Using Single-Molecule Chemo-Mechanical Unfolding to Simultaneously Probe Multiple Structural Parameters in Protein Folding
Abstract
:1. Introduction
2. Experimental Design
2.1. Structural Parameters Used in Chemo-Mechanical Unfolding
2.2. Chemo-Mechanical Unfolding using Optical Tweezers
2.3. Collecting and Fitting Chemo-Mechanical Unfolding Data for Folding Thermodynamics
2.4. Collecting and Fitting Chemo-Mechanical Unfolding Data for Folding Kinetics
3. Applications of Chemo-Mechanical Unfolding
3.1. Chemo-Mechanical Analysis of Unfolding Thermodynamics in ACBP to Probe the Denatured State
3.2. Chemo-Mechanical Unfolding Reveals Parallel Unfolding Pathways in the Src SH3 Domain
3.3. Chemo-Mechanical Unfolding Probes the Effect of the Ribosome on Folding Pathways
3.4. Chemo-Mechanical Analysis to Characterize Folding Intermediate in T4-Lysozyme
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jagannathan, B.; Marqusee, S. Protein folding and unfolding under force. Biopolymers 2013, 99, 860–869. [Google Scholar] [CrossRef]
- Bustamante, C.J.; Kaiser, C.M.; Maillard, R.A.; Goldman, D.H.; Wilson, C.A. Mechanisms of cellular proteostasis: Insights from single-molecule approaches. Annu. Rev. Biophys. 2014, 43, 119–140. [Google Scholar] [CrossRef]
- Oberhauser, A.F.; Carrión-Vázquez, M. Mechanical biochemistry of proteins one molecule at a time. J. Biol. Chem. 2008, 283, 6617–6621. [Google Scholar] [CrossRef]
- Woodside, M.T.; Block, S.M. Reconstructing folding energy landscapes by single-molecule force spectroscopy. Annu. Rev. Biophys. 2014, 43, 19–39. [Google Scholar] [CrossRef] [PubMed]
- Street, T.O.; Courtemanche, N.; Barrick, D. Protein folding and stability using denaturants. Methods Cell Biol. 2008, 84, 295–325. [Google Scholar] [PubMed]
- Sosnick, T.R.; Barrick, D. The folding of single domain proteins—Have we reached a consensus? Curr. Opin. Struct. Biol. 2011, 21, 12–24. [Google Scholar] [CrossRef]
- Fersht, A. Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis and Protein Folding; World Scientific: Hackensack, NJ, USA, 2017; 631p. [Google Scholar]
- Dill, K.A.; MacCallum, J.L. The protein-folding problem, 50 years on. Science 2012, 338, 1042–1046. [Google Scholar] [CrossRef] [PubMed]
- Onuchic, J.N.; Wolynes, P.G. Theory of protein folding. Curr. Opin. Struct. Biol. 2004, 14, 70–75. [Google Scholar] [CrossRef]
- Wolynes, P.G. Evolution, energy landscapes and the paradoxes of protein folding. Biochimie 2015, 119, 218–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stigler, J.; Ziegler, F.; Gieseke, A.; Gebhardt, J.C.; Rief, M. The complex folding network of single calmodulin molecules. Science 2011, 334, 512–516. [Google Scholar] [CrossRef] [PubMed]
- Guinn, E.J.; Marqusee, S. Exploring the Denatured State Ensemble by Single-Molecule Chemo-Mechanical Unfolding: The Effect of Force, Temperature, and Urea. J. Mol. Biol 2018, 430, 450–464. [Google Scholar] [CrossRef] [PubMed]
- Guinn, E.J.; Jagannathan, B.; Marqusee, S. Single-molecule chemo-mechanical unfolding reveals multiple transition state barriers in a small single-domain protein. Nat. Commun. 2015, 6, 6861. [Google Scholar] [CrossRef] [Green Version]
- Yu, H.; Liu, X.; Neupane, K.; Gupta, A.N.; Brigley, A.M.; Solanki, A.; Sosova, I.; Woodside, M.T. Direct observation of multiple misfolding pathways in a single prion protein molecule. Proc. Natl. Acad. Sci. USA 2012, 109, 5283–5288. [Google Scholar] [CrossRef] [Green Version]
- Jagannathan, B.; Elms, P.J.; Bustamante, C.; Marqusee, S. Direct observation of a force-induced switch in the anisotropic mechanical unfolding pathway of a protein. Proc. Natl. Acad. Sci. USA 2012, 109, 17820–17825. [Google Scholar] [CrossRef]
- Shank, E.A.; Cecconi, C.; Dill, J.W.; Marqusee, S.; Bustamante, C. The folding cooperativity of a protein is controlled by its chain topology. Nature 2010, 465, 637–640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaiser, C.M.; Goldman, D.H.; Chodera, J.D.; Tinoco, I.; Bustamante, C. The ribosome modulates nascent protein folding. Science 2011, 334, 1723–1727. [Google Scholar] [CrossRef]
- Maillard, R.A.; Chistol, G.; Sen, M.; Righini, M.; Tan, J.; Kaiser, C.M.; Hodges, C.; Martin, A.; Bustamante, C. ClpX(P) generates mechanical force to unfold and translocate its protein substrates. Cell 2011, 145, 459–469. [Google Scholar] [CrossRef]
- Bustamante, C.; Chemla, Y.R.; Forde, N.R.; Izhaky, D. Mechanical processes in biochemistry. Annu. Rev. Biochem. 2004, 73, 705–748. [Google Scholar] [CrossRef]
- Myers, J.K.; Pace, C.N.; Scholtz, J.M. Denaturant m values and heat capacity changes: Relation to changes in accessible surface areas of protein unfolding. Protein Sci. 1995, 4, 2138–2148. [Google Scholar] [CrossRef]
- Guinn, E.J.; Pegram, L.M.; Capp, M.W.; Pollock, M.N.; Record, M.T. Quantifying why urea is a protein denaturant whereas glycine betaine is a protein stabilizer. Proc. Nat. Acad. Sci. USA 2011, 108, 16932. [Google Scholar] [CrossRef] [PubMed]
- Guinn, E.J.; Tian, P.; Shin, M.; Best, R.B.; Marqusee, S. A small single-domain protein folds through the same pathway on and off the ribosome. Proc. Natl. Acad. Sci. USA 2018, 115, 12206–12211. [Google Scholar] [CrossRef]
- Pace, C.N.; Shaw, K.L. Linear extrapolation method of analyzing solvent denaturation curves. Proteins 2000, 41 (Suppl. 4), 1–7. [Google Scholar] [CrossRef]
- Guinn, E.J.; Kontur, W.S.; Tsodikov, O.V.; Shkel, I.; Record, M.T. Probing the protein-folding mechanism using denaturant and temperature effects on rate constants. Proc. Natl. Acad. Sci. USA 2013, 110, 16784–16789. [Google Scholar] [CrossRef] [PubMed]
- Fersht, A.R.; Matouschek, A.; Serrano, L. The folding of an enzyme. I. Theory of protein engineering analysis of stability and pathway of protein folding. J. Mol. Biol. 1992, 224, 771–782. [Google Scholar] [CrossRef]
- Bell, G.I. Models for the specific adhesion of cells to cells. Science 1978, 200, 618–627. [Google Scholar] [CrossRef]
- Dudko, O.K.; Hummer, G.; Szabo, A. Theory, analysis, and interpretation of single-molecule force spectroscopy experiments. Proc. Natl. Acad. Sci. USA 2008, 105, 15755–15760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cecconi, C.; Shank, E.A.; Marqusee, S.; Bustamante, C. DNA molecular handles for single-molecule protein-folding studies by optical tweezers. Methods Mol. Biol. 2011, 749, 255–271. [Google Scholar] [CrossRef]
- Smith, S.B.; Cui, Y.; Bustamante, C. Optical-trap force transducer that operates by direct measurement of light momentum. Methods Enzymol. 2003, 361, 134–162. [Google Scholar]
- Jiao, J.; Rebane, A.A.; Ma, L.; Zhang, Y. Single-Molecule Protein Folding Experiments Using High-Precision Optical Tweezers. Methods Mol. Biol. 2017, 1486, 357–390. [Google Scholar] [CrossRef] [PubMed]
- Min, D.; Arbing, M.A.; Jefferson, R.E.; Bowie, J.U. A simple DNA handle attachment method for single molecule mechanical manipulation experiments. Protein Sci. 2016, 25, 1535–1544. [Google Scholar] [CrossRef] [Green Version]
- Motlagh, H.N.; Toptygin, D.; Kaiser, C.M.; Hilser, V.J. Single-Molecule Chemo-Mechanical Spectroscopy Provides Structural Identity of Folding Intermediates. Biophys. J. 2016, 110, 1280–1290. [Google Scholar] [CrossRef]
- Dudko, O.K. Decoding the mechanical fingerprints of biomolecules. Q. Rev. Biophys. 2016, 49, e3. [Google Scholar] [CrossRef]
- Tinoco, I.; Li, P.T.; Bustamante, C. Determination of thermodynamics and kinetics of RNA reactions by force. Q. Rev. Biophys. 2006, 39, 325–360. [Google Scholar] [CrossRef] [Green Version]
- Chodera, J.D.; Elms, P.J.; Swope, W.C.; Prinz, J.; Marqusee, S.; Bustamante, C.; Now, F.; Pande, V.S. A robust approach to estimating rates from time-correlation functions. arXiv 2011, arXiv:1108.2304. [Google Scholar]
- McKinney, S.A.; Joo, C.; Ha, T. Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophys. J. 2006, 91, 1941–1951. [Google Scholar] [CrossRef] [PubMed]
- Heidarsson, P.O.; Valpapuram, I.; Camilloni, C.; Imparato, A.; Tiana, G.; Poulsen, F.M.; Kragelund, B.B.; Cecconi, C. A highly compliant protein native state with a spontaneous-like mechanical unfolding pathway. J. Am. Chem. Soc. 2012, 134, 17068–17075. [Google Scholar] [CrossRef]
- Zhuravlev, P.I.; Hinczewski, M.; Chakrabarti, S.; Marqusee, S.; Thirumalai, D. Force-dependent switch in protein unfolding pathways and transition-state movements. Proc. Natl. Acad. Sci. USA 2016, 113, E715–724. [Google Scholar] [CrossRef]
- Pierse, C.A.; Dudko, O.K. Distinguishing Signatures of Multipathway Conformational Transitions. Phys. Rev. Lett. 2017, 118, 088101. [Google Scholar] [CrossRef] [PubMed]
- Gabizon, R.; Lee, A.; Vahedian-Movahed, H.; Ebright, R.H.; Bustamante, C.J. Pause sequences facilitate entry into long-lived paused states by reducing RNA polymerase transcription rates. Nat. Commun. 2018, 9, 2930. [Google Scholar] [CrossRef]
- Goldman, D.H.; Kaiser, C.M.; Milin, A.; Righini, M.; Tinoco, I.; Bustamante, C. Mechanical force releases nascent chain-mediated ribosome arrest in vitro and in vivo. Science 2015, 348, 457–460. [Google Scholar] [CrossRef] [Green Version]
- Liu, K.; Rehfus, J.E.; Mattson, E.; Kaiser, C.M. The ribosome destabilizes native and non-native structures in a nascent multidomain protein. Protein Sci. 2017, 26, 1439–1451. [Google Scholar] [CrossRef] [PubMed]
- Record, M.T.J.; Guinn, E.; Pegram, L.; Capp, M. Introductory Lecture: Interpreting and predicting Hofmeister salt ion and solute effects on biopolymer and model processes using the solute partitioning model. Faraday Discuss. 2013, 160, 9–44. [Google Scholar] [CrossRef] [PubMed]
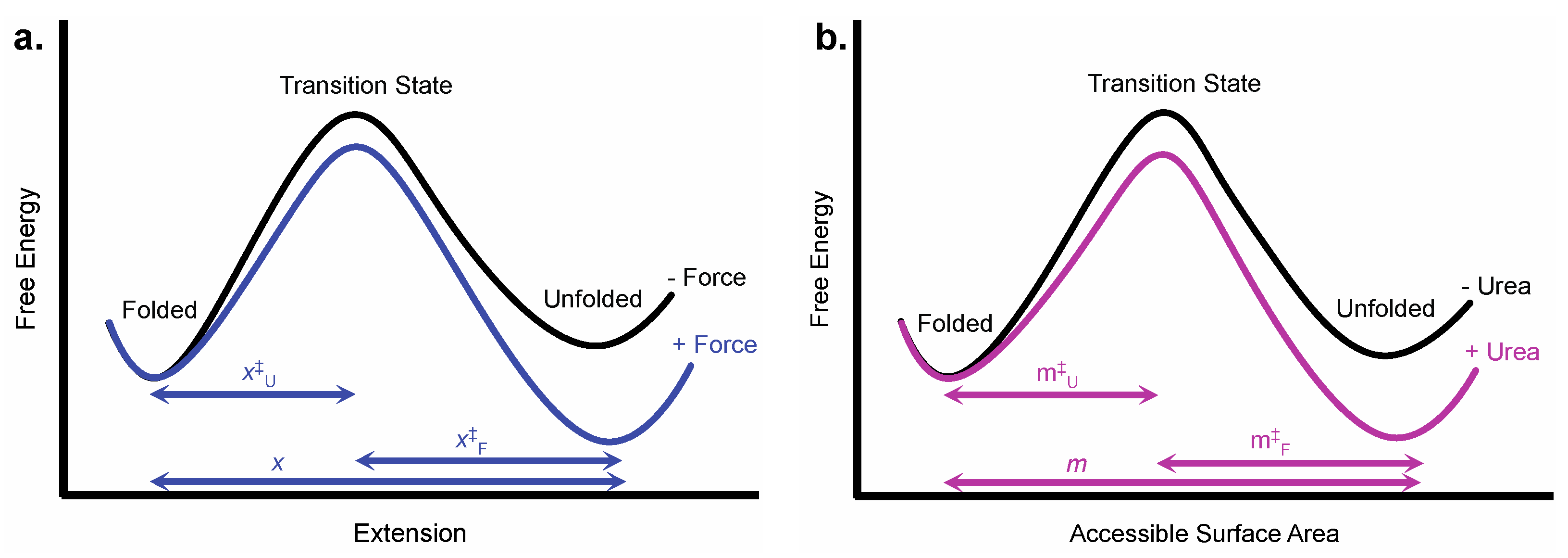
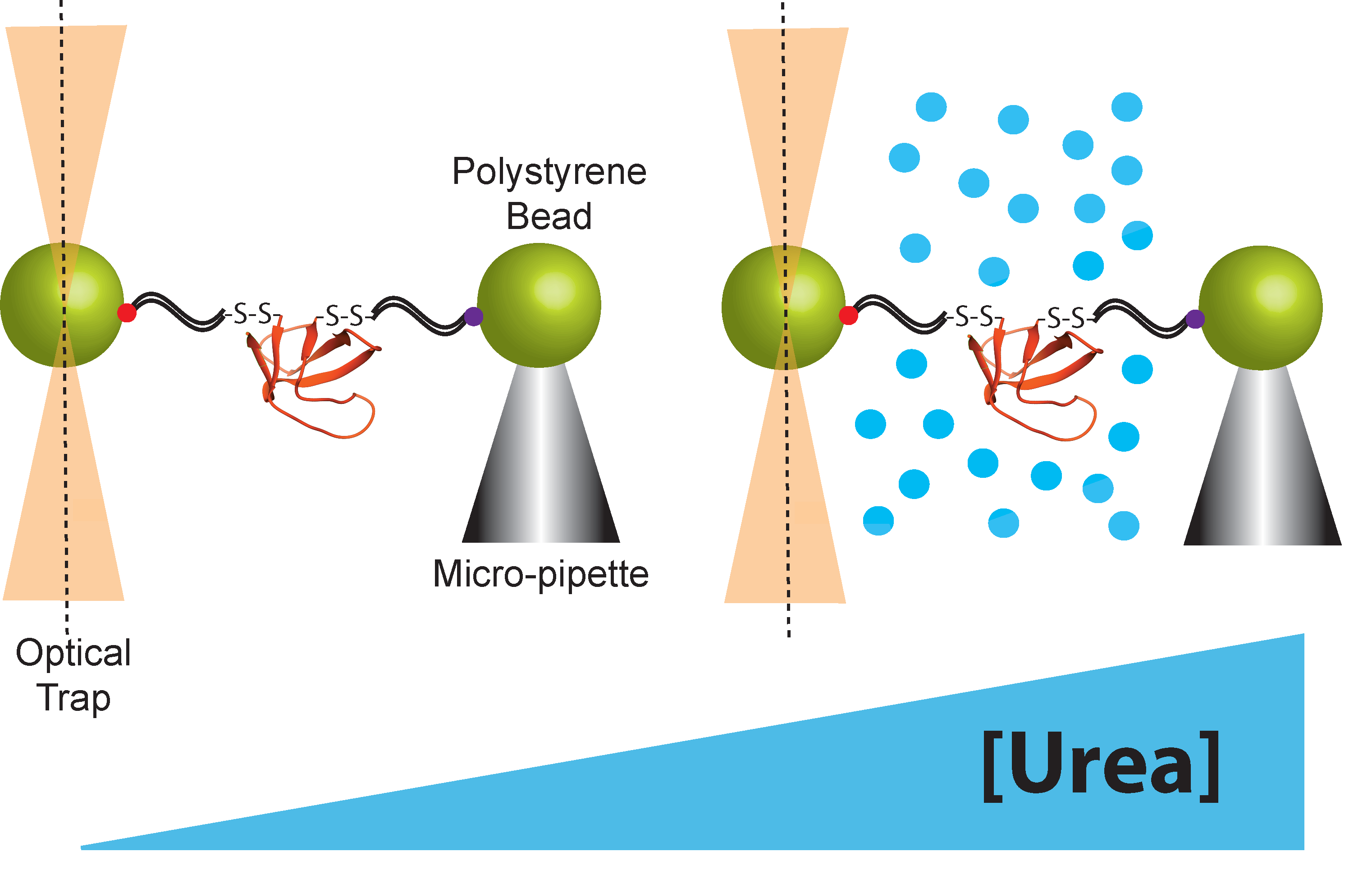
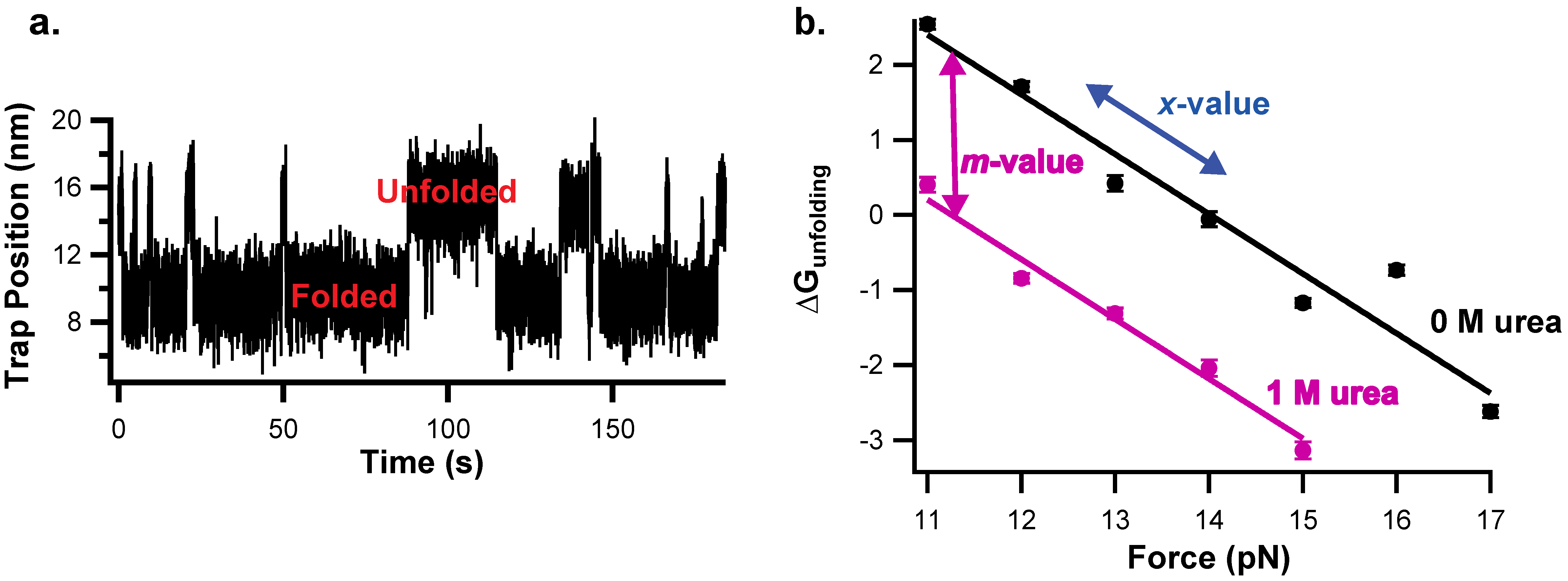
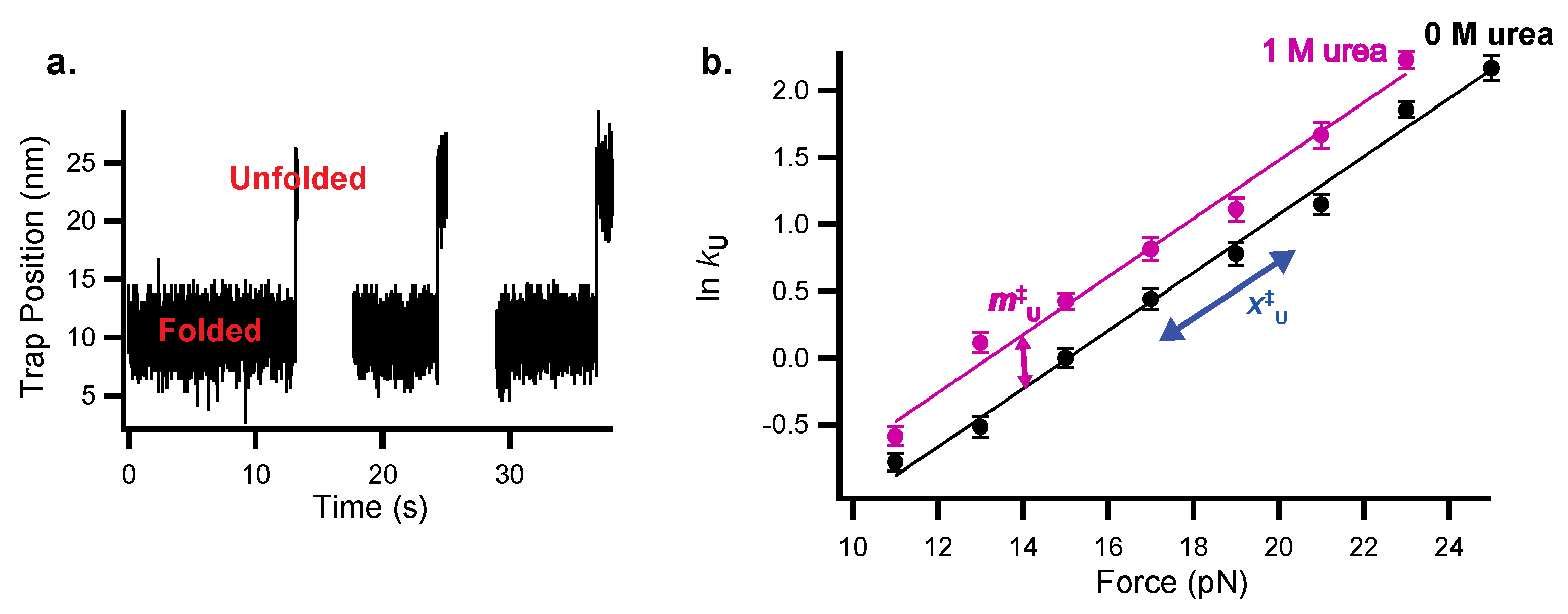
| Parameter | Experimental Origin | Structural Information |
|---|---|---|
| Urea m-value | Urea effect on ΔGunfolding | Change in accessible surface area between folded and unfolded state |
| Urea mU‡-value | Urea effect on unfolding rate constant | Change in accessible surface area between folded state and transition state |
| Urea mF‡-value | Urea effect on folding rate constant | Change in accessible surface area between unfolded state and transition state |
| Force x-value | Force effect on ΔGunfolding | Change in extension between folded and unfolded state |
| Force xU‡-value | Force effect on unfolding rate constant | Change in extension between folded state and transition state |
| Force xF‡-value | Force effect on folding rate constant | Change in extension between unfolded state and transition state |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guinn, E.J.; Marqusee, S. Using Single-Molecule Chemo-Mechanical Unfolding to Simultaneously Probe Multiple Structural Parameters in Protein Folding. Methods Protoc. 2019, 2, 32. https://doi.org/10.3390/mps2020032
Guinn EJ, Marqusee S. Using Single-Molecule Chemo-Mechanical Unfolding to Simultaneously Probe Multiple Structural Parameters in Protein Folding. Methods and Protocols. 2019; 2(2):32. https://doi.org/10.3390/mps2020032
Chicago/Turabian StyleGuinn, Emily J., and Susan Marqusee. 2019. "Using Single-Molecule Chemo-Mechanical Unfolding to Simultaneously Probe Multiple Structural Parameters in Protein Folding" Methods and Protocols 2, no. 2: 32. https://doi.org/10.3390/mps2020032
APA StyleGuinn, E. J., & Marqusee, S. (2019). Using Single-Molecule Chemo-Mechanical Unfolding to Simultaneously Probe Multiple Structural Parameters in Protein Folding. Methods and Protocols, 2(2), 32. https://doi.org/10.3390/mps2020032




