Enhanced Light Sheet Elastic Scattering Microscopy by Using a Supercontinuum Laser
Abstract
1. Introduction
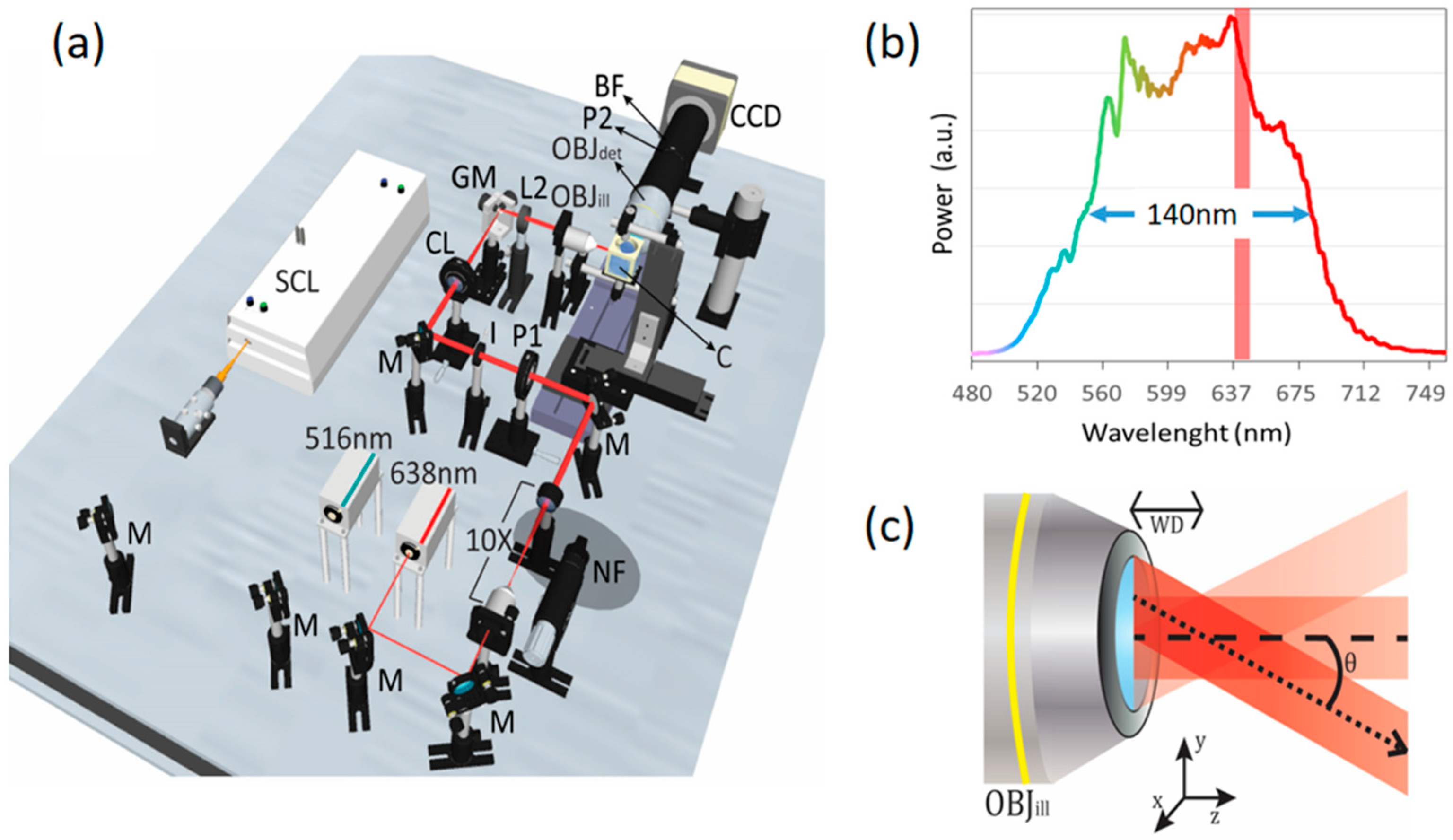
2. Experimental Setup
3. Results and Discussion
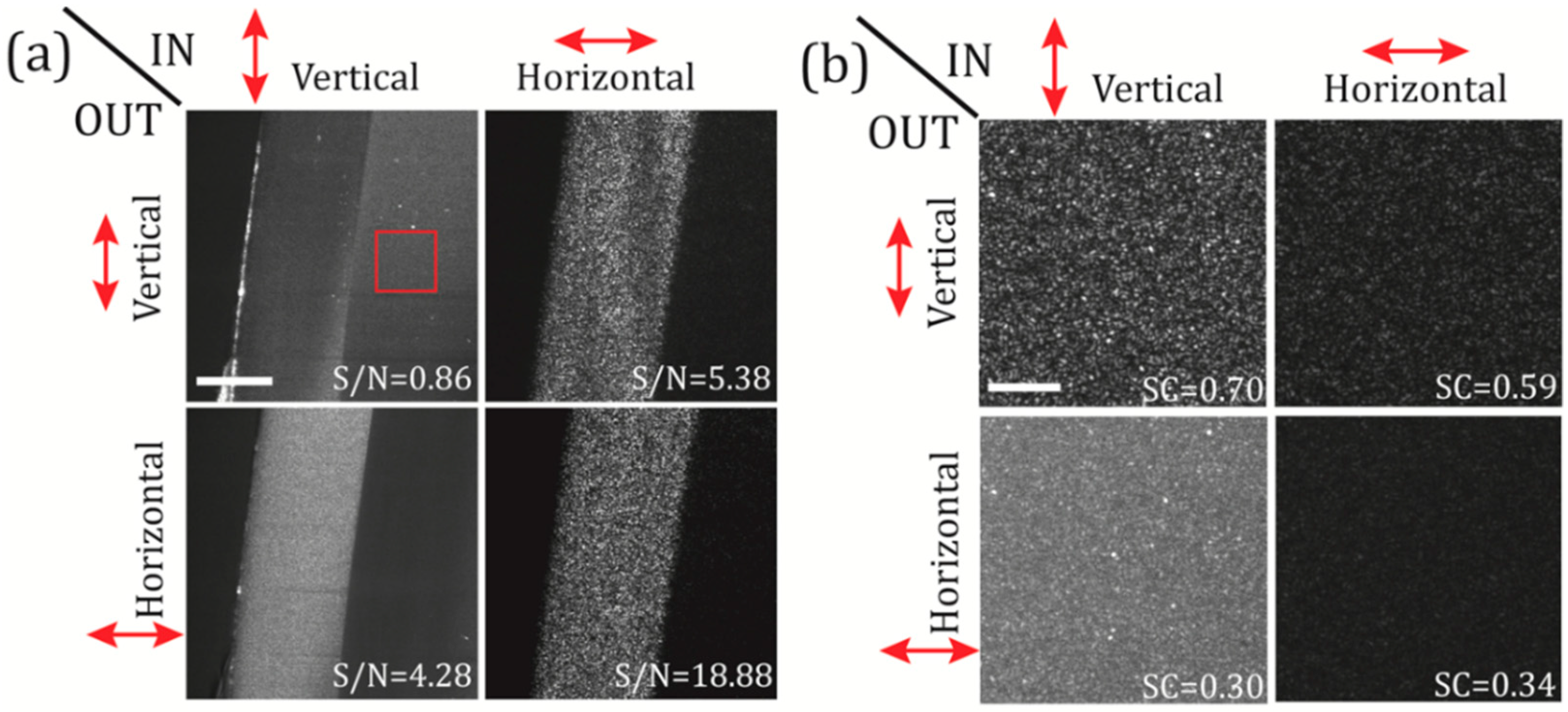
3.1. Polarization Filtering
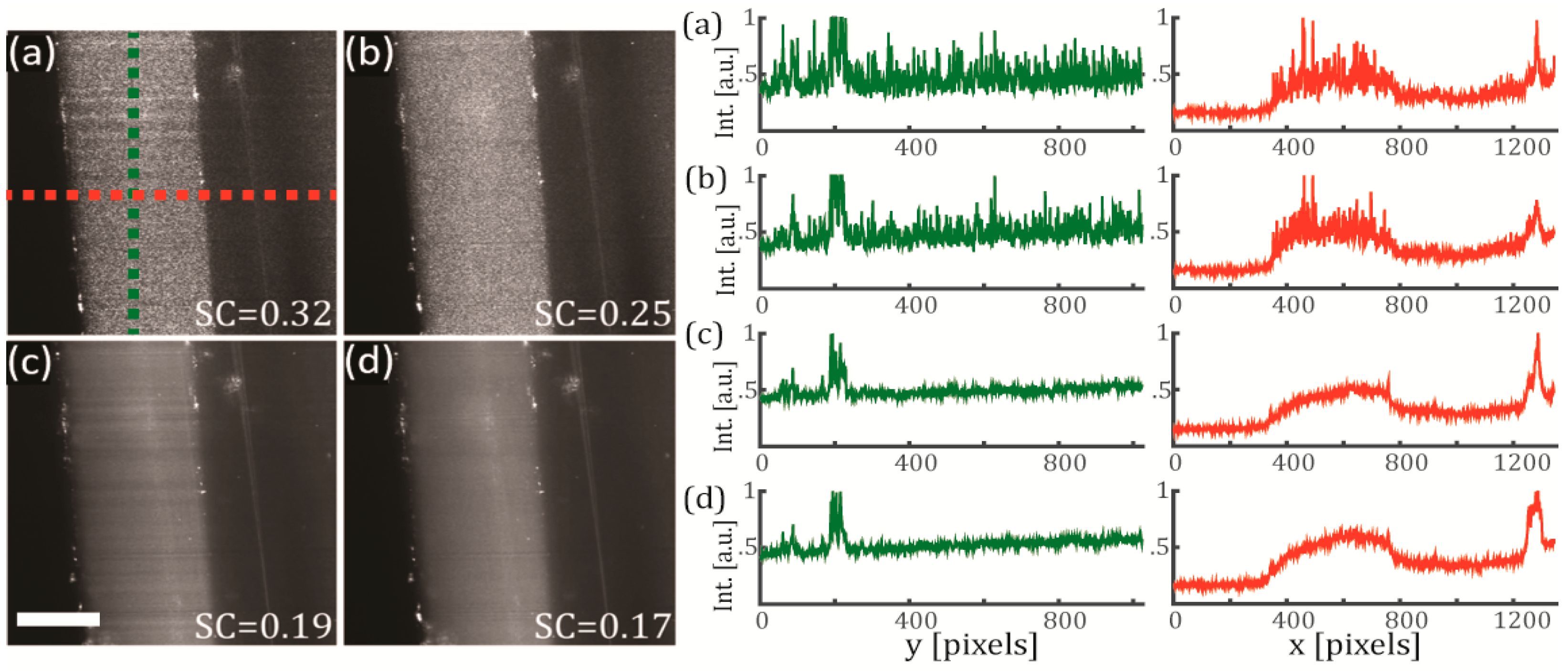
3.2. Effects of the Temporal and the Spatial Coherence on the Speckle Noise
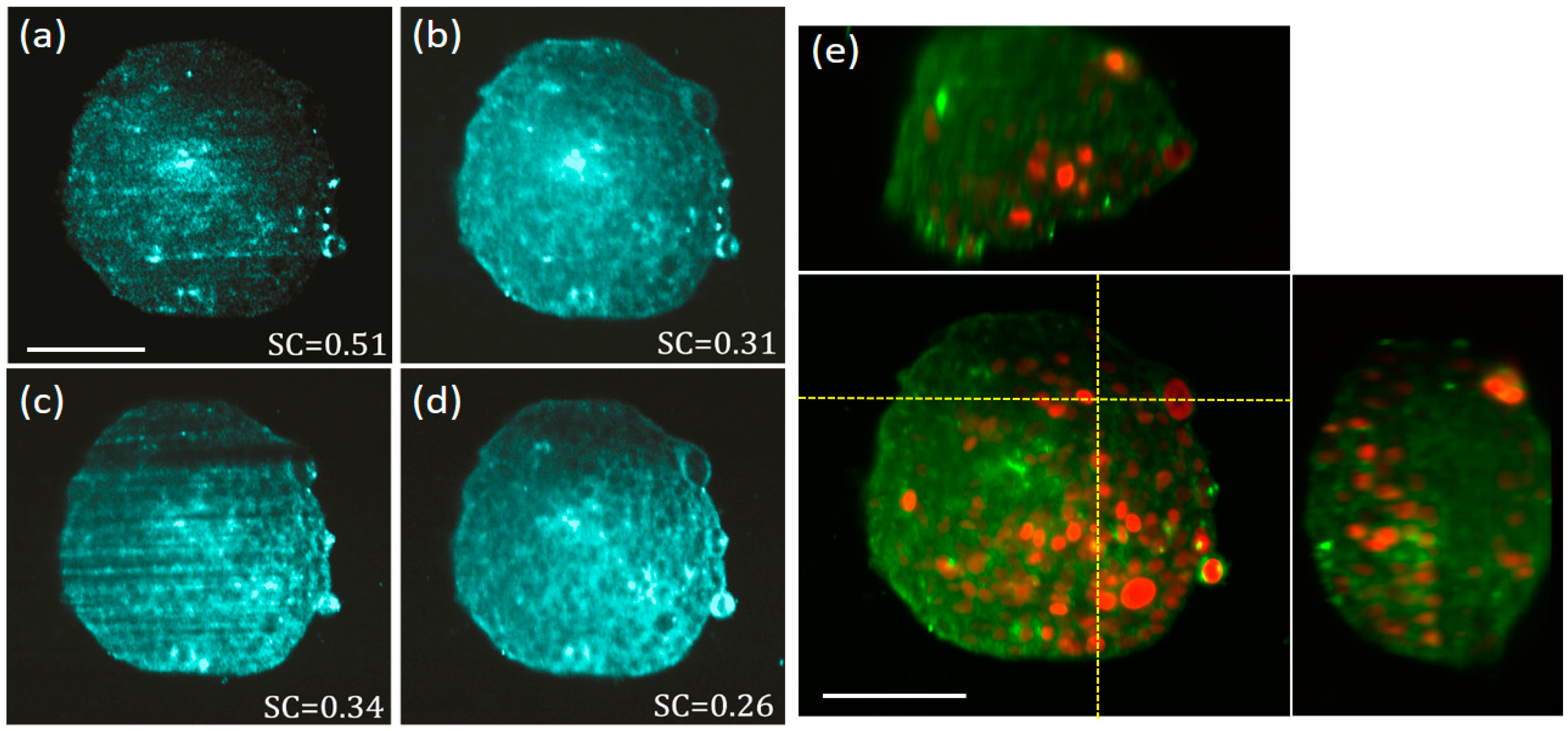
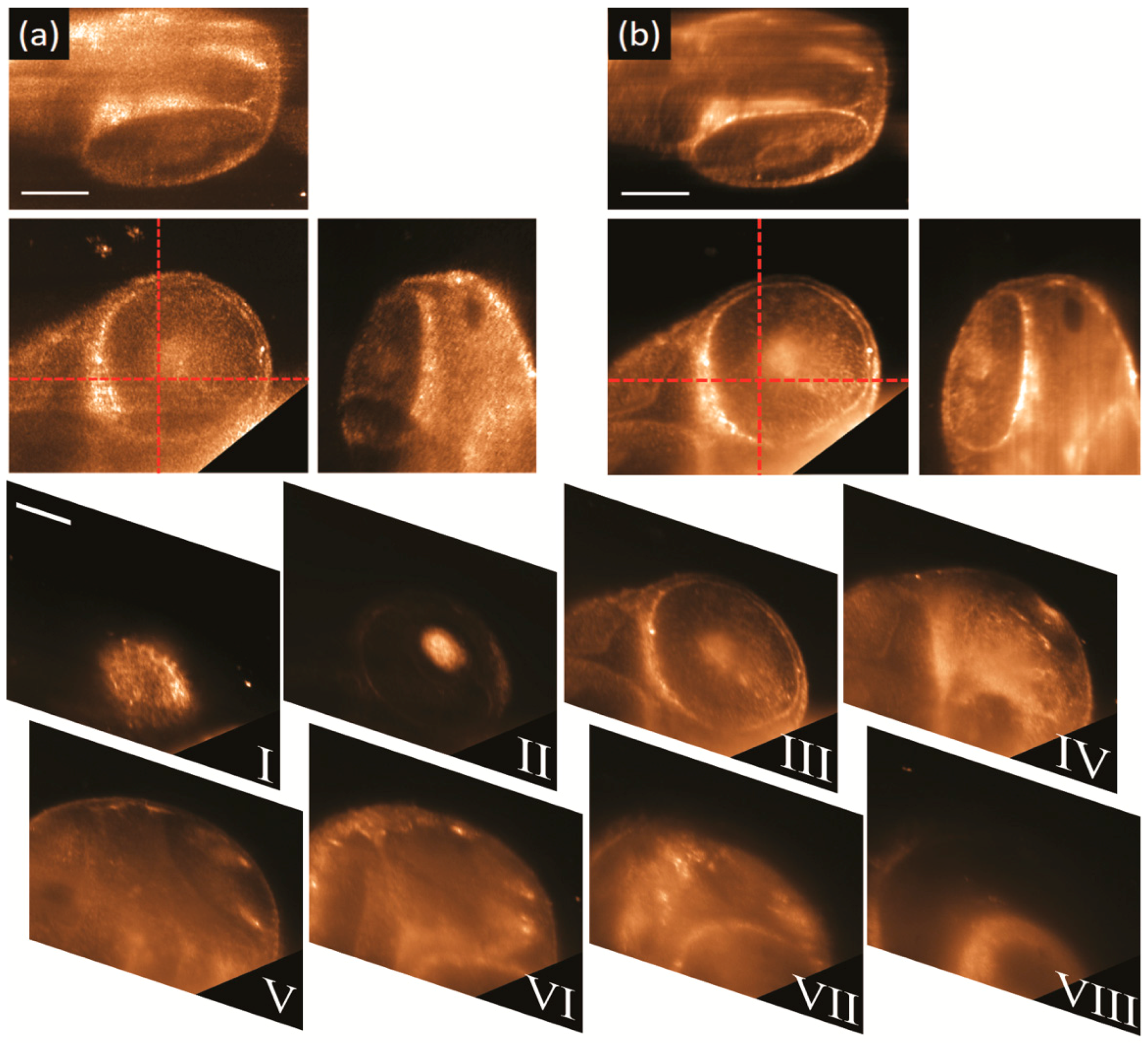
3.3. Results Obtained by Using Biological Samples
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Huisken, J.; Swoger, J.; Del Bene, F.; Wittbrodt, J.; Stelzer, E.H.K. Optical Sectioning Deep Inside Live Embryos by Selective Plane Illumination Microscopy. Science 2004, 305, 1007–1009. [Google Scholar] [CrossRef] [PubMed]
- Olarte, O.E.; Andilla, J.; Gualda, E.J.; Loza-Alvarez, P. Light-sheet microscopy: A tutorial. Adv. Opt. Photon. 2018, 10, 111–179. [Google Scholar] [CrossRef]
- Rocha-Mendoza, I.; Licea-Rodriguez, J.; Marro, M.; Olarte, O.E.; Plata-Sanchez, M.; Loza-Alvarez, P. Rapid spontaneous Raman light sheet microscopy using cw-lasers and tunable filters. Biomed. Opt. Express 2015, 6, 3449–3461. [Google Scholar] [CrossRef] [PubMed]
- Boustany, N.N.; Boppart, S.A.; Backman, V. Microscopic Imaging and Spectroscopy with Scattered Light. Annu. Rev. Biomed. Eng. 2010, 12, 285–314. [Google Scholar] [CrossRef] [PubMed]
- Jacques, S.L. Optical properties of biological tissues: A review. Phys. Med. Biol. 2013, 58, R37–R61. [Google Scholar] [CrossRef] [PubMed]
- Fercher, A.F.; Drexler, W.; Hitzenberger, C.K.; Lasser, T. Optical coherence tomography-principles and applications. Rep. Prog. Phys. 2003, 66, 239–303. [Google Scholar] [CrossRef]
- Drexler, W.; Fujimoto, J.G. (Eds.) Optical Coherence Tomography: Technology and Applications, 2nd ed.; Springer International Publishing: Basel, Switzerland, 2015; ISBN 978-3-319-06418-5. [Google Scholar]
- Goodman, J.W. Some fundamental properties of speckle. J. Opt. Soc. Am. 1976, 66, 1145–1150. [Google Scholar] [CrossRef]
- Ancora, D.; Battista, D.D.; Giasafaki, G.; Psycharakis, S.E.; Liapis, E.; Ripoll, J.; Zacharakis, G. Phase-Retrieved Tomography enables Mesoscopic imaging of Opaque Tumor Spheroids. Sci. Rep. 2017, 7, 11854. [Google Scholar] [CrossRef]
- Di Battista, D.; Ancora, D.; Zhang, H.; Lemonaki, K.; Marakis, E.; Liapis, E.; Tzortzakis, S.; Zacharakis, G. Tailored light sheets through opaque cylindrical lenses. Optica 2016, 3, 1237–1240. [Google Scholar] [CrossRef]
- Yang, Z.; Downie, H.; Rozbicki, E.; Dupuy, L.X.; MacDonald, M.P. Light Sheet Tomography (LST) for in situ imaging of plant roots. Opt. Express 2013, 21, 16239–16247. [Google Scholar] [CrossRef]
- Reidt, S.L.; O’Brien, D.J.; Wood, K.; MacDonald, M.P. Polarised light sheet tomography. Opt. Express 2016, 24, 11239–11249. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Holy, T.E. Development of Low-Coherence Light Sheet Illumination Microscope for Fluorescence-Free Bioimaging. In Proceedings of the SPIE 8129, San Diego, CA, USA, 21–25 August 2011; Koshel, R.J., Gregory, G.G., Eds.; 2011; pp. 812908-1–812908-10. [Google Scholar]
- Dudley, J.M.; Genty, G.; Coen, S. Supercontinuum generation in photonic crystal fiber. Rev. Mod. Phys. 2006, 78, 1135–1184. [Google Scholar] [CrossRef]
- Merino, D.; Olarte, O.E.; Cruz, J.L.; Diez, J.; Barmenkov, Y.; Andres, M.; Perez, P.; Loza-Alvarez, P. Use of A Supercontinuum Laser Source with Low Temporal Coherence for Elastic Scattering Light Sheet Microscopy. In Proceedings of the CLEO/Europe-EQEC 2015, Munich, Germany, 21–25 June 2015; OSA: Zurich, Germany, 2015; Volume CL-P.7 SUN. [Google Scholar]
- Tran, T.T.K.; Svensen, Ø.; Chen, X.; Akram, M.N. Speckle reduction in laser projection displays through angle and wavelength diversity. Appl. Opt. 2016, 55, 1267–1274. [Google Scholar] [CrossRef] [PubMed]
- Shu, X.; Beckmann, L.J.; Zhang, H.F. Visible-light optical coherence tomography: A review. J. Biomed. Opt. 2017, 22, 121707. [Google Scholar] [CrossRef] [PubMed]
- Huisken, J.; Stainier, D.Y.R. Even fluorescence excitation by multidirectional selective plane illumination microscopy (mSPIM). Opt. Lett. 2007, 32, 2608–2610. [Google Scholar] [CrossRef] [PubMed]
- Garbellotto, C.; Taylor, J.M. Multi-purpose SLM-light-sheet microscope. Biomed. Opt. Express 2018, 9, 5419–5436. [Google Scholar] [CrossRef] [PubMed]
- Olarte, O.E.; Licea-Rodriguez, J.; Palero, J.A.; Gualda, E.J.; Artigas, D.; Mayer, J.; Swoger, J.; Sharpe, J.; Rocha-Mendoza, I.; Rangel-Rojo, R.; et al. Image formation by linear and nonlinear digital scanned light-sheet fluorescence microscopy with Gaussian and Bessel beam profiles. Biomed. Opt. Express 2012, 3, 1492–1505. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, A.; Mickoleit, M.; Weber, M.; Huisken, J. Multilayer mounting enables long-term imaging of zebrafish development in a light sheet microscope. Development 2012, 139, 3242–3247. [Google Scholar] [CrossRef]
- De Luis Balaguer, M.A.; Ramos-Pezzotti, M.; Rahhal, M.B.; Melvin, C.E.; Johannes, E.; Horn, T.J.; Sozzani, R. Multi-sample Arabidopsis Growth and Imaging Chamber (MAGIC) for long term imaging in the ZEISS Lightsheet, Z.1. Dev. Biol. 2016, 419, 19–25. [Google Scholar] [CrossRef]
- Hecht, E. Optics, 4th ed.; Addison-Wesley: San Francisco, CA, USA, 2010; ISBN 978-0-321-18878-6. [Google Scholar]
- Trisnadi, J.I. Speckle contrast reduction in laser projection displays. Proc. SPIE 4657 2002, 4657, 131–138. [Google Scholar]
- Lin, R.Z.; Lin, R.Z.; Chang, H.Y. Recent advances in three-dimensional multicellular spheroid culture for biomedical research. Biotechnol. J. 2008, 3, 1172–1184. [Google Scholar] [CrossRef] [PubMed]
- Lorenzo, C.; Frongia, C.; Jorand, R.; Fehrenbach, J.; Weiss, P.; Maandhui, A.; Gay, G.; Ducommun, B.; Lobjois, V. Live cell division dynamics monitoring in 3D large spheroid tumor models using light sheet microscopy. Cell Div. 2011, 6, 22. [Google Scholar] [CrossRef] [PubMed]
- Jorand, R.; Le Corre, G.; Andilla, J.; Maandhui, A.; Frongia, C.; Lobjois, V.; Ducommun, B.; Lorenzo, C. Deep and Clear Optical Imaging of Thick Inhomogeneous Samples. PLoS ONE 2012, 7, e35795. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Andilla, J.; Jorand, R.; Olarte, O.E.; Dufour, A.C.; Cazales, M.; Montagner, Y.L.E.; Ceolato, R.; Riviere, N.; Olivo-Marin, J.C.; Loza-Alvarez, P.; et al. Imaging tissue-mimic with light sheet microscopy: A comparative guideline. Sci. Rep. 2017, 7, 44939. [Google Scholar] [CrossRef]
- McGann, L.E.; Walterson, M.L.; Hogg, L.M. Light scattering and cell volumes in osmotically stressed and frozen-thawed cells. Cytometry 1988, 9, 33–38. [Google Scholar] [CrossRef]
- Videen, G.; Ngo, D. Light scattering multipole solution for a cell. J. Biomed. Opt. 1998, 3, 212–220. [Google Scholar] [CrossRef] [PubMed]
- Keller, P.J.; Schmidt, A.D.; Wittbrodt, J.; Stelzer, E.H.K. Reconstruction of zebrafish early embryonic development by scanned light sheet microscopy. Science 2008, 322, 1065–1069. [Google Scholar] [CrossRef]
- Richardson, R.; Tracey-White, D.; Webster, A.; Moosajee, M. The zebrafish eye-a paradigm for investigating human ocular genetics. Eye 2017, 31, 68–86. [Google Scholar] [CrossRef]
- Icha, J.; Schmied, C.; Sidhaye, J.; Tomancak, P.; Preibisch, S.; Norden, C. Using Light Sheet Fluorescence Microscopy to Image Zebrafish Eye Development. J. Vis. Exp. 2016, 1102, e53966. [Google Scholar] [CrossRef]
- Olarte, O.E.; Andilla, J.; Artigas, D.; Loza-Alvarez, P. Decoupled illumination detection in light sheet microscopy for fast volumetric imaging. Optica 2015, 2, 702–705. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Battista, D.; Merino, D.; Zacharakis, G.; Loza-Alvarez, P.; Olarte, O.E. Enhanced Light Sheet Elastic Scattering Microscopy by Using a Supercontinuum Laser. Methods Protoc. 2019, 2, 57. https://doi.org/10.3390/mps2030057
Di Battista D, Merino D, Zacharakis G, Loza-Alvarez P, Olarte OE. Enhanced Light Sheet Elastic Scattering Microscopy by Using a Supercontinuum Laser. Methods and Protocols. 2019; 2(3):57. https://doi.org/10.3390/mps2030057
Chicago/Turabian StyleDi Battista, Diego, David Merino, Giannis Zacharakis, Pablo Loza-Alvarez, and Omar E. Olarte. 2019. "Enhanced Light Sheet Elastic Scattering Microscopy by Using a Supercontinuum Laser" Methods and Protocols 2, no. 3: 57. https://doi.org/10.3390/mps2030057
APA StyleDi Battista, D., Merino, D., Zacharakis, G., Loza-Alvarez, P., & Olarte, O. E. (2019). Enhanced Light Sheet Elastic Scattering Microscopy by Using a Supercontinuum Laser. Methods and Protocols, 2(3), 57. https://doi.org/10.3390/mps2030057






