Dedicated Cone-Beam Breast CT: Reproducibility of Volumetric Glandular Fraction with Advanced Image Reconstruction Methods
Abstract
1. Introduction
2. Materials and Methods
2.1. Human Subjects
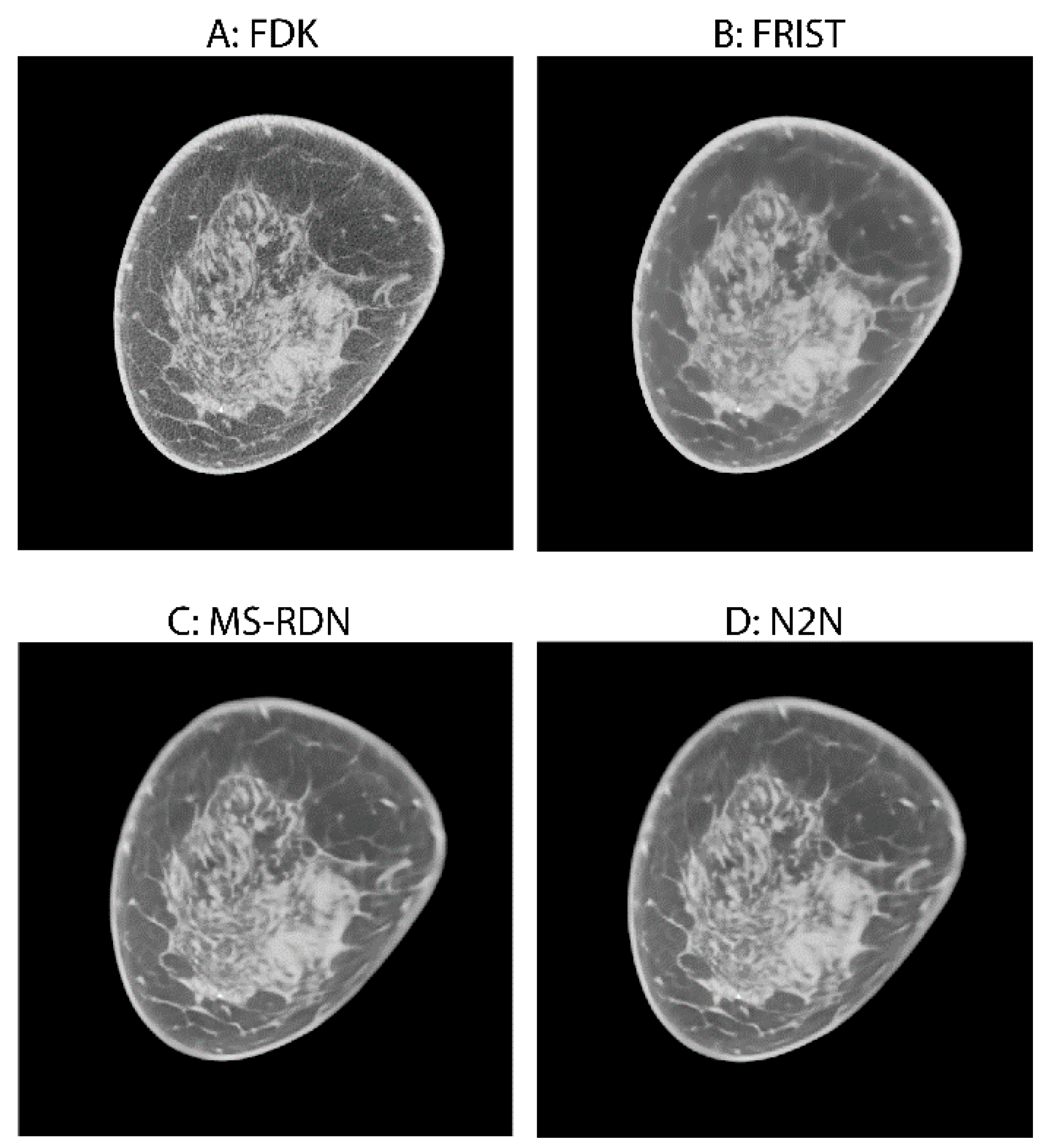
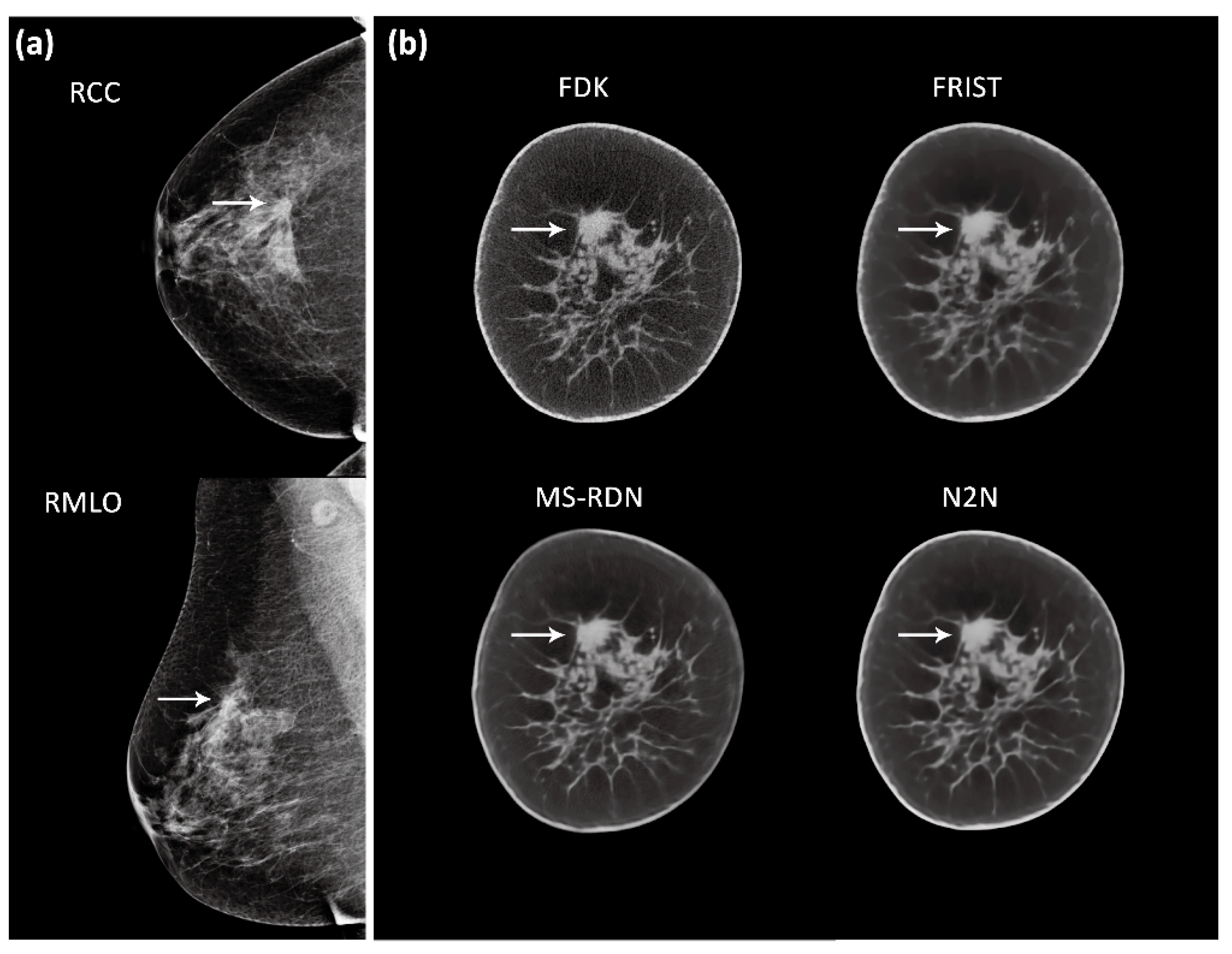
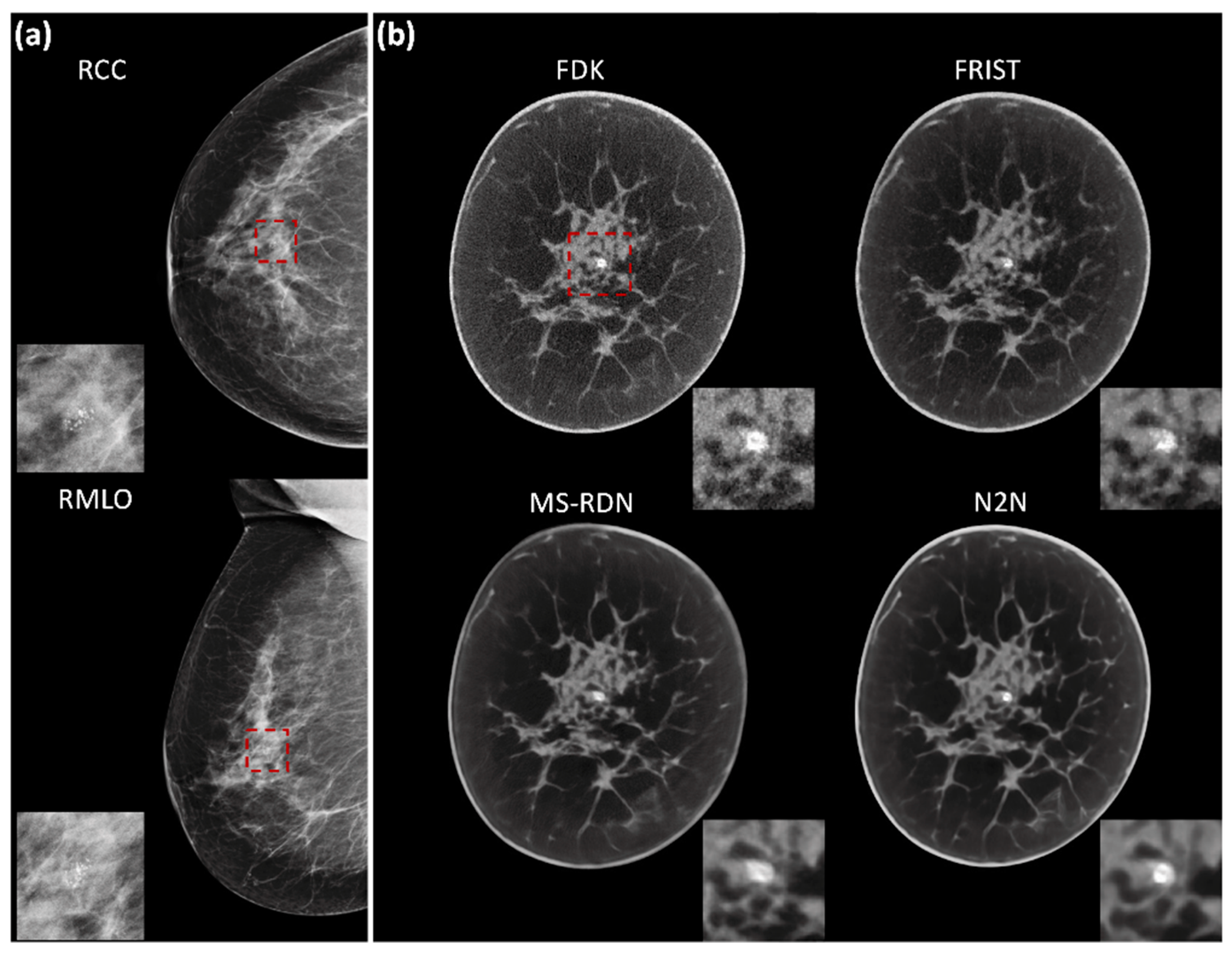
2.2. Image Reconstruction
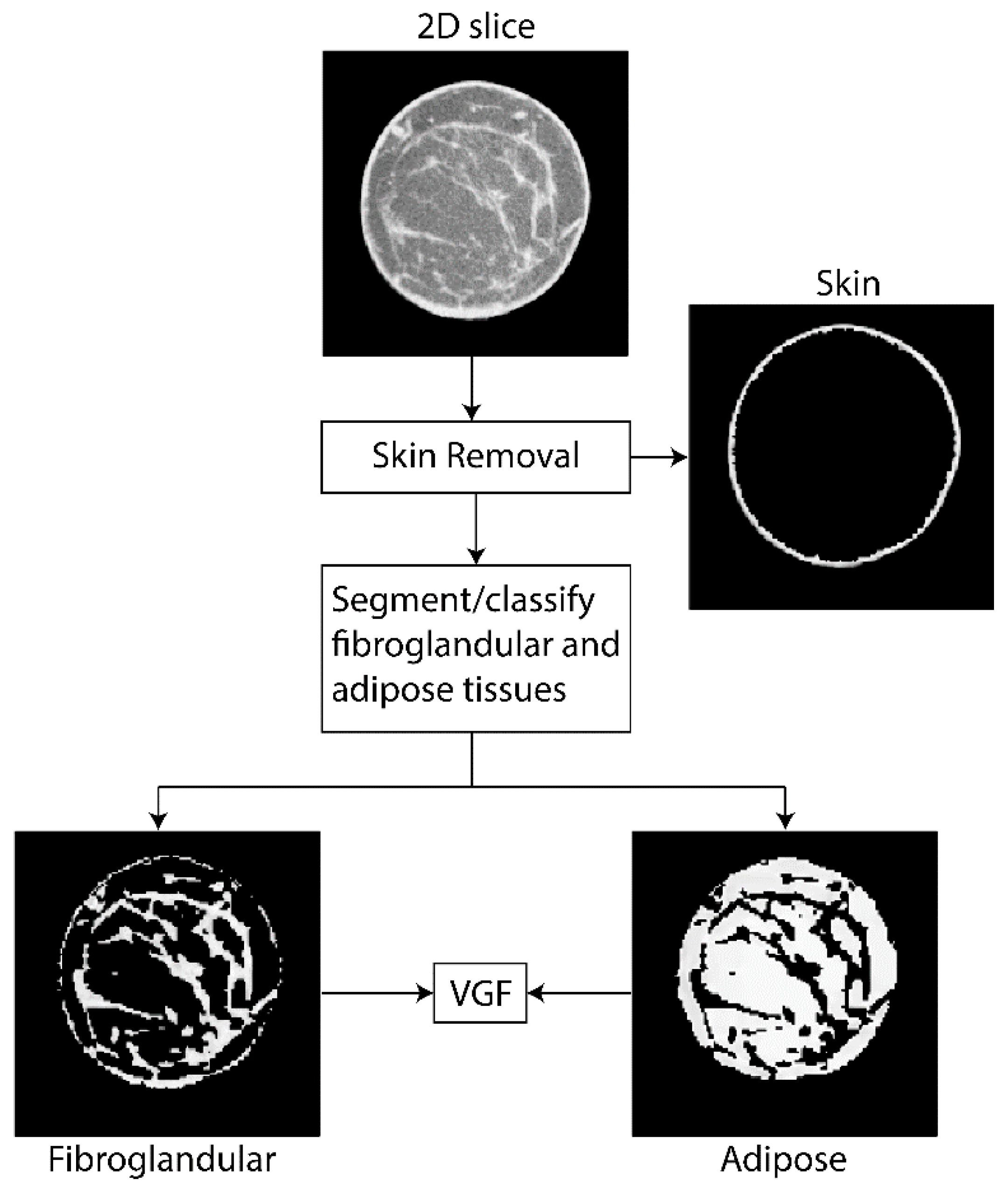
2.3. VGF Computation
2.4. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tabar, L.; Vitak, B.; Chen, H.H.; Yen, M.F.; Duffy, S.W.; Smith, R.A. Beyond randomized controlled trials: Organized mammographic screening substantially reduces breast carcinoma mortality. Cancer 2001, 91, 1724–1731. [Google Scholar] [CrossRef] [PubMed]
- Berry, D.A.; Cronin, K.A.; Plevritis, S.K.; Fryback, D.G.; Clarke, L.; Zelen, M.; Mandelblatt, J.S.; Yakovlev, A.Y.; Habbema, J.D.F.; Feuer, E.J. Effect of Screening and Adjuvant Therapy on Mortality from Breast Cancer. N. Engl. J. Med. 2005, 353, 1784–1792. [Google Scholar] [CrossRef] [PubMed]
- Pisano, E.D.; Gatsonis, C.; Hendrick, E.; Yaffe, M.; Baum, J.K.; Acharyya, S.; Conant, E.F.; Fajardo, L.L.; Bassett, L.; D’Orsi, C.; et al. Diagnostic Performance of Digital versus Film Mammography for Breast-Cancer Screening. N. Engl. J. Med. 2005, 353, 1773–1783. [Google Scholar] [CrossRef] [PubMed]
- Kolb, T.M.; Lichy, J.; Newhouse, J.H. Occult cancer in women with dense breasts: Detection with screening US—Diagnostic yield and tumor characteristics. Radiology 1998, 207, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Kelly, K.M.; Dean, J.; Comulada, W.S.; Lee, S.-J. Breast cancer detection using automated whole breast ultrasound and mammography in radiographically dense breasts. Eur. Radiol. 2009, 20, 734–742. [Google Scholar] [CrossRef]
- Duric, N.; Littrup, P.; Poulo, L.; Babkin, A.; Pevzner, R.; Holsapple, E.; Rama, O.; Glide, C. Detection of breast cancer with ultrasound tomography: First results with the Computed Ultrasound Risk Evaluation (CURE) prototype. Med. Phys. 2007, 34, 773–785. [Google Scholar] [CrossRef]
- Berg, W.A.; Blume, J.D.; Cormack, J.B.; Mendelson, E.B.; Lehrer, D.; Böhm-Vélez, M.; Pisano, E.D.; Jong, R.A.; Evans, W.P.; Morton, M.J.; et al. Combined Screening With Ultrasound and Mammography vs. Mammography Alone in Women at Elevated Risk of Breast Cancer. JAMA 2008, 299, 2151–2163. [Google Scholar] [CrossRef]
- Niklason, L.T.; Christian, B.T.; Niklason, L.E.; Kopans, D.B.; Castleberry, D.E.; Opsahl-Ong, B.H.; Landberg, C.E.; Slanetz, P.J.; Giardino, A.A.; Moore, R.; et al. Digital tomosynthesis in breast imaging. Radiology 1997, 205, 399–406. [Google Scholar] [CrossRef]
- Friedewald, S.M.; Rafferty, E.A.; Rose, S.L.; Durand, M.A.; Plecha, D.M.; Greenberg, J.S.; Hayes, M.K.; Copit, D.S.; Carlson, K.L.; Cink, T.M.; et al. Breast Cancer Screening Using Tomosynthesis in Combination With Digital Mammography. JAMA 2014, 311, 2499–2507. [Google Scholar] [CrossRef]
- Peters, N.H.; Borel Rinkes, I.H.; Zuithoff, N.P.; Mali, W.P.; Moons, K.G.; Peeters, P.H. Meta-analysis of MR imaging in the diagnosis of breast lesions. Radiology 2008, 246, 116–124. [Google Scholar] [CrossRef]
- Lindfors, K.K.; Boone, J.M.; Nelson, T.R.; Yang, K.; Kwan, A.L.; Miller, D.F. Dedicated breast CT: Initial clinical experience. Radiology 2008, 246, 725–733. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, A.M.; Karellas, A.; Vedantham, S. The Potential Role of Dedicated 3D Breast CT as a Diagnostic Tool: Review and Early Clinical Examples. Breast J. 2014, 20, 592–605. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; O’connell, A.M.; Ma, Y.; Liu, A.; Li, H.; Zhang, Y.; Zhang, X.; Ye, Z. Dedicated breast CT: State of the art—Part I. Historical evolution and technical aspects. Eur. Radiol. 2021, 32, 1579–1589. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.H.; Sibala, J.L.; Fritz, S.L.; Dwyer, S.J.; Templeton, A.W., 3rd; Lin, F.; Jewell, W.R. Computed tomography in detection and diagnosis of breast cancer. Cancer 1980, 46 (Suppl. 4), 939–946. [Google Scholar] [CrossRef] [PubMed]
- Raptopoulos, V.; Baum, J.K.; Hochman, M.; Karellas, A.; Houlihan, M.-J.; D’Orsi, C.J. High Resolution CT Mammography of Surgical Biopsy Specimens. J. Comput. Assist. Tomogr. 1996, 20, 179–184. [Google Scholar] [CrossRef]
- Boone, J.M.; Nelson, T.R.; Lindfors, K.K.; Seibert, J.A. Dedicated Breast CT: Radiation Dose and Image Quality Evaluation. Radiology 2001, 221, 657–667. [Google Scholar] [CrossRef]
- Geiser, W.R.; Einstein, S.A.; Yang, W.-T. Artifacts in Digital Breast Tomosynthesis. Am. J. Roentgenol. 2018, 211, 926–932. [Google Scholar] [CrossRef]
- O’Connell, A.; Conover, D.L.; Zhang, Y.; Seifert, P.; Logan-Young, W.; Lin, C.-F.L.; Sahler, L.; Ning, R. Cone-Beam CT for Breast Imaging: Radiation Dose, Breast Coverage, and Image Quality. Am. J. Roentgenol. 2010, 195, 496–509. [Google Scholar] [CrossRef]
- Wienbeck, S.; Uhlig, J.; Luftner-Nagel, S.; Zapf, A.; Surov, A.; von Fintel, E.; Stahnke, V.; Lotz, J.; Fischer, U. The role of cone-beam breast-CT for breast cancer detection relative to breast density. Eur. Radiol. 2017, 27, 5185–5195. [Google Scholar] [CrossRef]
- Madhav, P.; Crotty, D.J.; McKinley, R.L.; Tornai, M.P. Evaluation of tilted cone-beam CT orbits in the development of a dedicated hybrid mammotomograph. Phys. Med. Biol. 2009, 54, 3659–3676. [Google Scholar] [CrossRef][Green Version]
- Berger, N.; Marcon, M.; Saltybaeva, N.; Kalender, W.A.; Alkadhi, H.; Frauenfelder, T.; Boss, A. Dedicated Breast Computed Tomography With a Photon-Counting Detector. Investig. Radiol. 2019, 54, 409–418. [Google Scholar] [CrossRef]
- Ghazi, P.; Youssefian, S.; Ghazi, T. A novel hardware duo of beam modulation and shielding to reduce scatter acquisition and dose in cone-beam breast CT. Med. Phys. 2021, 49, 169–185. [Google Scholar] [CrossRef]
- KONING BREAST CT (MODEL CBCT1000): PMA Number P130025 [Internet]. US Food and Drug Administration. 2015. Available online: https://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfpma/pma.cfm?id=P130025 (accessed on 5 May 2023).
- Cole, E.B.; Campbell, A.S.; Vedantham, S.; Pisano, E.D.; Karellas, A. Clinical Performance of Dedicated Breast Computed Tomography in Comparison to Diagnostic Digital Mammography [abstract # SSA01-09]. In Proceedings of the 101st Scientific Assembly and Annual Meeting of the Radiological Society of North America (RSNA 2015), Chicago, IL, USA, 29 November–4 December 2015. [Google Scholar]
- Gazi, P.M.; Yang, K.; Burkett, G.W.; Aminololama-Shakeri, S.; Seibert, J.A.; Boone, J.M. Evolution of spatial resolution in breast CT at UC Davis. Med. Phys. 2015, 42, 1973–1981. [Google Scholar] [CrossRef]
- Tseng, H.W.; Karellas, A.; Vedantham, S. Cone-beam breast CT using an offset detector: Effect of detector offset and image reconstruction algorithm. Phys. Med. Biol. 2022, 67, 085008. [Google Scholar] [CrossRef]
- Tseng, H.W.; Karellas, A.; Vedantham, S. Sparse-view, short-scan, dedicated cone-beam breast computed tomography: Image quality assessment. Biomed. Phys. Eng. Express 2020, 6, 065015. [Google Scholar] [CrossRef] [PubMed]
- Tseng, H.W.; Karellas, A.; Vedantham, S. Dedicated cone-beam breast CT: Data acquisition strategies based on projection angle-dependent normalized glandular dose coefficients. Med. Phys. 2022, 50, 1406–1417. [Google Scholar] [CrossRef] [PubMed]
- Kalender, W.A.; Beister, M.; Boone, J.M.; Kolditz, D.; Vollmar, S.V.; Weigel, M.C.C. High-resolution spiral CT of the breast at very low dose: Concept and feasibility considerations. Eur. Radiol. 2011, 22, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Hammerstein, G.R.; Miller, D.W.; White, D.R.; Masterson, M.E.; Woodard, H.Q.; Laughlin, J.S. Absorbed Radiation Dose in Mammography. Radiology 1979, 130, 485–491. [Google Scholar] [CrossRef] [PubMed]
- FDA US. Mammography Quality Standards Act Regulations. In Sec. 900.12 Quality Standards.(e) Quality Assurance-Equipment (5) Annual Quality Control Tests; U.S. Food and Drug Administration: Rockville, MD, USA, 2002. [Google Scholar]
- Hendrick, R.E.; Pisano, E.D.; Averbukh, A.; Moran, C.; Berns, E.A.; Yaffe, M.J.; Herman, B.; Acharyya, S.; Gatsonis, C. Comparison of Acquisition Parameters and Breast Dose in Digital Mammography and Screen-Film Mammography in the American College of Radiology Imaging Network Digital Mammographic Imaging Screening Trial. Am. J. Roentgenol. 2010, 194, 362–369. [Google Scholar] [CrossRef]
- Svahn, T.; Houssami, N.; Sechopoulos, I.; Mattsson, S. Review of radiation dose estimates in digital breast tomosynthesis relative to those in two-view full-field digital mammography. Breast 2014, 24, 93–99. [Google Scholar] [CrossRef]
- Vedantham, S.; Shi, L.; Karellas, A.; O’Connell, A.M.; Conover, D.L. Personalized estimates of radiation dose from dedicated breast CT in a diagnostic population and comparison with diagnostic mammography. Phys. Med. Biol. 2013, 58, 7921–7936. [Google Scholar] [CrossRef] [PubMed]
- O’connell, A.M.; Kawakyu-O’connor, D. Dedicated Cone-beam Breast Computed Tomography and Diagnostic Mammography: Comparison of Radiation Dose, Patient Comfort, And Qualitative Review of Imaging Findings in BI-RADS 4 and 5 Lesions. J. Clin. Imaging Sci. 2012, 2, 7. [Google Scholar] [CrossRef] [PubMed]
- Di Maria, S.; Vedantham, S.; Vaz, P. Breast dosimetry in alternative X-ray-based imaging modalities used in current clinical practices. Eur. J. Radiol. 2022, 155, 110509. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, A.M.; Becker, A.E.; Lyu, S.H.; Abbey, C.K.; Boone, J.M. High-resolution μCT imaging for characterizing microcalcification detection performance in breast CT. J. Med. Imaging 2021, 8, 052107. [Google Scholar] [CrossRef]
- Boyd, N.F.; Guo, H.; Martin, L.J.; Sun, L.; Stone, J.; Fishell, E.; Jong, R.A.; Hislop, G.; Chiarelli, A.; Minkin, S.; et al. Mammographic Density and the Risk and Detection of Breast Cancer. N. Engl. J. Med. 2007, 356, 227–236. [Google Scholar] [CrossRef] [PubMed]
- McCormack, V.A.; dos Santos Silva, I. Breast Density and Parenchymal Patterns as Markers of Breast Cancer Risk: A Meta-analysis. Cancer Epidemiol. Biomark. Prev. 2006, 15, 1159–1169. [Google Scholar] [CrossRef] [PubMed]
- Warner, E.; Lockwood, G.; Tritchler, D.; Boyd, N.F. The risk of breast cancer associated with mammographic parenchymal patterns: A meta-analysis of the published literature to examine the effect of method of classification. Cancer Detect. Prev. 1992, 16, 67–72. [Google Scholar]
- Alonzo-Proulx, O.; Mawdsley, G.E.; Patrie, J.T.; Yaffe, M.J.; Harvey, J.A. Reliability of automated breast density measurements. Radiology 2015, 275, 366–376. [Google Scholar] [CrossRef]
- Gastounioti, A.; Pantalone, L.; Scott, C.G.; Cohen, E.A.; Wu, F.F.; Winham, S.J.; Jensen, M.R.; Maidment, A.D.A.; Vachon, C.M.; Conant, E.F.; et al. Fully Automated Volumetric Breast Density Estimation from Digital Breast Tomosynthesis. Radiology 2021, 301, 561–568. [Google Scholar] [CrossRef]
- Maghsoudi, O.H.; Gastounioti, A.; Scott, C.; Pantalone, L.; Wu, F.-F.; Cohen, E.A.; Winham, S.; Conant, E.F.; Vachon, C.; Kontos, D. Deep-LIBRA: An artificial-intelligence method for robust quantification of breast density with independent validation in breast cancer risk assessment. Med. Image Anal. 2021, 73, 102138. [Google Scholar] [CrossRef]
- Vedantham, S.; Shi, L.; Michaelsen, K.E.; Krishnaswamy, V.; Pogue, B.W.; Poplack, S.P.; Karellas, A.; Paulsen, K.D. Digital breast tomosynthesis guided near infrared spectroscopy: Volumetric estimates of fibroglandular fraction and breast density from tomosynthesis reconstructions. Biomed. Phys. Eng. Express 2015, 1, 045202. [Google Scholar] [CrossRef] [PubMed]
- Yala, A.; Lehman, C.; Schuster, T.; Portnoi, T.; Barzilay, R. A Deep Learning Mammography-based Model for Improved Breast Cancer Risk Prediction. Radiology 2019, 292, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Vedantham, S.; Shi, L.; Karellas, A.; O’Connell, A.M. Dedicated breast CT: Fibroglandular volume measurements in a diagnostic population. Med. Phys. 2012, 39, 7317–7328. [Google Scholar] [CrossRef] [PubMed]
- Yaffe, M.J.; Boone, J.M.; Packard, N.; Alonzo-Proulx, O.; Huang, S.-Y.; Peressotti, C.L.; Al-Mayah, A.; Brock, K. The myth of the 50-50 breast. Med. Phys. 2009, 36, 5437–5443. [Google Scholar] [CrossRef]
- Feldkamp, L.A.; Davis, L.C.; Kress, J.W. Practical Cone-Beam Algorithm. J. Opt. Soc. Am. Opt. Image Sci. Vis. 1984, 1, 612–619. [Google Scholar] [CrossRef]
- Bian, J.; Yang, K.; Boone, J.M.; Han, X.; Sidky, E.Y.; Pan, X. Investigation of iterative image reconstruction in low-dose breast CT. Phys. Med. Biol. 2014, 59, 2659–2685. [Google Scholar] [CrossRef]
- Tseng, H.W.; Vedantham, S.; Karellas, A. Cone-beam breast computed tomography using ultra-fast image reconstruction with constrained, total-variation minimization for suppression of artifacts. Phys. Med. 2020, 73, 117–124. [Google Scholar] [CrossRef]
- Xie, H.; Shan, H.; Cong, W.; Liu, C.; Zhang, X.; Liu, S.; Ning, R.; Wang, G. Deep Efficient End-to-End Reconstruction (DEER) Network for Few-View Breast CT Image Reconstruction. IEEE Access 2020, 8, 196633–196646. [Google Scholar] [CrossRef]
- Fu, Z.; Tseng, H.W.; Vedantham, S.; Karellas, A.; Bilgin, A. A residual dense network assisted sparse view reconstruction for breast computed tomography. Sci. Rep. 2020, 10, 21111. [Google Scholar] [CrossRef]
- Wu, D.; Kim, K.; Li, Q. Low-dose CT reconstruction with Noise2Noise network and testing-time fine-tuning. Med. Phys. 2021, 48, 7657–7672. [Google Scholar] [CrossRef]
- Sidky, E.Y.; Pan, X. Image reconstruction in circular cone-beam computed tomography by constrained, total-variation minimization. Phys. Med. Biol. 2008, 53, 4777–4807. [Google Scholar] [CrossRef] [PubMed]
- Zbijewski, W.; Beekman, F.J. Characterization and suppression of edge and aliasing artefacts in iterative x-ray CT reconstruction. Phys. Med. Biol. 2003, 49, 145–157. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zhang, Y.; Kalra, M.K.; Lin, F.; Chen, Y.; Liao, P.; Zhou, J.; Wang, G. Low-Dose CT With a Residual Encoder-Decoder Convolutional Neural Network. IEEE Trans. Med. Imaging 2017, 36, 2524–2535. [Google Scholar] [CrossRef]
- Yuan, N.; Zhou, J.; Qi, J. Half2Half: Deep neural network based CT image denoising without independent reference data. Phys. Med. Biol. 2020, 65, 215020. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Vedantham, S.; Karellas, A.; O’Connell, A.M. Technical Note: Skin thickness measurements using high-resolution flat-panel cone-beam dedicated breast CTa). Med. Phys. 2013, 40, 031913. [Google Scholar] [CrossRef]
- Koetzier, L.R.; Mastrodicasa, D.; Szczykutowicz, T.P.; van der Werf, N.R.; Wang, A.S.; Sandfort, V.; van der Molen, A.J.; Fleischmann, D.; Willemink, M.J. Deep Learning Image Reconstruction for CT: Technical Principles and Clinical Prospects. Radiology 2023, 306, e221257. [Google Scholar] [CrossRef]
- Tseng, H.W.; Karellas, A.; Vedantham, S. Radiation dosimetry of a clinical prototype dedicated cone-beam breast CT system with offset detector. Med. Phys. 2021, 48, 1079–1088. [Google Scholar] [CrossRef]

| Diameter of breast at chest-wall | 13.1 ± 2.3 cm |
| Chest-wall to nipple length | 9.7 ± 2.8 cm |
| BI-RADS breast density categories from mammography | |
| A: Almost entirely fatty | 7/104 (7%) |
| B: Scattered areas of fibroglandular density | 39/104 (38%) |
| C: Heterogeneously dense | 35/104 (34%) |
| D: Extremely dense | 23/104 (22%) |
| Reconstruction Method | Median (IQR) [Range] | p-Value |
|---|---|---|
| FDK | 0.186 (0.122, 0.239) [0.04–0.505] | NA |
| FRIST | 0.18 (0.131, 0.235) [0.07–0.417] | 0.936 |
| MS-RDN | 0.187 (0.129, 0.235) [0.043–0.409] | 0.862 |
| N2N | 0.193 (0.133, 0.235) [0.047–0.487] | >0.999 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vedantham, S.; Tseng, H.W.; Fu, Z.; Chow, H.-H.S. Dedicated Cone-Beam Breast CT: Reproducibility of Volumetric Glandular Fraction with Advanced Image Reconstruction Methods. Tomography 2023, 9, 2039-2051. https://doi.org/10.3390/tomography9060160
Vedantham S, Tseng HW, Fu Z, Chow H-HS. Dedicated Cone-Beam Breast CT: Reproducibility of Volumetric Glandular Fraction with Advanced Image Reconstruction Methods. Tomography. 2023; 9(6):2039-2051. https://doi.org/10.3390/tomography9060160
Chicago/Turabian StyleVedantham, Srinivasan, Hsin Wu Tseng, Zhiyang Fu, and Hsiao-Hui Sherry Chow. 2023. "Dedicated Cone-Beam Breast CT: Reproducibility of Volumetric Glandular Fraction with Advanced Image Reconstruction Methods" Tomography 9, no. 6: 2039-2051. https://doi.org/10.3390/tomography9060160
APA StyleVedantham, S., Tseng, H. W., Fu, Z., & Chow, H.-H. S. (2023). Dedicated Cone-Beam Breast CT: Reproducibility of Volumetric Glandular Fraction with Advanced Image Reconstruction Methods. Tomography, 9(6), 2039-2051. https://doi.org/10.3390/tomography9060160







