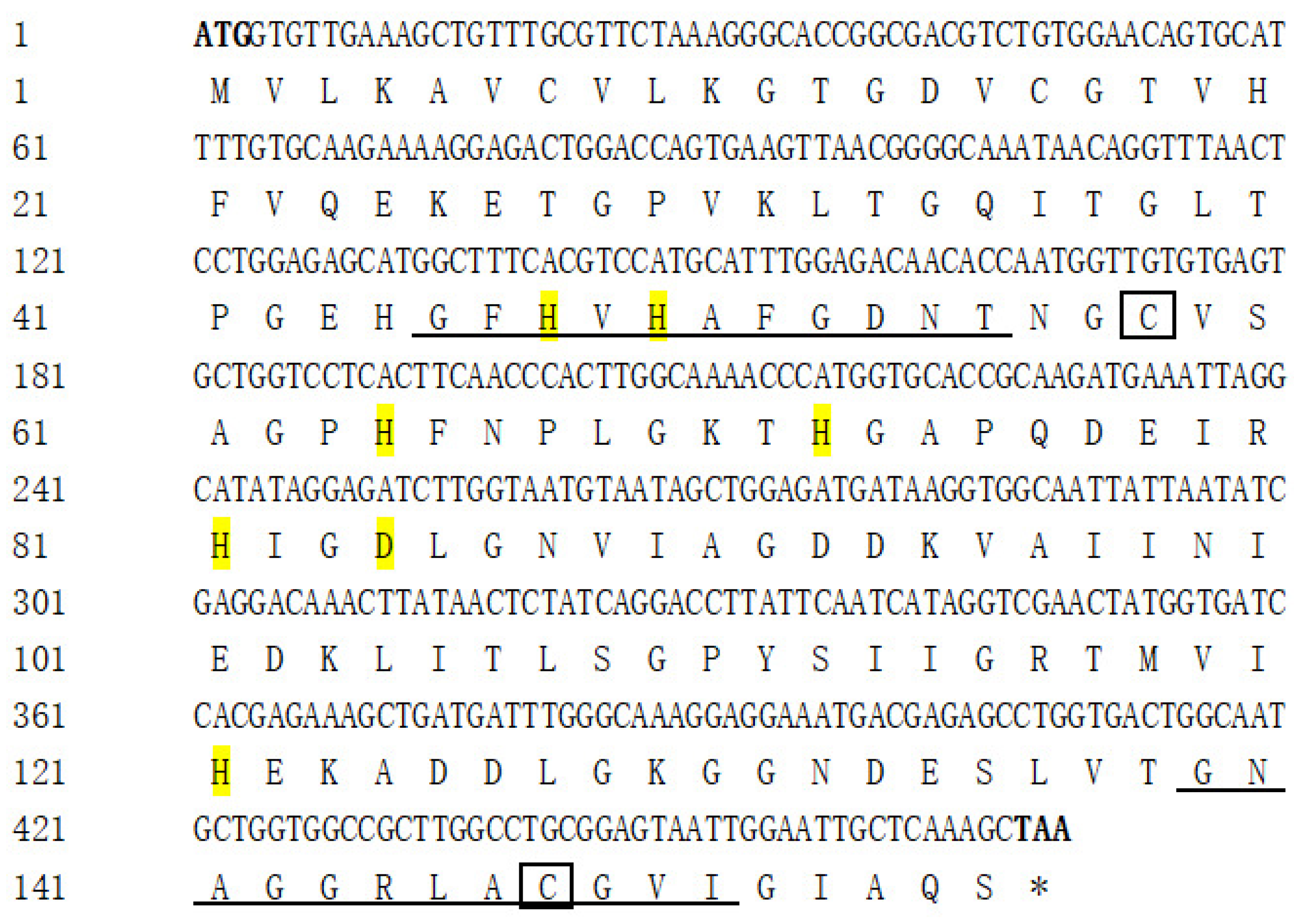
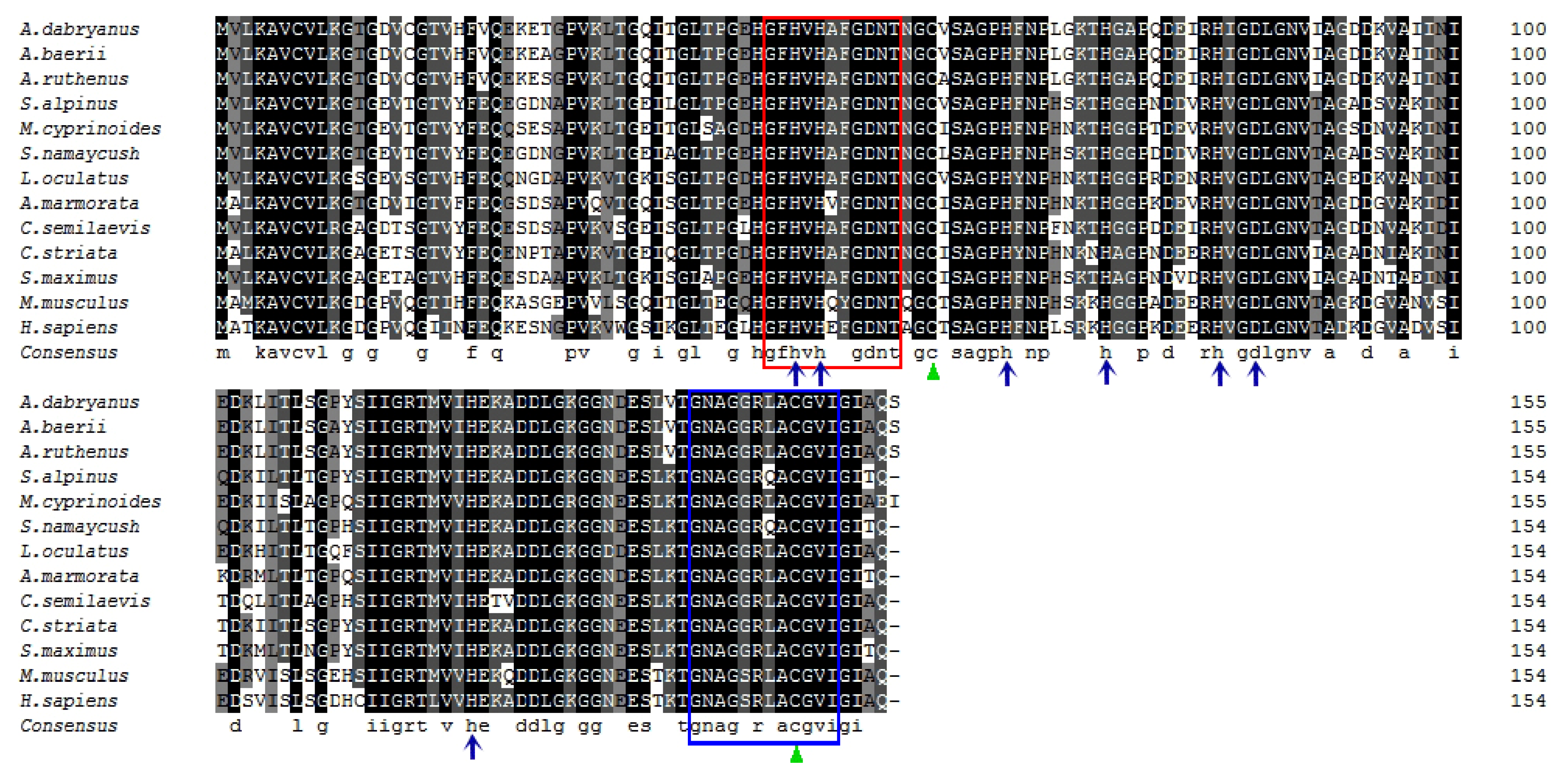
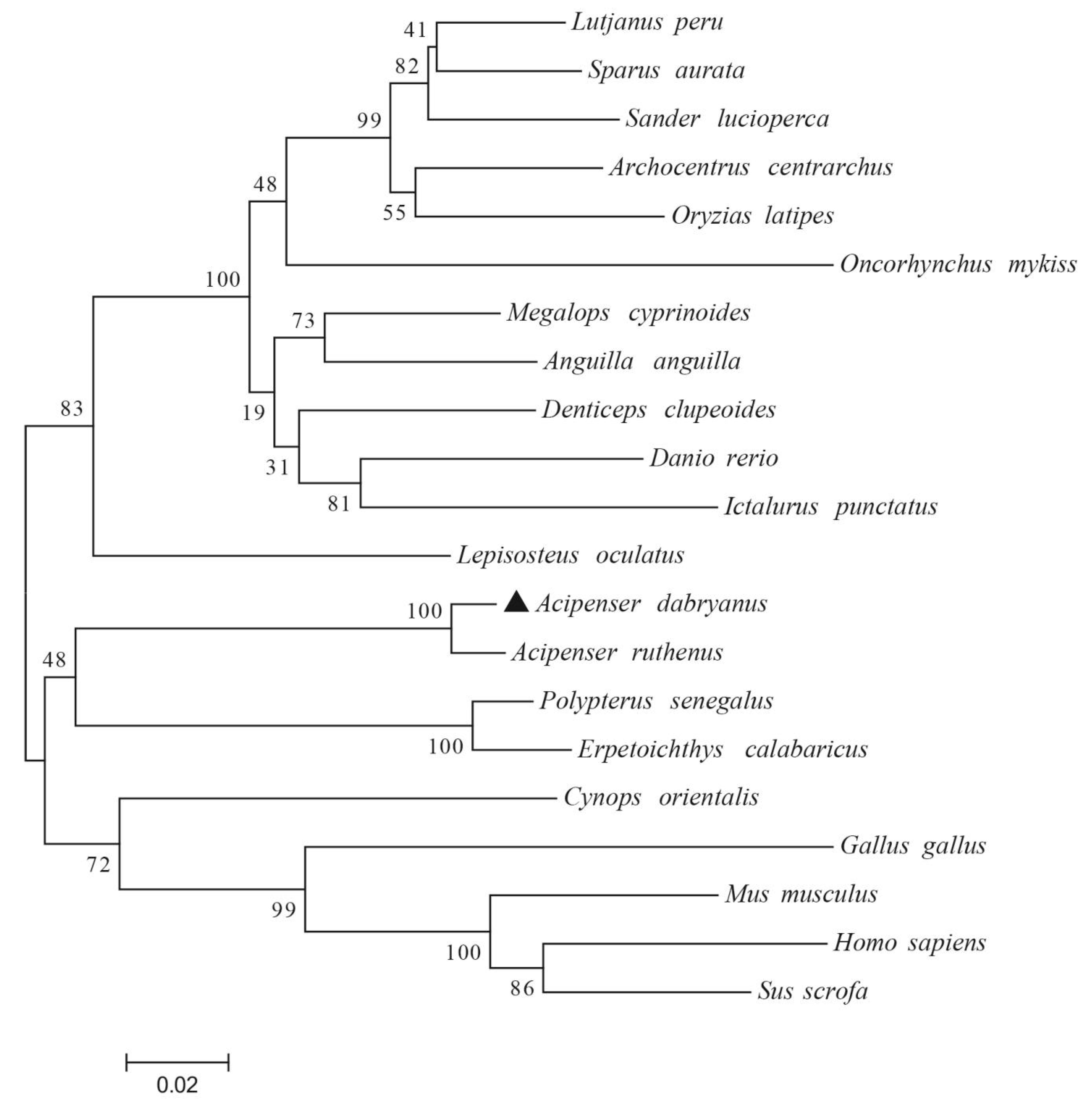
3.1. Cloning, Characterization, and Phylogenetic Analysis of AdCu/Zn-SOD and AdCAT
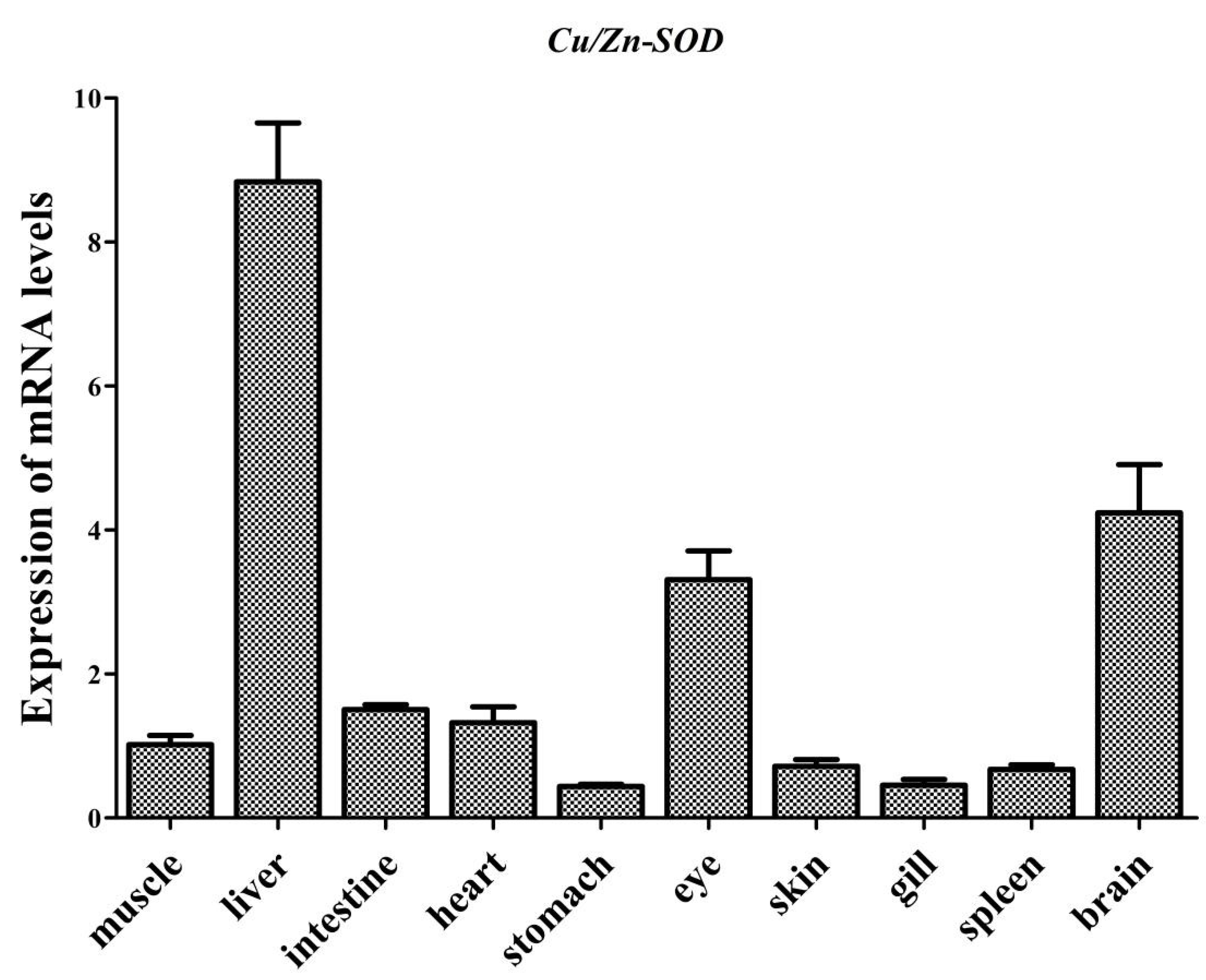
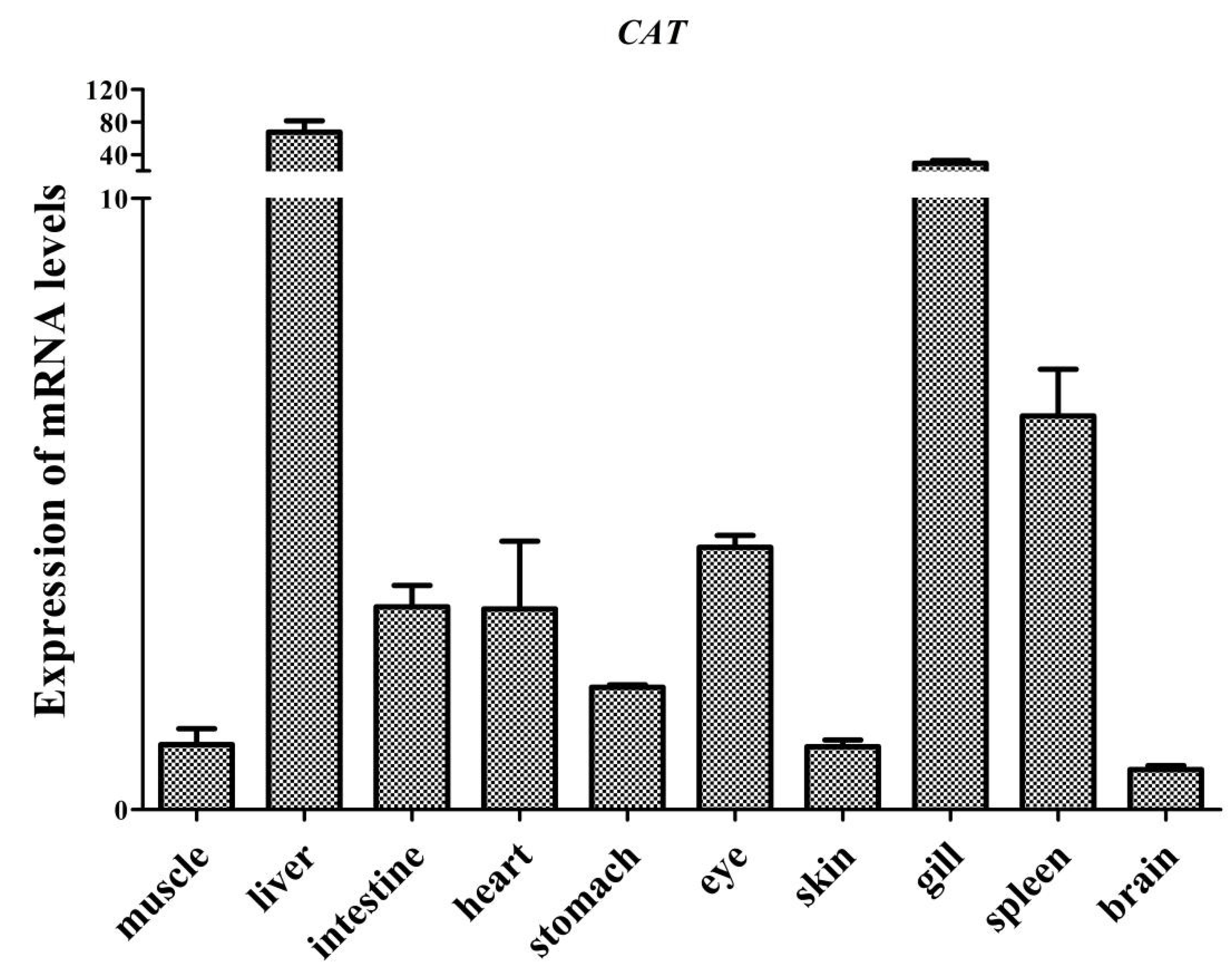
3.2. Tissue Distribution of AdCu/Zn-SOD and AdCAT mRNA Expression
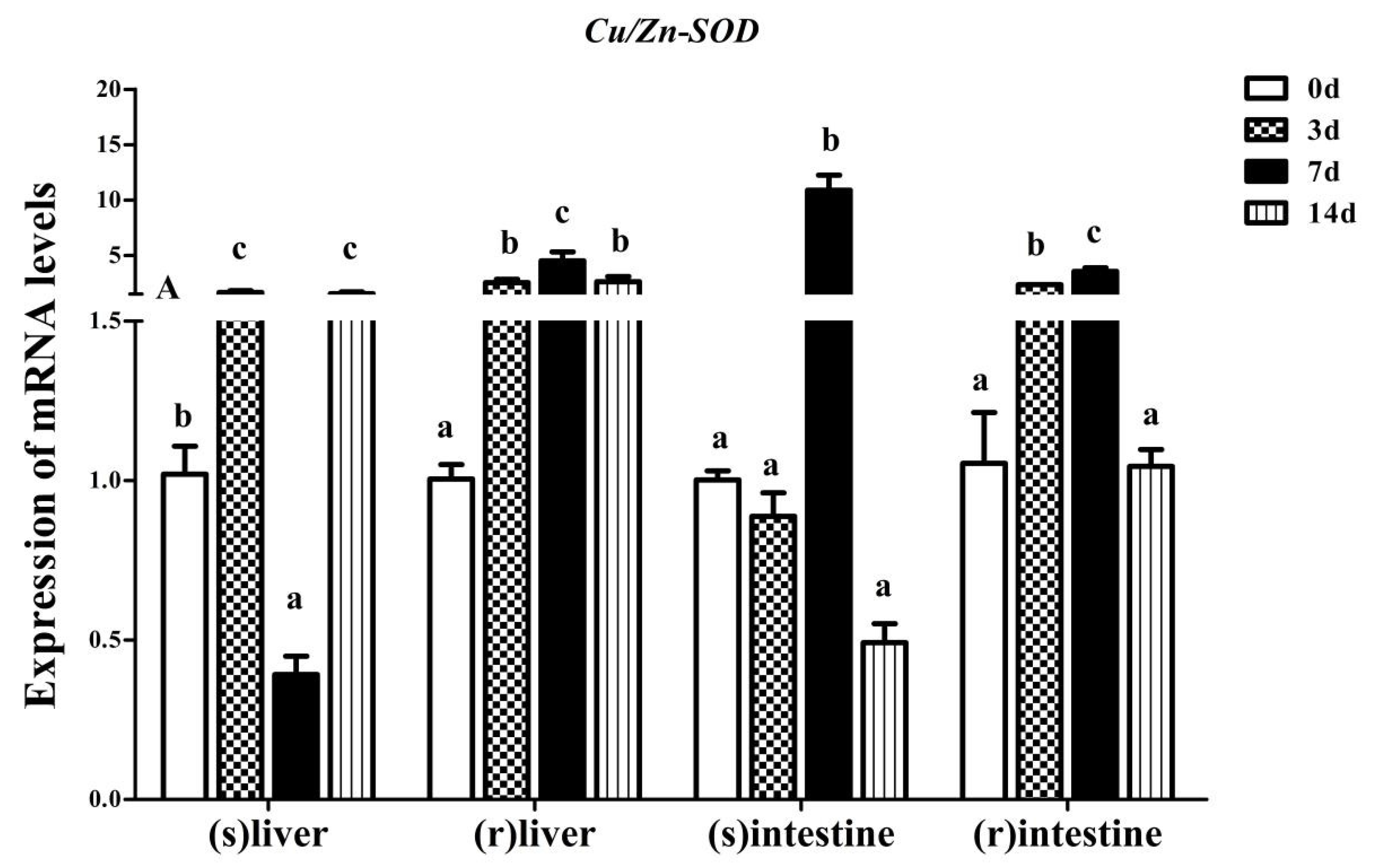
3.3. Effect of Fasting and Refeeding on Yangtze Sturgeon Liver and Intestine AdCu/Zn-SOD Expression
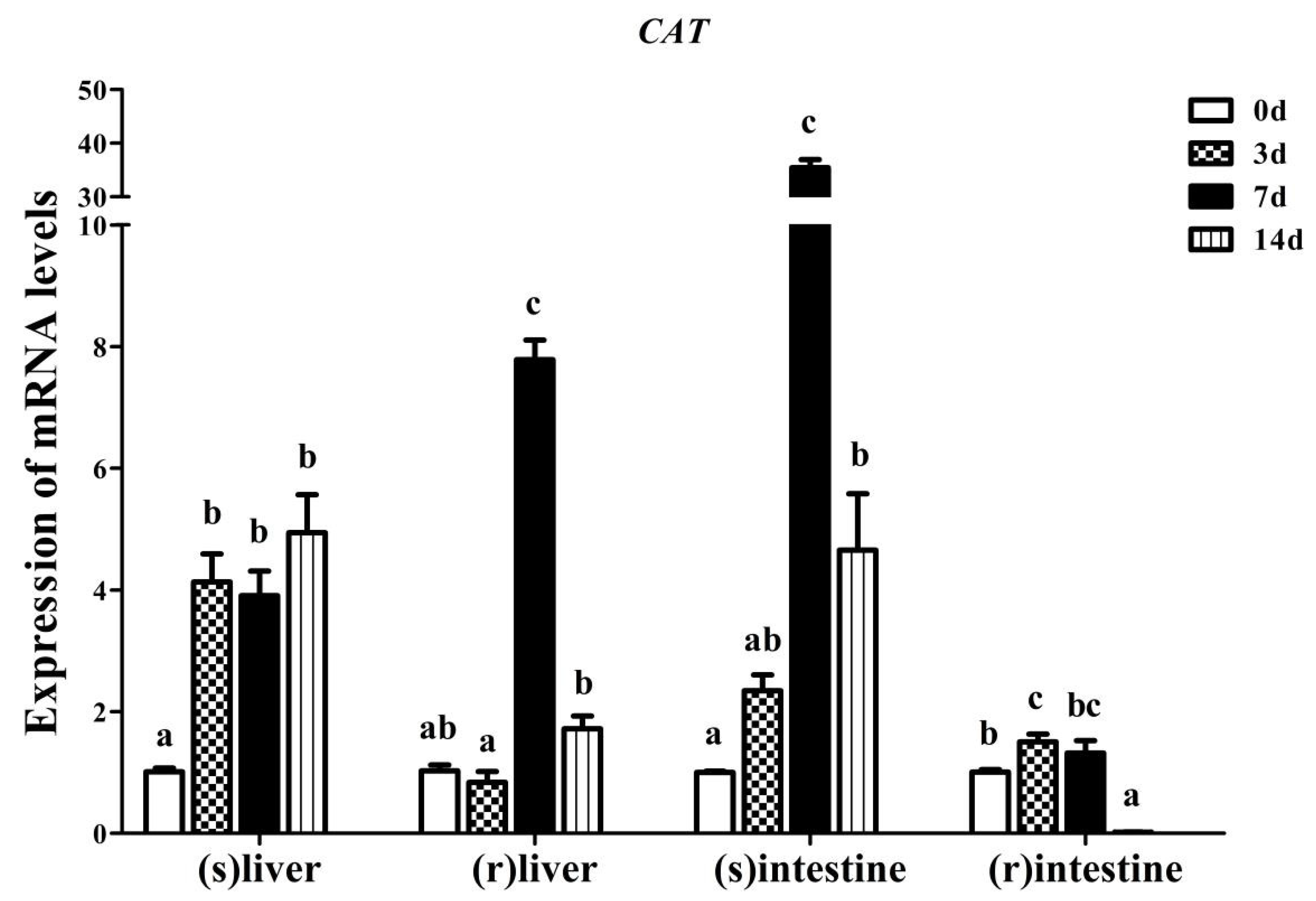
3.4. Effect of Fasting and Refeeding on Yangtze Sturgeon Liver and Intestine AdCAT Expression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Dar, S.A.; Srivastava, P.P.; Varghese, T.; Nazir, M.I.; Gupta, S.; Krishna, G. Temporal changes in superoxide dismutase, catalase, and heat shock protein 70 gene expression, cortisol and antioxidant enzymes activity of Labeo rohita fingerlings subjected to starvation and refeeding. Gene 2019, 692, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Furné, M.; Sanz, A.; García-Gallego, M.; Hidalgo, M.C.; Domezain, A.; Domezain, J.; Morales, A.E. Metabolic organization of the sturgeon Acipenser naccarii: A comparative study with rainbow trout Oncorhynchus mykiss. Aquaculture 2009, 289, 161–166. [Google Scholar] [CrossRef]
- Bayir, A.; Sirkecioglu, A.N.; Bayir, M.; Haliloglu, H.I.; Kocaman, E.M.; Aras, N.M. Metabolic responses to prolonged starvation, food restriction, and refeeding in the brown trout, Salmo trutta: Oxidative stress and antioxidant defenses. Comp. Biochem. Physiol. B 2011, 159, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Rios, F.S.; Kalinin, A.L.; Rantin, F.T. The effects of long-term food deprivation on respiration and haematology of the neotropical fish Hoplias malabaricus. J. Fish Biol. 2002, 61, 85–95. [Google Scholar] [CrossRef]
- Furné, M.; Morales, A.E.; Trenzado, C.E.; García-Gallego, M.; Carmen Hidalgo, M.; Domezain, A.; Sanz Rus, A. The metabolic effects of prolonged starvation and refeeding in sturgeon and rainbow trout. J. Comp. Physiol. B 2012, 182, 63–76. [Google Scholar] [CrossRef]
- Slater, J.J.; Lankford, T.E.; Buckel, J.A. Overwintering ability of young-of-the-year bluefish Pomatomus saltatrix: Effect of ration and cohort of origin on survival. Mar. Ecol. Prog. Ser 2007, 339, 259–269. [Google Scholar] [CrossRef] [Green Version]
- Azodi, M.; Ebrahimi, E.; Motaghi, E.; Morshedi, V. Metabolic responses to short starvation and re-feeding in rainbow trout (Oncorhynchus mykiss). Ichthyol. Res 2015, 62, 177–183. [Google Scholar] [CrossRef]
- Naisbitt, C.; Davies, S. Starvation, exercise and the stress response. Anaesth. Intensive Care 2017, 18, 508–512. [Google Scholar] [CrossRef]
- Wu, X.Y.; Chen, Y.Y.; Lai, J.S.; Liu, Y.; Long, Z.H. Effects of starvation and refeeding on growth performace, appetite, GH-IGFs axis levels and digestive function of Acipenser dabryanus. Brit. J. Nutr. 2020, 126, 1–39. [Google Scholar]
- Yang, S.; He, K.; Yan, T.; Wu, H.; Zhou, J.; Zhao, L.L.; Wang, Y.; Gong, Q. Effect of starvation and refeeding on oxidative stress and antioxidant defenses in Yangtze sturgeon (Acipenser dabryanus). Fish Physiol. Biochem 2019, 45, 987–995. [Google Scholar] [CrossRef]
- Choi, C.Y.; Choi, J.Y.; Choi, Y.J.; Yoo, J.H. Physiological effects of various light spectra on oxidative stress by starvation in olive flounder, Paralichthys olivaceus. Mol. Cell. Toxicol. 2018, 14, 399–408. [Google Scholar] [CrossRef]
- Regoli, F.; Giuliani, M.E. Oxidative pathways of chemical toxicity and oxidative stress biomarkers in marine organisms. Mar. Environ. Res. 2014, 93, 106–117. [Google Scholar] [CrossRef]
- Nikahval, B.; Hosseini, Z.; Nazifi, S.; Foroud, M. Evaluation of antioxidant enzymes changes in synovial fluid and blood, following experimental osteoarthritis in dogs. Comp. Clin. Path. 2015, 24, 23–27. [Google Scholar] [CrossRef]
- Tiwari, M.K.; Hägglund, P.M.; Møller, I.M.; Davies, M.J.; Bjerrum, M.J. Copper ion/H2O2 oxidation of Cu/Zn-Superoxide dismutase: Implications for enzymatic activity and antioxidant action. Redox. Biol. 2019, 26, 101262. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Anduro, G.A.; Barillas-Mury, C.V.; Peregrino-Uriarte, A.B.; Gupta, L.; Gollas-Galván, T.; Hernández-López, J.; Yepiz-Plascencia, G. The cytosolic manganese superoxide dismutase from the shrimp Litopenaeus vannamei: Molecular cloning and expression. Dev. Comp. Immunol. 2006, 30, 893–900. [Google Scholar] [CrossRef] [PubMed]
- Martins, D.; English, A.M. SOD1 oxidation and formation of soluble aggregates in yeast: Relevance to sporadic ALS development. Redox. Biol. 2014, 2, 632–639. [Google Scholar] [CrossRef] [Green Version]
- Olson, K.R.; Gao, Y.; DeLeon, E.R.; Arif, M.; Arif, F.; Arora, N.; Straub, K.D. Catalase as a sulfide-sulfur oxido-reductase: An ancient (and modern?) regulator of reactive sulfur species (RSS). Redox. Biol. 2017, 12, 325–339. [Google Scholar] [CrossRef]
- Yu, H.; Deng, W.; Zhang, D.; Gao, Y.; Ji, H. Antioxidant defenses of Onychostoma macrolepis in response to thermal stress: Insight from mRNA expression and activity of superoxide dismutase and catalase. Fish Shellfish Immunol. 2017, 66, 50–61. [Google Scholar] [CrossRef]
- Li, M.; Zheng, Y.; Liang, H.; Zou, L.; Sun, J.; Zhang, Y.; Qin, F.; Liu, S.; Wang, Z. Molecular cloning and characterization of cat, gpx1 and Cu/Zn-sod genes in pengze crucian carp (Carassius auratus var. Pengze) and antioxidant enzyme modulation induced by hexavalent chromium in juveniles. Comp. Biochem. Physiol. C 2013, 157, 310–321. [Google Scholar] [CrossRef]
- Wang, X.; Wang, L.; Ren, Q.; Yin, S.; Liang, F.; Jia, Y. Two superoxide dismutases (SODs) respond to bacterial challenge identified in the marbled eel Anguilla marmorata. Aquaculture 2016, 451, 316–325. [Google Scholar] [CrossRef]
- Liu, H.; He, J.; Chi, C.; Gu, Y. Identification and analysis of icCu/Zn-SOD, Mn-SOD and ecCu/Zn-SOD in superoxide dismutase multigene family of Pseudosciaena crocea. Fish Shellfish Immunol. 2015, 43, 491–501. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.; Li, J.; Liu, P.; Ren, X.; Song, T.; Gao, G.; Li, D.; Liu, S. Cloning of catalase gene and antioxidant genes in Scophthalmus maximus response to metalloprotease of Vibrio anguillarum stress. J. Oceanol. Limnol. 2021. [Google Scholar] [CrossRef]
- Elvitigala, D.A.S.; Premachandra, H.K.A.; Whang, I.; Priyathilaka, T.T.; Kim, E.; Lim, B.S.; Jung, H.B.; Yeo, S.Y.; Park, H.C.; Lee, J. Marine teleost ortholog of catalase from rock bream (Oplegnathus fasciatus): Molecular perspectives from genomic organization to enzymatic behavior with respect to its potent antioxidant properties. Fish Shellfish Immunol. 2013, 35, 1086–1096. [Google Scholar] [CrossRef] [PubMed]
- Shen, B.; Wei, K.; Ding, Y.; Zhang, J. Molecular cloning, mRNA expression and functional characterization of a catalase from Chinese black sleeper (Bostrychus sinensis). Fish Shellfish Immunol. 2020, 103, 310–320. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wei, Q.W.; Du, H.; Li, L.X. Present status and risk for extinction of the Dabry’s sturgeon (Acipenser dabryanus) in the Yangtze River watershed: A concern for intensified rehabilitation needs. J. Appl. Ichthyol. 2011, 27, 181–185. [Google Scholar] [CrossRef]
- Wu, J.M.; Wei, Q.W.; Du, H.; Wang, C.Y.; Zhang, H. Initial evaluation of the release programme for Dabry’s sturgeon (Acipenser dabryanus Duméril, 1868) in the upper Yangtze River. J. Appl. Ichthyol. 2014, 30, 1423–1427. [Google Scholar] [CrossRef]
- Zhuang, P.; Ke, F.E.; Wei, Q.; He, X.; Cen, Y. Biology and life history of Dabry’s sturgeon, Acipenser dabryanus, in the Yangtze River. Environ. Biol. Fish 1997, 48, 257–264. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, Y.; Quan, G.; Lai, J.; Deng, X. Paternity assignment in the polyploid Acipenser dabryanus based on a novel microsatellite marker system. PLoS ONE 2017, 12, e0185280. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Liu, X.; Yang, J.; Xiao, K.; Wang, B.; Du, H. Development of 17 novel cross-Species microsatellite markers for Dabry’s Sturgeon (Acipenser dabryanus) from Chinese sturgeon (Acipenser sinensis) via next-generation sequencing. Pak. J. Zool. 2019, 51, 2381–2384. [Google Scholar] [CrossRef]
- Ruan, R.; Feng, T.; Li, Y.; Yue, H.; Ye, H.; Du, H.; Liu, Q.; Ruan, L.; Li, H.; Wei, Q. Screening and identification of female-specific DNA sequences in octaploid sturgeon using comparative genomics with high-throughput sequencing. Genomics 2021, 113, 4237–4244. [Google Scholar] [CrossRef]
- Chen, Y.; Gong, Q.; Song, M.; Lai, J.; Sun, J.; Liu, Y. Identification and characterization of three novel antimicrobial peptides from Acipenser dabryanus. Fish Shellfish Immunol. 2019, 88, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Liu, Y.; Du, J.; Song, M.; Lai, J.; Gong, Q. Isolation and characterization of 47 SNP markers in the critically endangered Acipenser dabryanus. Conserv. Genet. Resour 2020, 12, 9–12. [Google Scholar] [CrossRef]
- Chen, Y.; Wu, X.; Lai, J.; Liu, Y.; Song, M.; Li, F.; Gong, Q. Full-length transcriptome sequencing and identification and immune response of TRIM genes in Dabry’s sturgeon (Acipenser dabryanus). Aquaculture 2021, 538, 736599. [Google Scholar] [CrossRef]
- Han, P.; Wang, S.; Zhang, Q.; Zhang, S.; Shao, R.; Xu, W.; Zhang, W.; Xu, Q.; Wei, Q.; Qi, Z. Molecular characterization and expression analysis of TLR1 and TLR4 from the endangered fish Dabry’s sturgeon (Acipenser dabryanus). Dev. Comp. Immunol. 2018, 86, 180–188. [Google Scholar] [CrossRef]
- Wu, X.Y.; Chen, Y.Y.; Lai, J.S.; Liu, Y.; Song, M.J.; Gong, Q.; Long, Z.H. Effects of starvation and re-feeding on antioxidative function of liver, intestine and muscle in Acipenser dabryanus. J. South. Agric. 2020, 17, 100370. [Google Scholar]
- Qin, C.J.; Wen, Z.Y.; Wang, J.; He, Y.; Yuan, D.Y.; Li, R. Uncoupling protein 1 in snakehead (Channa argus): Cloning, tissue distribution, and its expression in response to fasting and refeeding. Comp. Biochem. Physiol. A 2018, 225, 1–6. [Google Scholar] [CrossRef]
- Zhang, Z.W.; Li, Z.; Liang, H.W.; Li, L.; Luo, X.Z.; Zou, G.W. Molecular cloning and differential expression patterns of copper/zinc superoxide dismutase and manganese superoxide dismutase in Hypophthalmichthys molitrix. Fish Shellfish Immunol. 2011, 30, 473–479. [Google Scholar] [CrossRef]
- Ren, H.; Li, J.; Li, J.; Ying, Y.; Ge, H.; Li, D.; Yu, T. Cloning of catalase and expression patterns of catalase and selenium-dependent glutathione peroxidase from Exopalaemon carinicauda in response to low salinity stress. Acta. Oceanol. Sin. 2015, 34, 52–61. [Google Scholar] [CrossRef]
- Shi, Q.; Rong, H.; Hao, M.; Zhu, D.; Aweya, J.J.; Li, S.; Wen, X. Effects of dietary Sargassum horneri on growth performance, serum biochemical parameters, hepatic antioxidant status, and immune responses of juvenile black sea bream Acanthopagrus schlegelii. J. Appl. Phycol. 2019, 31, 2103–2113. [Google Scholar] [CrossRef]
- Choi, C.Y.; Shin, H.S.; Choi, Y.J.; Kim, N.N.; Lee, J.; Kil, G.S. Effect of LED light spectra on starvation-induced oxidative stress in the cinnamon clownfish Amphiprion melanopus. Comp. Biochem. Physiol. A 2012, 163, 357–363. [Google Scholar] [CrossRef]
- Martínez-Álvarez, R.M.; Morales, A.E.; Sanz, A. Antioxidant defenses in fish: Biotic and abiotic factors. Rev. Fish Biol. Fish. 2005, 15, 75–88. [Google Scholar] [CrossRef]
- Bouayed, J.; Rammal, H.; Soulimani, R. Oxidative stress and anxiety: Relationship and cellular pathways. Oxid. Med. Cell. Longev. 2009, 2, 63–67. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.L.; Zhu, Q.L.; Shen, B.; Zeng, L.; Zhu, A.Y.; Wu, C.W. Effects of starvation on lipid accumulation and antioxidant response in the right and left lobes of liver in large yellow croaker Pseudosciaena crocea. Ecol. Indic. 2016, 66, 269–274. [Google Scholar] [CrossRef]
- Sakyi, M.E.; Cai, J.; Tang, J.; Abarike, E.D.; Xia, L.; Li, P.; Kuebutornye, F.K.A.; Zou, Z.; Liang, Z.; Jian, J. Effects of starvation and subsequent re-feeding on intestinal microbiota, and metabolic responses in Nile tilapia, Oreochromis niloticus. Aquacult. Rep. 2020, 17, 100370. [Google Scholar] [CrossRef]
- Wang, C.; Liang, Y.; Fang, Y.; Chang, X. Effects of cyclical short-term food deprivation and refeeding on compensatory growth and gene expression of SOD, GPX and HSP70 in Schizothorax wangchiachii. Fish Shellfish Immunol. 2019, 94, 628–633. [Google Scholar] [CrossRef]
- Feng, G.; Shi, X.; Huang, X.; Zhuang, P. Oxidative stress and antioxidant defenses after long-term fasting in blood of Chinese sturgeon (Acipenser sinensis). Procedia Environ. Sci. 2011, 8, 469–475. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.C.; Chen, J.C.; Man, S.C.; Morni, W.W.; Shahrir, N.; Cheng, S.Y.; Hu, C.H. Modulation of innate immunity and gene expressions in white shrimp Litopenaeus vannamei following long-term starvation and re-feeding. Res. Immunol. 2012, 2, 148–156. [Google Scholar] [CrossRef] [Green Version]
- Florescu, I.E.; Georgescu, S.E.; Dudu, A.; Balaș, M.; Voicu, S.; Grecu, I.; Dediu, L.; Dinischiotu, A.; Costache, M. Oxidative stress and antioxidant defense mechanisms in response to starvation and refeeding in the intestine of stellate sturgeon (Acipenser stellatus) Juveniles from Aquaculture. Animals 2021, 11, 76. [Google Scholar] [CrossRef]
- Ticho, A.L.; Malhotra, P.; Dudeja, P.K.; Gill, R.K.; Alrefai, W.A. Bile acid receptors and gastrointestinal functions. Liver Res. 2019, 3, 31–39. [Google Scholar] [CrossRef]
- Datta, S.; Kundu, S.; Ghosh, P.; De, S.; Ghosh, A.; Chatterjee, M. Correlation of oxidant status with oxidative tissue damage in patients with rheumatoid arthritis. Clin. Rheumatol. 2014, 33, 1557–1564. [Google Scholar] [CrossRef]
- Florescu, I.E.; Burcea, A.; Popa, G.O.; Dudu, A.; Georgescu, S.E.; Balas, M.; Dinescu, S.; Voicu, S.; Grecu, I.; Dediu, L.; et al. Effects of starvation and refeeding on growth performance and stress defense mechanisms of stellate sturgeon Acipenser stellatus juveniles from aquaculture. Acta Biochim. Pol. 2019, 66, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antonopoulou, E.; Kentepozidou, E.; Feidantsis, K.; Roufidou, C.; Despoti, S.; Chatzifotis, S. Starvation and re-feeding affect Hsp expression, MAPK activation and antioxidant enzymes activity of European sea bass (Dicentrarchus labrax). Comp. Biochem. Physiol. A 2013, 165, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.L.; Ge, X.P.; Sun, S.M.; Zhu, J.; Zhang, W.X.; Yu, H. Effects of compensatory feeding after starvation on growth performance, serum biochemical indexes, intestinal digestive enzyme activities and hepatic antioxidant enzyme activities of juvenile blunt snout bream (Megalobrama amblycephal) in Summer. Chinese. J. Animal. Nutr. 2017, 29, 4198–4206. [Google Scholar]



| Primer | Sequence (5′ to 3′) | OAT (°C) |
|---|---|---|
| AdCu/Zn-SOD-F | ATGGTGTTGAAAGCTGTTTGCG | 51 |
| AdCu/Zn-SOD-R | GACAGAAACACTGAAGATTAGC | |
| AdCAT-F | ATGGCGGGAAACCGAGAC | 51 |
| AdCAT-R | TCACATCTTGGATTCACGTGCA | |
| AdCu/Zn-SOD-qF | AAACTTATAACTCTATCAGGACCTTATTCA | 54.9 |
| AdCu/Zn-SOD-qR | CAGTCACCAGGCTCTCGTCAT | |
| AdCAT-qF | CCTGTGAACTGCCCCTAT | 60.4 |
| AdCAT-qR | ACATTGTCATCGTCGGAG | |
| β-actin-qF | GACCGAGGCACCCCTGAAC | 54.9 |
| β-actin-qR | GATGGGCACTGTGTGTGTGAC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, Q.; Xiong, X.; Wen, Z.; Qin, C.; Li, R.; Zhang, Z.; Gong, Q.; Wu, X. Cu/Zn Superoxide Dismutase and Catalase of Yangtze Sturgeon, Acipenser dabryanus: Molecular Cloning, Tissue Distribution and Response to Fasting and Refeeding. Fishes 2022, 7, 35. https://doi.org/10.3390/fishes7010035
Shi Q, Xiong X, Wen Z, Qin C, Li R, Zhang Z, Gong Q, Wu X. Cu/Zn Superoxide Dismutase and Catalase of Yangtze Sturgeon, Acipenser dabryanus: Molecular Cloning, Tissue Distribution and Response to Fasting and Refeeding. Fishes. 2022; 7(1):35. https://doi.org/10.3390/fishes7010035
Chicago/Turabian StyleShi, Qingchao, Xiaoqin Xiong, Zhengyong Wen, Chuanjie Qin, Rui Li, Zhiyong Zhang, Quan Gong, and Xiaoyun Wu. 2022. "Cu/Zn Superoxide Dismutase and Catalase of Yangtze Sturgeon, Acipenser dabryanus: Molecular Cloning, Tissue Distribution and Response to Fasting and Refeeding" Fishes 7, no. 1: 35. https://doi.org/10.3390/fishes7010035
APA StyleShi, Q., Xiong, X., Wen, Z., Qin, C., Li, R., Zhang, Z., Gong, Q., & Wu, X. (2022). Cu/Zn Superoxide Dismutase and Catalase of Yangtze Sturgeon, Acipenser dabryanus: Molecular Cloning, Tissue Distribution and Response to Fasting and Refeeding. Fishes, 7(1), 35. https://doi.org/10.3390/fishes7010035







