Cell-Free Metabolic Engineering: Recent Developments and Future Prospects
Abstract
1. Introduction
2. Cell-Free Protein Synthesis Systems
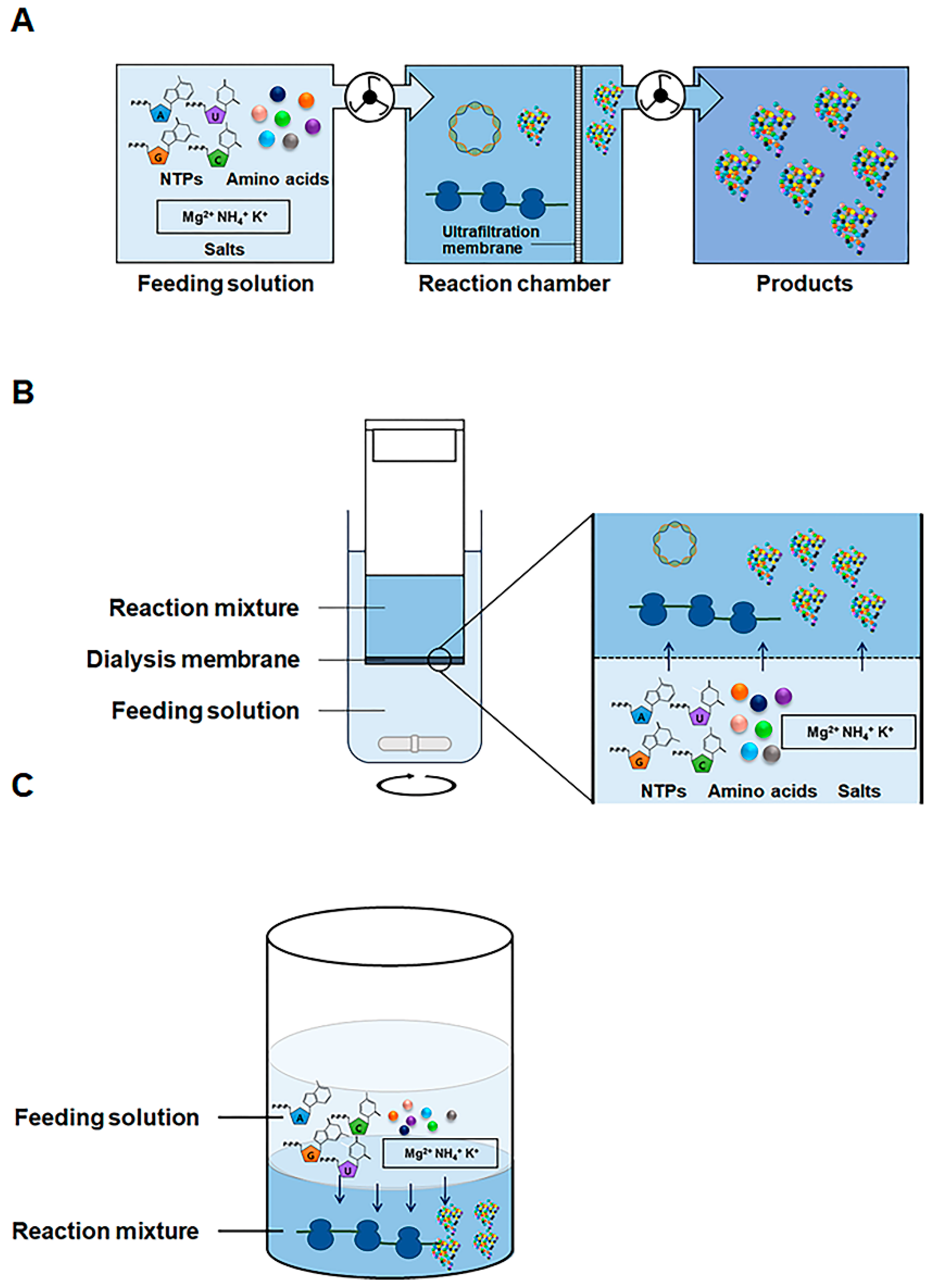
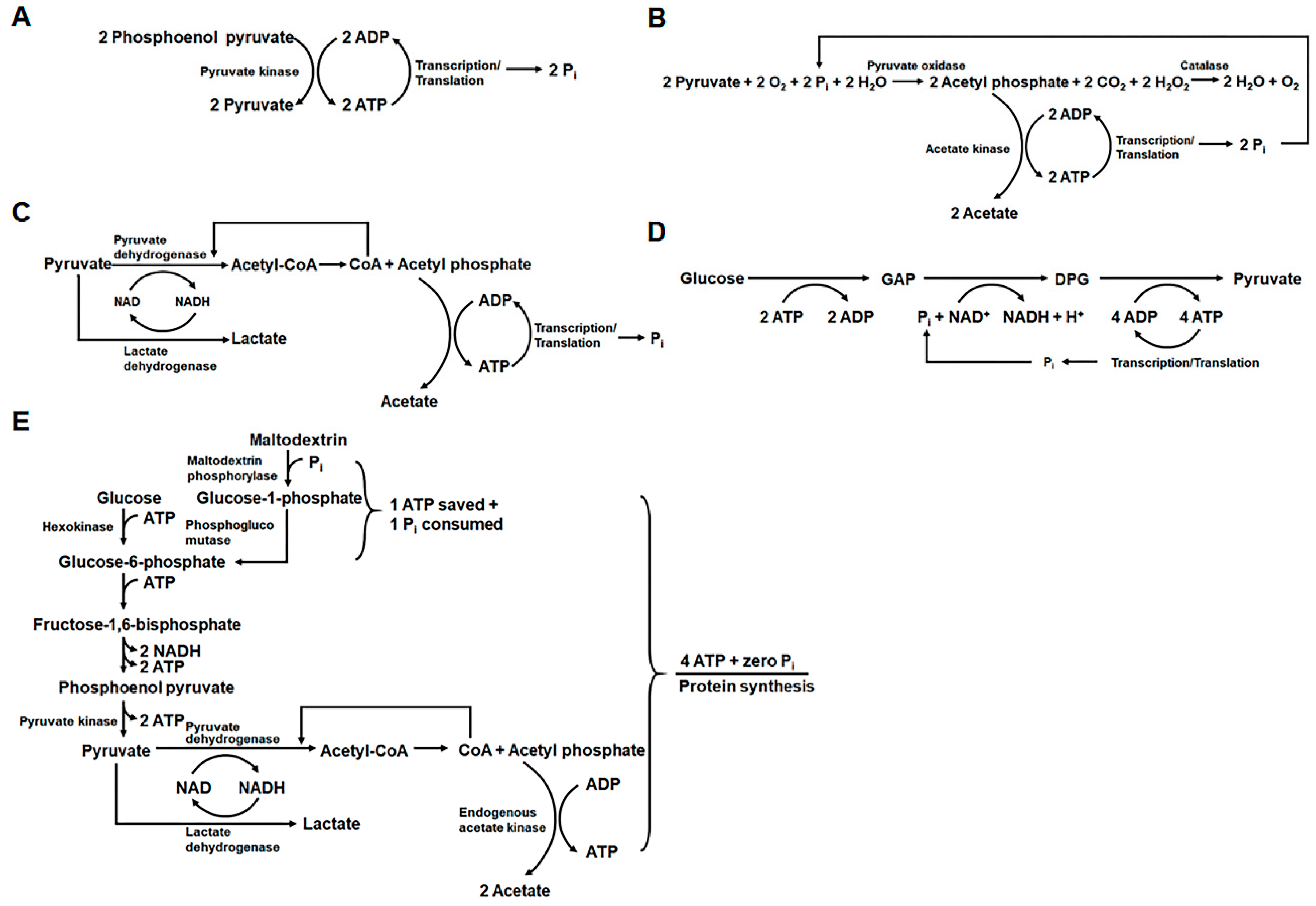
2.1. Development of Highly Productive Cell-Free Protein Synthesis Systems
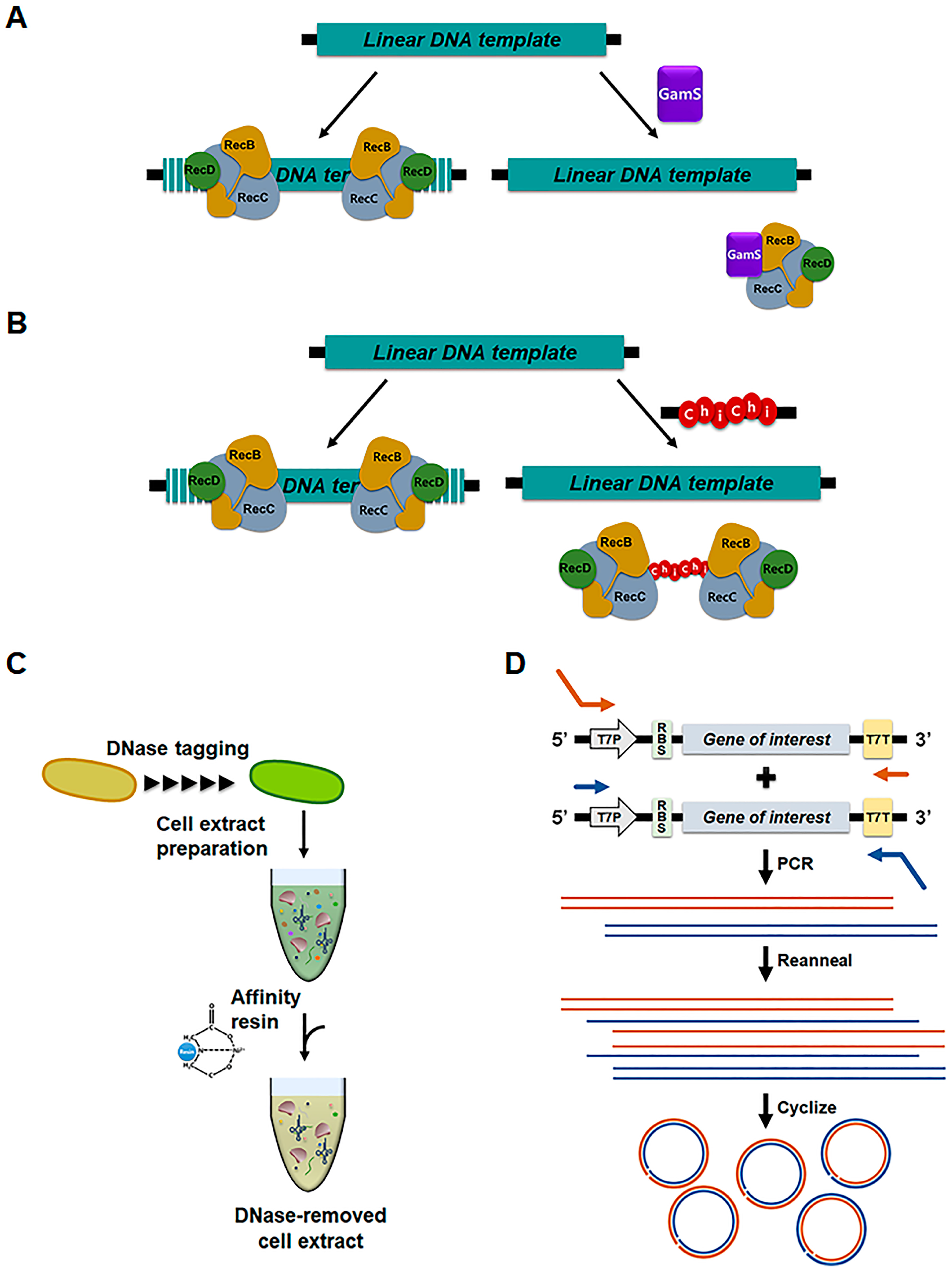
2.2. Direct Programming of Cell-Free Protein Synthesis with Linear DNA Templates
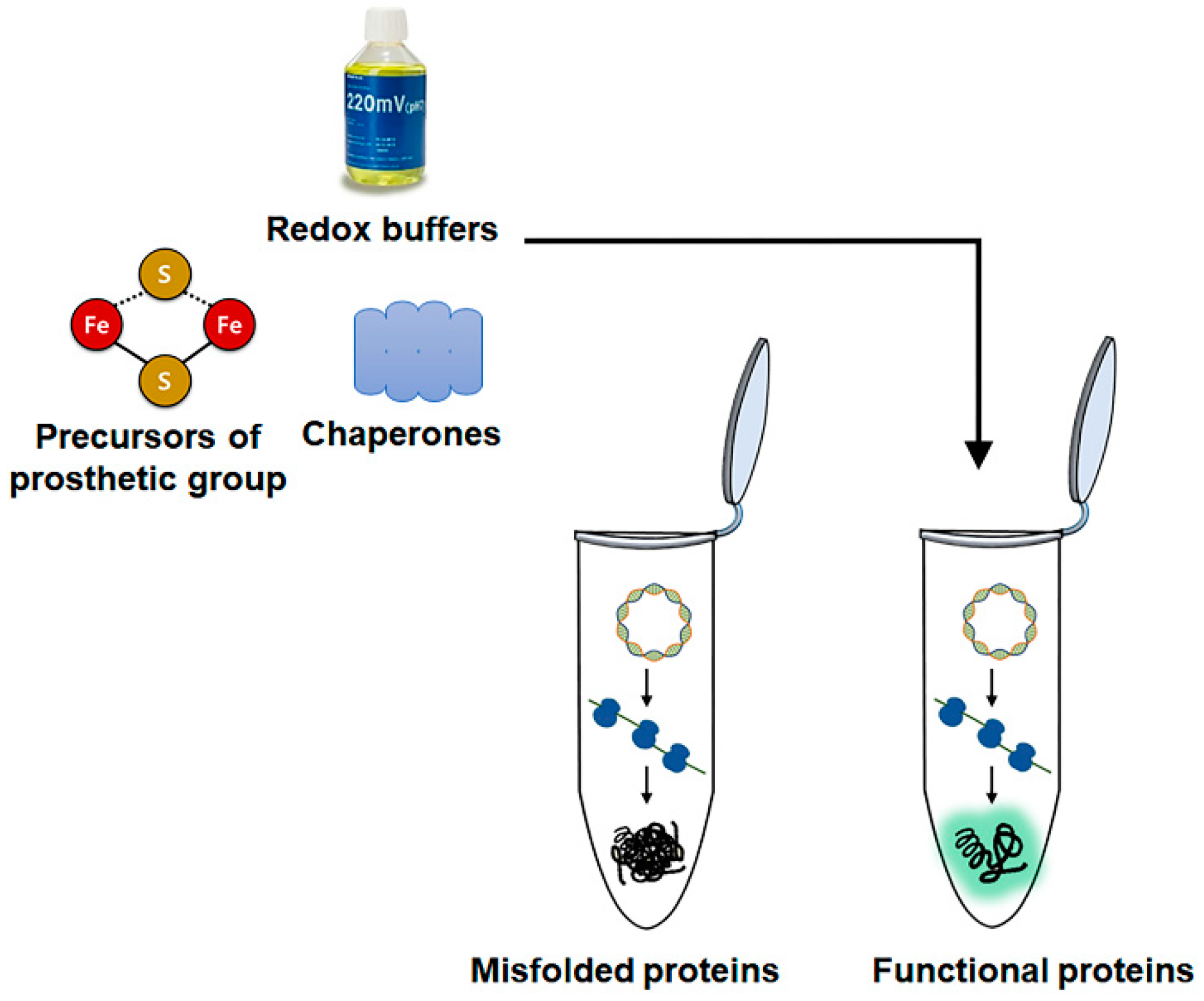
2.3. Cell-Free Enzyme Synthesis
3. Cell-Free Metabolic Engineering
3.1. Purified Protein-Based Cell-Free Metabolic Engineering
3.2. Cell Extract-Based Cell-Free Metabolic Engineering
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- McCarty, N.S.; Ledesma-Amaro, R. Synthetic biology tools to engineer microbial communities for biotechnology. Trends Biotechnol. 2019, 37, 181–197. [Google Scholar] [CrossRef] [PubMed]
- Vervoort, Y.; Linares, A.G.; Roncoroni, M.; Liu, C.; Steensels, J.; Verstrepen, K.J. High-throughput system-wide engineering and screening for microbial biotechnology. Curr. Opin. Biotechnol. 2017, 46, 120–125. [Google Scholar] [CrossRef]
- Dudley, Q.M.; Karim, A.S.; Jewett, M.C. Cell-free metabolic engineering: Biomanufacturing beyond the cell. Biotechnol. J. 2015, 10, 69–82. [Google Scholar] [CrossRef]
- Morley, A.A. Digital PCR: A brief history. Biomol. Detect. Quantif. 2014, 1, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, Y.; Inoue, A.; Tomari, Y.; Suzuki, T.; Yokogawa, T.; Nishikawa, K.; Ueda, T. Cell-free translation reconstituted with purified components. Nat. Biotechnol. 2001, 19, 751–755. [Google Scholar] [CrossRef]
- Kuruma, Y.; Ueda, T. The PURE system for the cell-free synthesis of membrane proteins. Nat. Protoc. 2015, 10, 1328–1344. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Gu, L.; Aach, J.A.; Church, G.M. Improved cell-free RNA and protein synthesis system. PLoS ONE 2014, 9, e106232. [Google Scholar] [CrossRef]
- Carlson, E.D.; Gan, R.; Hodgman, C.E.; Jewett, M.C. Cell-free protein synthesis: Applications come of age. Biotechnol. Adv. 2012, 30, 1185–1194. [Google Scholar] [CrossRef]
- Ahn, J.H.; Hwang, M.Y.; Lee, K.H.; Choi, C.Y.; Kim, D.M. Use of signal sequences as an in situ removable sequence element to stimulate protein synthesis in cell-free extracts. Nucleic Acids Res. 2007, 35, e21. [Google Scholar] [CrossRef]
- Korman, T.P.; Opgenorth, P.H.; Bowie, J.U. A synthetic biochemistry platform for cell free production of monoterpenes from glucose. Nat. Commun. 2017, 8, 15526. [Google Scholar] [CrossRef] [PubMed]
- Kay, J.E.; Jewett, M.C. Lysate of engineered Escherichia coli supports high-level conversion of glucose to 2,3-butanediol. Metab. Eng. 2015, 32, 133–142. [Google Scholar] [CrossRef]
- Buchner, E. Alkoholische gärung ohne hefezellen. Ber. Chem. Ges. 1897, 30, 117–124. [Google Scholar] [CrossRef]
- Dopp, B.J.L.; Tamiev, D.D.; Reuel, N.F. Cell-free supplement mixtures: Elucidating the history and biochemical utility of additives used to support in vitro protein synthesis in E. coli extract. Biotechnol. Adv. 2019, 37, 246–258. [Google Scholar] [CrossRef] [PubMed]
- Jewett, M.C.; Swartz, J.R. Mimicking the Escherichia coli cytoplasmic environment activates long-lived and efficient cell-free protein synthesis. Biotechnol. Bioeng. 2004, 88, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.C.; Kim, D.M. Methods for energizing cell-free protein synthesis. J. Biosci. Bioeng. 2009, 1, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.M.; Swartz, J.R. Prolonging cell-free protein synthesis with a novel ATP regeneration system. Biotechnol. Bioeng. 1999, 66, 180–188. [Google Scholar] [CrossRef]
- Kim, D.M.; Choi, C.Y. A semicontinuous prokaryotic coupled transcription/translation system using a dialysis membrane. Biotechnol. Prog. 1996, 12, 645–649. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.W.; Kim, D.M.; Choi, C.Y. Rapid production of milligram quantities of proteins in a batch cell-free protein synthesis system. J. Biotechnol. 2006, 373–380. [Google Scholar] [CrossRef] [PubMed]
- Whittaker, J.W. Cell-free protein synthesis: The state of the art. Biotechnol. Lett. 2013, 35, 143–152. [Google Scholar] [CrossRef]
- Spirin, A.S.; Baranov, V.I.; Ryabova, L.A.; Ovodov, S.Y.; Alakhov, Y.B. A continuous cell-free translation system capable of producing polypeptides in high yield. Science 1988, 242, 1162–1164. [Google Scholar] [CrossRef]
- Endo, Y.; Otsuzuki, S.; Ito, K.; Miura, K.-I. Production of an enzymatic active protein using a continuous flow cell-free translation system. J. Biotechnol. 1992, 25, 221–230. [Google Scholar] [CrossRef]
- Chen, H.; Xu, Z.; Xu, N.; Cen, P. Efficient production of a soluble fusion protein containing human beta-defensin-2 in E. coli cell-free system. J. Biotechnol. 2005, 115, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Go, S.Y.; Lee, K.H.; Kim, D.M. Detergent-assisted enhancement of the translation rate during cell-free synthesis of peptides in an Escherichia coli extract. Biotechnol. Bioprocess Eng. 2019, 23, 679–685. [Google Scholar] [CrossRef]
- Spirin, A.S. High-throughput cell-free systems for synthesis of functionally active proteins. Trends Biotechnol. 2004, 22, 538–545. [Google Scholar] [CrossRef]
- Kim, D.M.; Swartz, J.R. Regeneration of adenosine triphosphate from glycolytic intermediates for cell-free protein synthesis. Biotechnol. Bioeng. 2001, 74, 309–316. [Google Scholar] [CrossRef]
- Calhoun, K.A.; Swartz, J.R. Energizing cell-free protein synthesis with glucose metabolism. Biotechnol. Bioeng. 2005, 90, 606–613. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Y.-H.P. Cell-free protein synthesis energized by slowly-metabolized maltodextrin. BMC Biotechnol. 2009, 9, 58. [Google Scholar] [CrossRef]
- Kim, H.C.; Kim, T.W.; Kim, D.M. Prolonged production of proteins in a cell-free protein synthesis system using polymeric carbohydrates as an energy source. Process Biochem. 2011, 46, 1366–1369. [Google Scholar] [CrossRef]
- Caschera, F.; Noireaux, V. A cost-effective polyphosphate-based metabolism fuels an all E. coli cell-free expression system. Metab. Eng. 2015, 27, 29–37. [Google Scholar] [CrossRef]
- Lesley, S.A.; Brow, M.A.; Burgess, R.R. Use of in vitro protein synthesis from polymerase chain reaction-generated templates to study interaction of Escherichia coli transcription factors with core RNA polymerase and for epitope mapping of monoclonal antibodies. J. Biol. Chem. 1991, 266, 2632–2638. [Google Scholar]
- Sitaraman, K.; Esposito, D.; Klarmann, G.; Le Grice, S.F.; Hartley, J.L.; Chatterjee, D.K. A novel cell-free protein synthesis system. J. Biotechnol. 2004, 110, 257–263. [Google Scholar] [CrossRef]
- Marshall, R.; Maxwell, C.S.; Collins, S.P.; Beisel, C.L.; Noireaux, V. Short DNA containing χ sites enhances DNA stability and gene expression in E. coli cell-free transcription-translation systems. Biotechnol. Bioeng. 2017, 114, 2137–2141. [Google Scholar] [CrossRef]
- Seki, E.; Matsuda, N.; Kigawa, T. Multiple inhibitory factor removal from an Escherichia coli cell extract improves cell-free protein synthesis. J. Biosci. Bioeng. 2009, 108, 30–35. [Google Scholar] [CrossRef]
- Wu, P.S.C.; Ozawa, K.; Lim, S.P.; Vasudevan, S.G.; Dixon, N.E.; Otting, G. Cell-free transcription/translation from PCR-amplified DNA for high-throughput NMR studies. Angew Chem. Int. Ed. 2007, 46, 3356–3358. [Google Scholar] [CrossRef]
- Ahn, J.H.; Chu, H.-S.; Kim, T.W.; Oh, I.S.; Choi, C.Y.; Hahn, G.H.; Park, C.G.; Kim, D.M. Cell-free synthesis of recombinant proteins from PCR-amplified genes at a comparable productivity to that of plasmid-based reactions. Biochem. Biophys. Res. Commun. 2005, 338, 1346–1352. [Google Scholar] [CrossRef]
- Hunt, J.P.; Yang, S.O.; Wilding, K.M.; Bundy, B.C. The growing impact of lyophilized cell-free expression systems. Bioengineered 2017, 8, 325–330. [Google Scholar] [CrossRef]
- Park, C.G.; Kim, T.W.; Oh, I.S.; Song, J.K.; Kim, D.M. Expression of functional Candida antarctica Lipase B in a cell-free protein synthesis system derived from Escherichia coli. Biotechnol. Prog. 2009, 25, 589–593. [Google Scholar] [CrossRef]
- Park, C.G.; Kwon, M.A.; Song, J.K.; Kim, D.M. Cell-free synthesis and multifold screening of Candida antarctica Lipase B variants after combinatorial mutagenesis of hot spots. Biotechnol. Prog. 2011, 27, 47–53. [Google Scholar] [CrossRef]
- Wu, J.C.Y.; Hutchings, C.H.; Lindsay, M.J.; Werner, C.J.; Bundy, B.C. Enhanced enzyme stability through site-directed covalent immobilization. J. Biotechnol. 2015, 193, 83–90. [Google Scholar] [CrossRef]
- Boyer, M.E.; Stapleton, J.A.; Kuchenreuther, J.M.; Wang, C.W.; Swartz, J.R. Cell-free synthesis and maturation of [FeFe] hydrogenases. Biotechnol. Bioeng. 2008, 99, 59–67. [Google Scholar] [CrossRef]
- Li, J.; Lawton, T.J.; Kostecki, J.S.; Nisthal, A.; Fang, J.; Mayo, S.L.; Rosenzweig, A.C.; Jewett, M.C. Cell-free protein synthesis enables high yielding synthesis of an active multicopper oxidase. Biotechnol. J. 2016, 11, 212–218. [Google Scholar] [CrossRef]
- Kwon, Y.C.; Oh, I.S.; Lee, N.; Lee, K.H.; Yoon, Y.J.; Lee, E.Y.; Kim, B.G.; Kim, D.M. Integrating cell-free biosynthesis of heme prosthetic group and apoenzyme for the synthesis of functional P450 monooxygenase. Biotechnol. Bioeng. 2013, 110, 1193–1200. [Google Scholar] [CrossRef]
- Lim, H.J.; Park, Y.J.; Jang, Y.J.; Choi, J.E.; Oh, J.Y.; Park, J.H.; Song, J.K.; Kim, D.M. Cell-free synthesis of functional phospholipase A1 from Serratia sp. Biotechnol. Biofuels 2016, 9, 159. [Google Scholar] [CrossRef]
- Price, P.B.; Sowers, T. Temperature dependence of metabolic rates for microbial growth, maintenance and survival. Proc. Natl. Acad. Sci. USA 2004, 10, 4631–4636. [Google Scholar] [CrossRef]
- Chubukov, V.; Mukhopadhyay, A.; Petzold, C.J.; Keasling, J.D.; Martín, H.G. Synthetic and systems biology for microbial production of commodity chemicals. NPJ Syst. Biol. Appl. 2016, 2, 16009. [Google Scholar] [CrossRef]
- Bujara, M.; Schümperli, M.; Pellaux, R.; Heinemann, M.; Panke, S. Optimization of a blueprint for in vitro glycolysis by metabolic real-time analysis. Nat. Chem. Biol. 2011, 7, 271–277. [Google Scholar] [CrossRef]
- Guterl, J.K.; Garbe, D.; Carsten, J.; Steffler, F.; Sommer, B.; Reiße, S.; Philipp, A.; Haack, M.; Rühmann, B.; Koltermann, A.; et al. Cell-free metabolic engineering: Production of chemicals by minimized reaction cascades. ChemSusChem 2012, 5, 2165–2172. [Google Scholar] [CrossRef]
- Martín del Campo, J.S.; Rollin, J.; Myung, S.; Chun, Y.; Chandrayan, S.; Patiño, R.; Adams, M.W.; Zhang, Y.-H.P. High-yield production of dihydrogen from xylose by using a synthetic enzyme cascade in a cell-free system. Angew. Chem. Int. Ed. Engl. 2013, 52, 4587–4590. [Google Scholar]
- Zhu, Z.; Tam, T.K.; Sun, F.; You, C.; Zhang, Y.-H.P. A high-energy-density sugar biobattery based on a synthetic enzymatic pathway. Nat. Commun. 2014, 5, 3026. [Google Scholar] [CrossRef]
- Dudley, Q.M.; Nash, C.J.; Jewett, M.C. Cell-free biosynthesis of limonene using enzyme-enriched Escherichia coli lysates. Synth. Biol. 2019, 4, ysz003. [Google Scholar] [CrossRef]
- Karim, A.S.; Jewett, M.C. Cell-free synthetic biology for pathway prototyping. Methods Enzymol. 2018, 608, 31–57. [Google Scholar]
- Casini, A.; Chang, F.Y.; Eluere, R.; King, A.M.; Young, E.M.; Dudley, Q.M.; Karim, A.; Pratt, K.; Bristol, C.; Foget, A.; et al. A pressure test to make 10 molecules in 90 days: External evaluation of methods to engineer biology. J. Am. Chem. Soc. 2018, 140, 4302–4316. [Google Scholar] [CrossRef]
- Karim, A.S.; Heggestad, J.T.; Crowe, S.A.; Jewett, M.C. Controlling cell-free metabolism through physiochemical perturbations. Metab. Eng. 2018, 45, 86–94. [Google Scholar] [CrossRef]
- Bujara, M.; Schümperli, M.; Billerbeck, S.; Heinemann, M.; Panke, S. Exploiting cell-free systems: Implementation and debugging of a system of biotransformations. Biotechnol. Bioeng. 2010, 106, 376–389. [Google Scholar] [CrossRef]
- Yi, T.; Lim, H.J.; Lee, S.J.; Lee, K.H.; Kim, D.M. Synthesis of (R,R)-2,3-butanediol from starch in a hybrid cell-free reaction system. J. Ind. Eng. Chem. 2018, 67, 231–235. [Google Scholar] [CrossRef]
- Karim, A.S.; Jewett, M.C. A cell-free framework for rapid biosynthetic pathway prototyping and enzyme discovery. Metab. Eng. 2016, 36, 116–126. [Google Scholar] [CrossRef]
- Lim, H.J.; Lee, K.H.; Kim, D.M. Rapid determination of effective folding agents by sequential cell-free protein synthesis. Biochem. Eng. J. 2018, 138, 106–110. [Google Scholar] [CrossRef]
- Ahn, J.H.; Keum, J.W.; Kim, D.M. High-throughput, combinatorial engineering of initial codons for tunable expression of recombinant proteins. J. Proteome Res. 2008, 7, 2107–2113. [Google Scholar] [CrossRef]
- Park, Y.J.; Lee, K.H.; Baek, M.S.; Kim, D.M. High-throughput engineering of initial coding regions for maximized production of recombinant proteins. Biotechnol. Bioprocess Eng. 2017, 22, 497–503. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lim, H.J.; Kim, D.-M. Cell-Free Metabolic Engineering: Recent Developments and Future Prospects. Methods Protoc. 2019, 2, 33. https://doi.org/10.3390/mps2020033
Lim HJ, Kim D-M. Cell-Free Metabolic Engineering: Recent Developments and Future Prospects. Methods and Protocols. 2019; 2(2):33. https://doi.org/10.3390/mps2020033
Chicago/Turabian StyleLim, Hye Jin, and Dong-Myung Kim. 2019. "Cell-Free Metabolic Engineering: Recent Developments and Future Prospects" Methods and Protocols 2, no. 2: 33. https://doi.org/10.3390/mps2020033
APA StyleLim, H. J., & Kim, D.-M. (2019). Cell-Free Metabolic Engineering: Recent Developments and Future Prospects. Methods and Protocols, 2(2), 33. https://doi.org/10.3390/mps2020033



