Artificial Intelligence for Image-Based Breast Cancer Risk Prediction Using Attention
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data
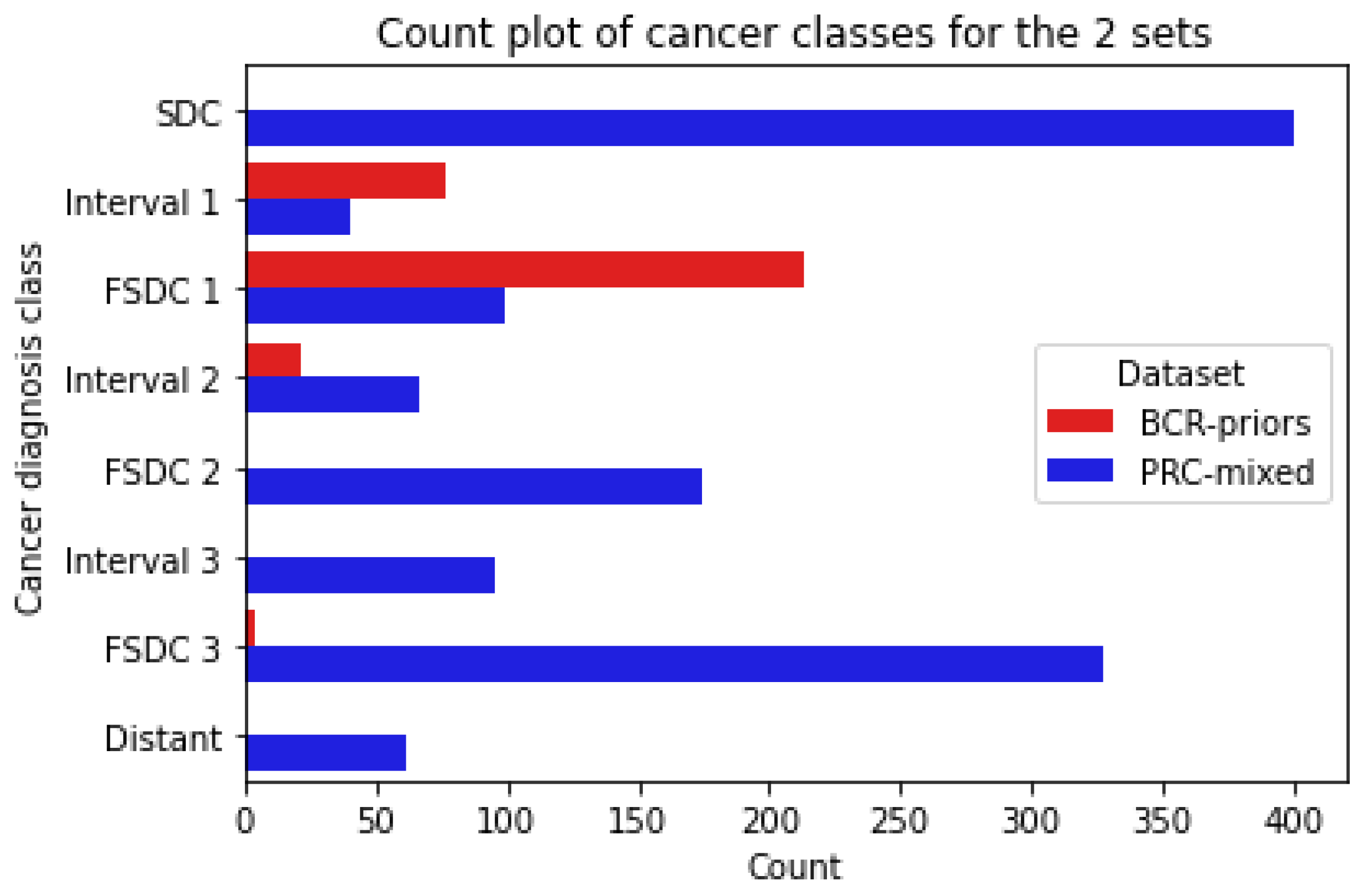
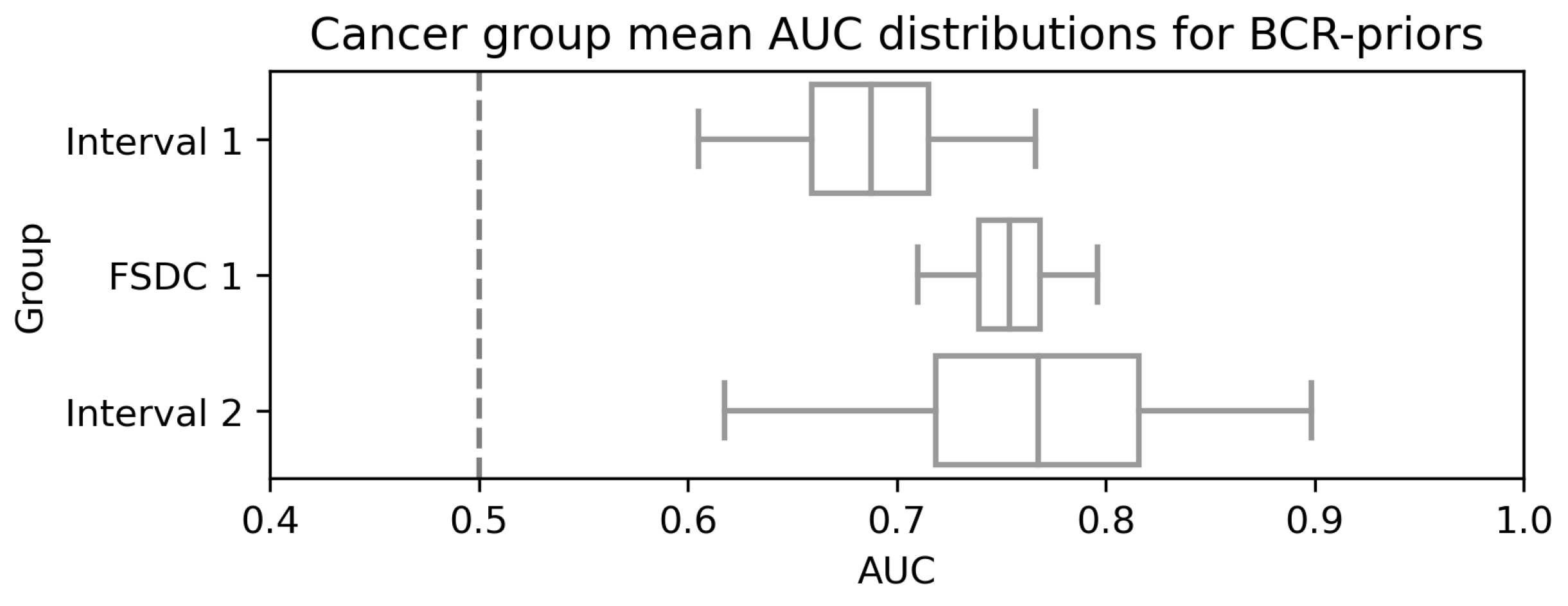
- Screen-Detected Cancers (SDC): women who were diagnosed with screen-detected breast cancer on study entry;
- Future Screen-Detected Cancers 1/2/3 (FSDC): women who were confirmed cancer-free on study entry but went on to develop screen-detected cancer. Numbers refer to which subsequent screen was used, so FSDC 1 refers to women who were diagnosed on the first subsequent screen and so on;
- Interval 1/2/3: women who were confirmed cancer-free on study entry but went on to develop an interval cancer after entry. The number refers to before which subsequent screen was the interval diagnosed, so Interval 1 refers to women who developed an interval cancer before the first subsequent screen.
2.2. Data Preparation
2.3. Model
2.4. Experimental Setup and Model Evaluation
3. Results
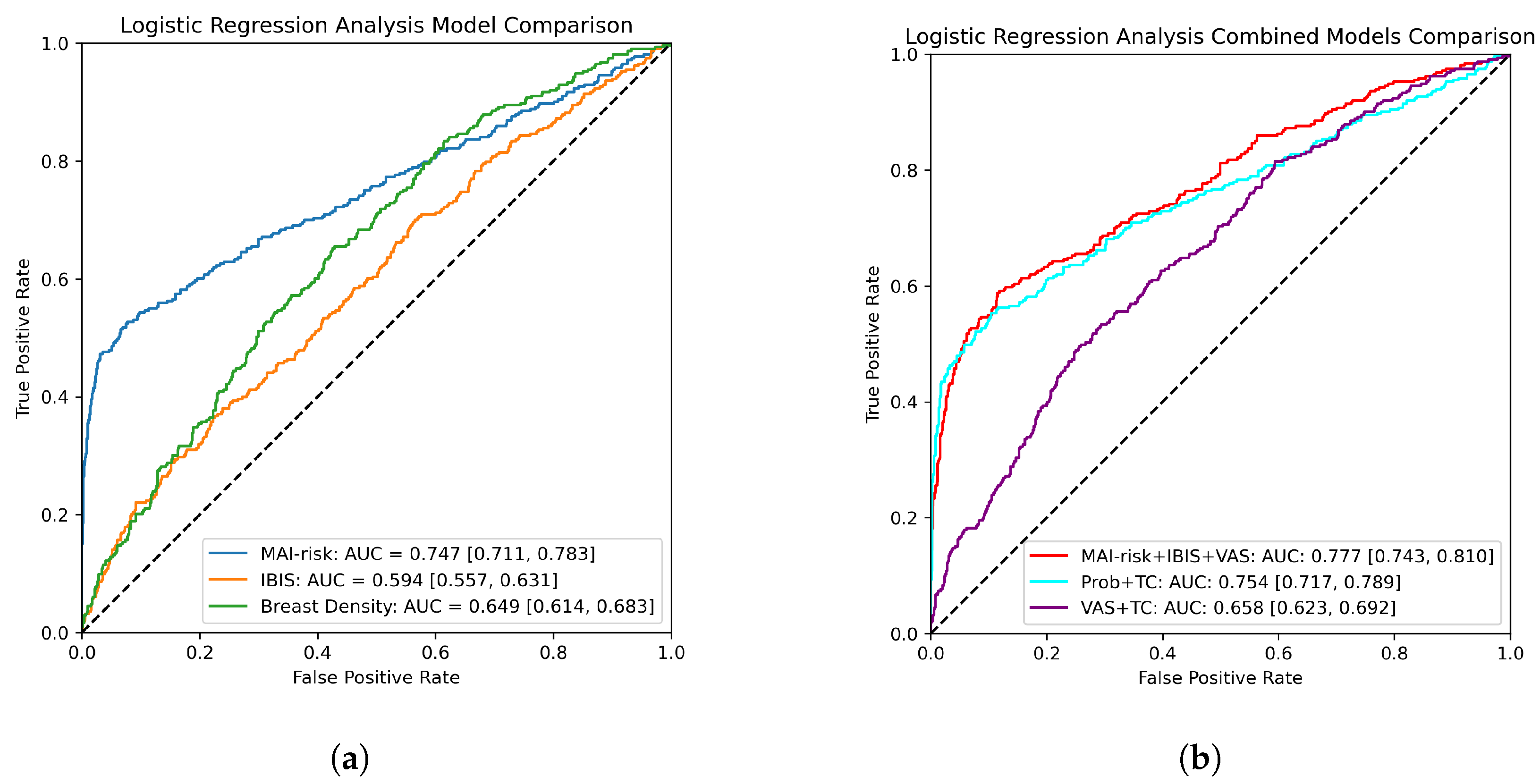
3.1. Logistic Regression Analysis
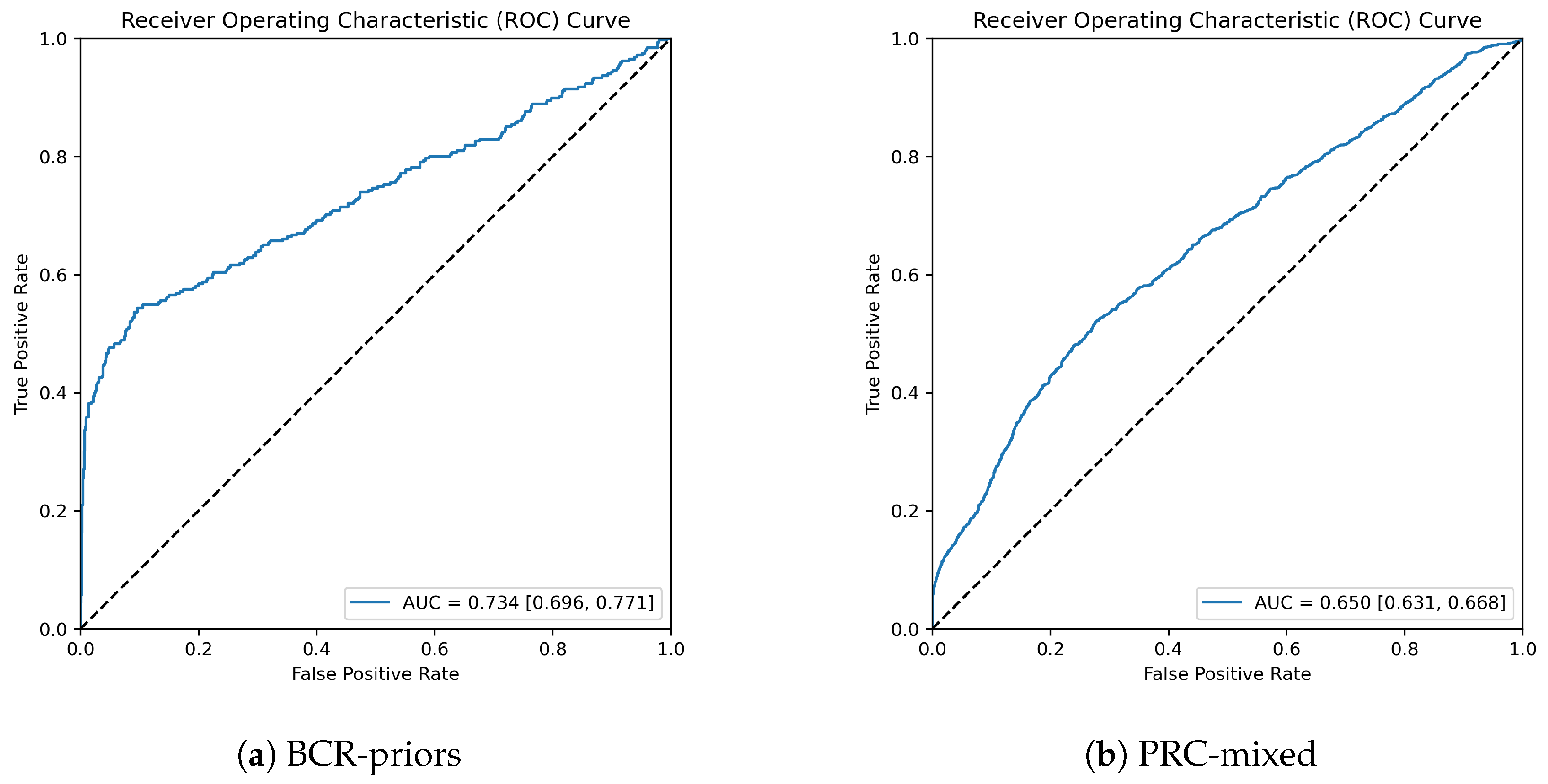
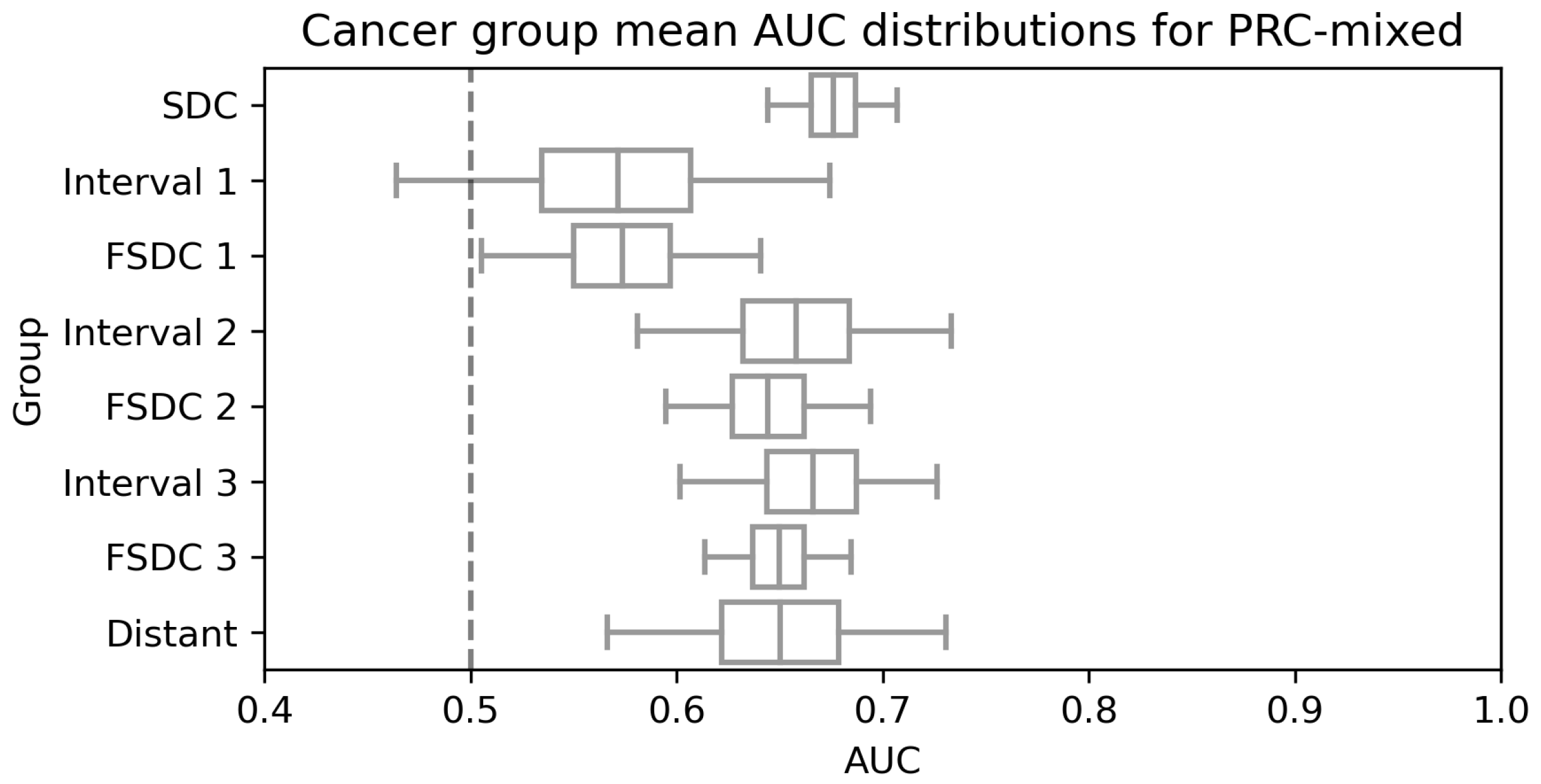
3.2. Model Performance for Different Sets
4. Discussion
Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| MAI-risk | Manchester Artificial Intelligence risk |
| PROCAS | Predicting Risk of Cancer At Screening |
| PRC-mixed | PROCAS Remaining Cancers - mixed |
| BCR-priors | Breast Cancer Research - priors |
| SDC | Screen-Detected Cancer |
| FSDC | Future Screen-Detected Cancer |
| MIL | Mulitple Instance Learning |
| CNN | Convolutional Neural Network |
| FFDM | Full-Field Digital Mammography |
| BMI | Body Mass Index |
| HRT | Hormone Replacement Therapy |
| CC | CranioCaudal |
| MLO | MedioLateral Oblique |
| AUC | Area Under the receiver operating characteristic Curve |
| FP | False Positive |
| TP | True Positive |
| OR | Odds Ratio |
| GE | General Electric |
Appendix A
| PRC-Mixed | BCR-Priors | ||||
|---|---|---|---|---|---|
| Cases (%) | Controls (%) | Cases (%) | Controls (%) | ||
| Age | <50 | 210 (5.5) | 129 (10.2) | 16 (5.1) | 47 (5.0) |
| 50–54 | 355 (28.0) | 948 (25.0) | 64 (20.3) | 196 (20.7) | |
| 55–59 | 245 (19.4) | 1156 (30.4) | 58 (18.4) | 171 (18.0) | |
| 60–64 | 303 (23.9) | 504 (13.3) | 99 (31.3) | 297 (31.3) | |
| 65–69 | 180 (14.2) | 756 (19.9) | 62 (19.6) | 186 (19.6) | |
| >70 | 54 (4.3) | 224 (5.9) | 17 (5.4) | 51 (5.4) | |
| Menopausal Status | Premenopausal | 165 (13.0) | 500 (13.2) | 22 (7.0) | 67 (7.1) |
| Perimenopausal | 209 (16.5) | 657 (17.3) | 46 (14.6) | 135 (14.2) | |
| Postmenopausal | 835 (66.0) | 2584 (68.0) | 237 (75.0) | 713 (75.2) | |
| Unknown | 57 (4.5) | 57 (1.5) | 11 (3.5) | 33 (3.5) | |
| HRT Status | Current/Previous | 470 (37.1) | 1480 (39.0) | 148 (46.8) | 456 (48.1) |
| Never | 779 (61.5) | 2316 (61.0) | 166 (52.5) | 485 (51.2) | |
| Unknown | 17 (1.3) | 2 (0.1) | 2 (0.6) | 7 (0.7) | |
| BMI | <25 kg/m | 396 (31.3) | 1193 (31.4) | 99 (31.3) | 292 (30.8) |
| 25–30 kg/m | 428 (33.8) | 1283 (33.8) | 111 (35.1) | 337 (35.5) | |
| >30 kg/m | 341 (26.9) | 1026 (27.0) | 88 (27.8) | 272 (28.7) | |
| Unknown | 101 (8.0) | 296 (7.8) | 18 (5.7) | 47 (5.0) | |
| Ethnic Origin | White | 1148 (90.7) | 3500 (92.2) | 282 (89.2) | 866 (91.4) |
| Other/Unknown | 118 (9.3) | 298 (7.8) | 34 (10.8) | 82 (8.6) | |
| Year of Entry | 2009 | 3 (0.2) | 23 (0.6) | 0 (0.0) | 0 (0.0) |
| 2010 | 261 (20.6) | 1267 (33.4) | 63 (19.9) | 178 (18.8) | |
| 2011 | 449 (35.5) | 1353 (35.6) | 163 (51.6) | 511 (53.9) | |
| 2012 | 375 (29.6) | 810 (21.3) | 85 (26.9) | 258 (27.2) | |
| 2013 | 85 (6.7) | 168 (4.4) | 3 (0.9) | 1 (0.1) | |
| 2014 | 78 (6.2) | 135 (3.6) | 2 (0.6) | 0 (0.0) | |
| 2015 | 15 (1.2) | 42 (1.1) | 0 (0.0) | 0 (0.0) | |
| Family History | FDR Only | 163 (12.9) | 403 (10.6) | 40 (12.7) | 103 (10.9) |
| SDR Only | 216 (17.1) | 606 (16.0) | 51 (16.1) | 153 (16.1) | |
| Both | 82 (6.5) | 163 (4.3) | 23 (7.3) | 46 (4.9) | |
| Neither | 805 (63.6) | 2626 (69.1) | 202 (63.9) | 646 (68.1) | |
| Alcohol Use | Yes | 961 (75.9) | 2677 (70.5) | 229 (72.5) | 673 (71.0) |
| No | 281 (22.2) | 1060 (27.9) | 85 (26.9) | 262 (27.6) | |
| Unknown | 24 (1.9) | 61 (1.6) | 2 (0.6) | 13 (1.4) | |
| Parity | Yes | 1085 (85.7) | 3261 (85.9) | 269 (85.1) | 852 (89.9) |
| No | 179 (14.1) | 537 (14.1) | 44 (13.9) | 95 (10.0) | |
| Unknown | 2 (0.2) | 0 (0.0) | 3 (0.9) | 1 (0.1) | |
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Carioli, G.; Malvezzi, M.; Rodriguez, T.; Bertuccio, P.; Negri, E.; La Vecchia, C. Trends and predictions to 2020 in breast cancer mortality in Europe. Breast 2017, 36, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Evans, D.G.; Astley, S.; Stavrinos, P.; Harkness, E.; Donnelly, L.S.; Dawe, S.; Jacob, I.; Harvie, M.; Cuzick, J.; Brentnall, A.; et al. Improvement in risk prediction, early detection and prevention of breast cancer in the NHS Breast Screening Programme and family history clinics: A dual cohort study. NIHR J. Libr. 2016. [Google Scholar] [CrossRef] [PubMed]
- Schünemann, H.J.; Lerda, D.; Quinn, C.; Follmann, M.; Alonso-Coello, P.; Rossi, P.G.; Lebeau, A.; Nyström, L.; Broeders, M.; Ioannidou-Mouzaka, L.; et al. Breast cancer screening and diagnosis: A synopsis of the European Breast Guidelines. Ann. Intern. Med. 2020, 172, 46–56. [Google Scholar] [CrossRef]
- Marmot, M.G.; Altman, D.; Cameron, D.; Dewar, J.; Thompson, S.; Wilcox, M. The benefits and harms of breast cancer screening: An independent review. Br. J. Cancer 2013, 108, 2205–2240. [Google Scholar] [CrossRef]
- Tyrer, J.; Duffy, S.W.; Cuzick, J. A breast cancer prediction model incorporating familial and personal risk factors. Stat. Med. 2004, 23, 1111–1130. [Google Scholar] [CrossRef] [PubMed]
- Brentnall, A.R.; Harkness, E.F.; Astley, S.M.; Donnelly, L.S.; Stavrinos, P.; Sampson, S.; Fox, L.; Sergeant, J.C.; Harvie, M.N.; Wilson, M.; et al. Mammographic density adds accuracy to both the Tyrer-Cuzick and Gail breast cancer risk models in a prospective UK screening cohort. Breast Cancer Res. 2015, 17, 147. [Google Scholar] [CrossRef] [PubMed]
- Quante, A.S.; Whittemore, A.S.; Shriver, T.; Strauch, K.; Terry, M.B. Breast cancer risk assessment across the risk continuum: Genetic and nongenetic risk factors contributing to differential model performance. Breast Cancer Res. 2012, 14, R144. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, A.M.; Guan, Z.; Welch, M.; Griffin, M.E.; Sippo, D.A.; Deng, Z.; Coopey, S.B.; Acar, A.; Semine, A.; Parmigiani, G.; et al. Performance of breast cancer risk-assessment models in a large mammography cohort. JNCI J. Natl. Cancer Inst. 2020, 112, 489–497. [Google Scholar] [CrossRef] [PubMed]
- Dembrower, K.; Liu, Y.; Azizpour, H.; Eklund, M.; Smith, K.; Lindholm, P.; Strand, F. Comparison of a deep learning risk score and standard mammographic density score for breast cancer risk prediction. Radiology 2020, 294, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Wanders, J.O.; van Gils, C.H.; Karssemeijer, N.; Holland, K.; Kallenberg, M.; Peeters, P.H.; Nielsen, M.; Lillholm, M. The combined effect of mammographic texture and density on breast cancer risk: A cohort study. Breast Cancer Res. 2018, 20, 36. [Google Scholar] [CrossRef] [PubMed]
- Azam, S.; Eriksson, M.; Sjölander, A.; Gabrielson, M.; Hellgren, R.; Czene, K.; Hall, P. Mammographic microcalcifications and risk of breast cancer. Br. J. Cancer 2021, 125, 759–765. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Wu, N.; Phang, J.; Park, J.; Liu, K.; Tyagi, S.; Heacock, L.; Kim, S.G.; Moy, L.; Cho, K.; et al. An interpretable classifier for high-resolution breast cancer screening images utilizing weakly supervised localization. Med. Image Anal. 2021, 68, 101908. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Lou, Q.; Vang, Y.S.; Xie, X. Deep multi-instance networks with sparse label assignment for whole mammogram classification. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Quebec, Canada, 10–14 September 2017; Springer: Berlin/Heidelberg, Germany, 2017; pp. 603–611. [Google Scholar]
- Quellec, G.; Lamard, M.; Cozic, M.; Coatrieux, G.; Cazuguel, G. Multiple-instance learning for anomaly detection in digital mammography. IEEE Trans. Med. Imaging 2016, 35, 1604–1614. [Google Scholar] [CrossRef] [PubMed]
- Astley, S.M.; Harkness, E.F.; Sergeant, J.C.; Warwick, J.; Stavrinos, P.; Warren, R.; Wilson, M.; Beetles, U.; Gadde, S.; Lim, Y.; et al. A comparison of five methods of measuring mammographic density: A case-control study. Breast Cancer Res. 2018, 20, 10. [Google Scholar] [CrossRef] [PubMed]
- Otsu, N. A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. 1979, 9, 62–66. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Ilse, M.; Tomczak, J.; Welling, M. Attention-based deep multiple instance learning. In Proceedings of the International Conference on Machine Learning, Stockholm, Sweden, 10–15 July 2018; PMLR, 2018; pp. 2127–2136. [Google Scholar]
- Dauphin, Y.N.; Fan, A.; Auli, M.; Grangier, D. Language modeling with gated convolutional networks. In Proceedings of the International Conference on Machine Learning, Sydney, Australia, 6–11 August 2017; PMLR, 2017; pp. 933–941. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Darbre, P.D. Recorded quadrant incidence of female breast cancer in Great Britain suggests a disproportionate increase in the upper outer quadrant of the breast. Anticancer Res. 2005, 25, 2543–2550. [Google Scholar] [PubMed]
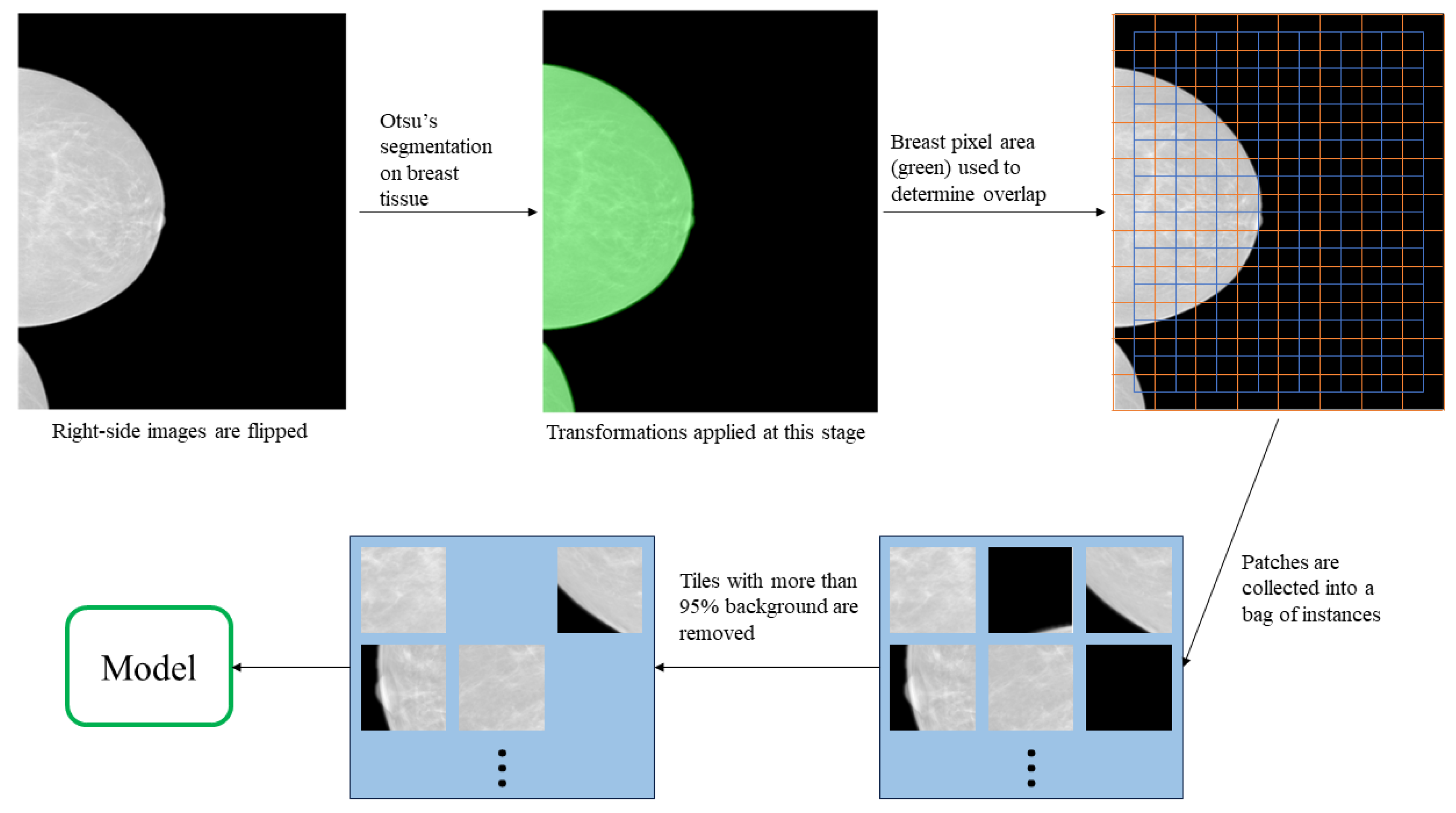
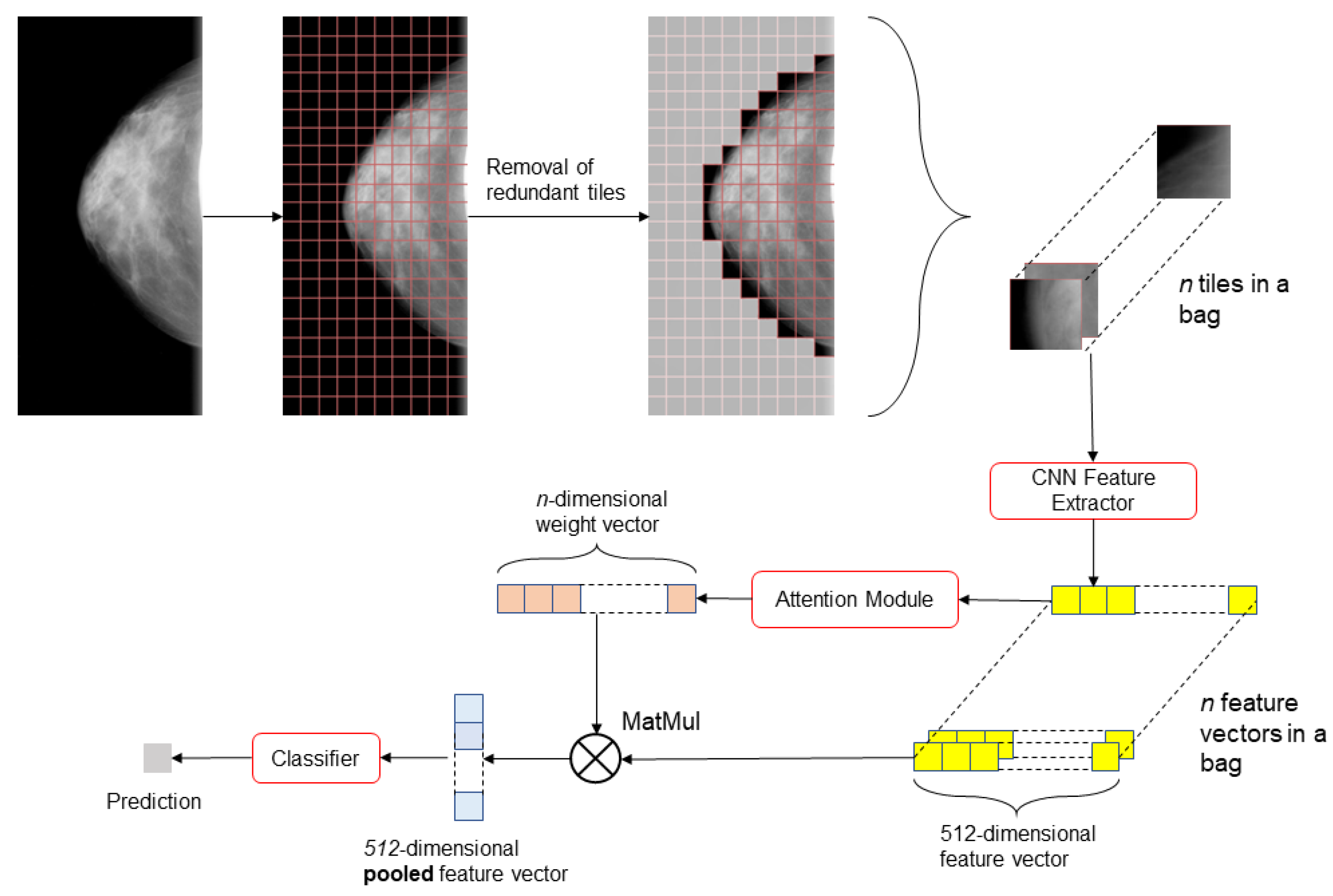
| Number of Pixels Corresponding to Breast Tissue | Overlap Step Size |
|---|---|
| >5,017,600 | 200 |
| 2,508,800–5,017,600 | 168 |
| 752,640–2,508,800 | 112 |
| <752,640 | 74 |
| Target Covariate | OR (Normalised) | OR (Representative) |
|---|---|---|
| VAS | 1.60 (1.37, 1.88) | 1.19 (1.05, 1.36) |
| IBIS | 1.34 (1.08, 1.65) | 1.03 (1.02, 1.04) |
| MAI-risk | 3.04 (2.58, 3.59) | 4.70 (3.74, 5.93) |
| Group | Number of Women | AUC |
|---|---|---|
| SDC | 1600 | 0.676 (0.645, 0.707) |
| Interval 1 | 160 | 0.571 (0.464, 0.679) |
| FSDC 1 | 396 | 0.573 (0.506, 0.641) |
| Interval 2 | 268 | 0.657 (0.582, 0.730) |
| FSDC 2 | 700 | 0.644 (0.594, 0.693) |
| Interval 3 | 380 | 0.666 (0.602, 0.727) |
| FSDC 3 | 1312 | 0.650 (0.614, 0.685) |
| Distant | 248 | 0.649 (0.565, 0.731) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Romanov, S.; Howell, S.; Harkness, E.; Bydder, M.; Evans, D.G.; Squires, S.; Fergie, M.; Astley, S. Artificial Intelligence for Image-Based Breast Cancer Risk Prediction Using Attention. Tomography 2023, 9, 2103-2115. https://doi.org/10.3390/tomography9060165
Romanov S, Howell S, Harkness E, Bydder M, Evans DG, Squires S, Fergie M, Astley S. Artificial Intelligence for Image-Based Breast Cancer Risk Prediction Using Attention. Tomography. 2023; 9(6):2103-2115. https://doi.org/10.3390/tomography9060165
Chicago/Turabian StyleRomanov, Stepan, Sacha Howell, Elaine Harkness, Megan Bydder, D. Gareth Evans, Steven Squires, Martin Fergie, and Sue Astley. 2023. "Artificial Intelligence for Image-Based Breast Cancer Risk Prediction Using Attention" Tomography 9, no. 6: 2103-2115. https://doi.org/10.3390/tomography9060165
APA StyleRomanov, S., Howell, S., Harkness, E., Bydder, M., Evans, D. G., Squires, S., Fergie, M., & Astley, S. (2023). Artificial Intelligence for Image-Based Breast Cancer Risk Prediction Using Attention. Tomography, 9(6), 2103-2115. https://doi.org/10.3390/tomography9060165








