A Quantitative Multiparametric MRI Analysis Platform for Estimation of Robust Imaging Biomarkers in Clinical Oncology
Abstract
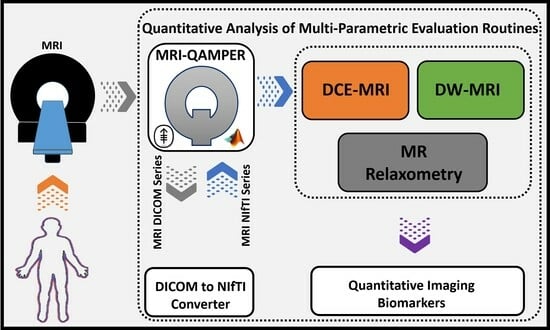
:1. Introduction
2. Materials and Methods
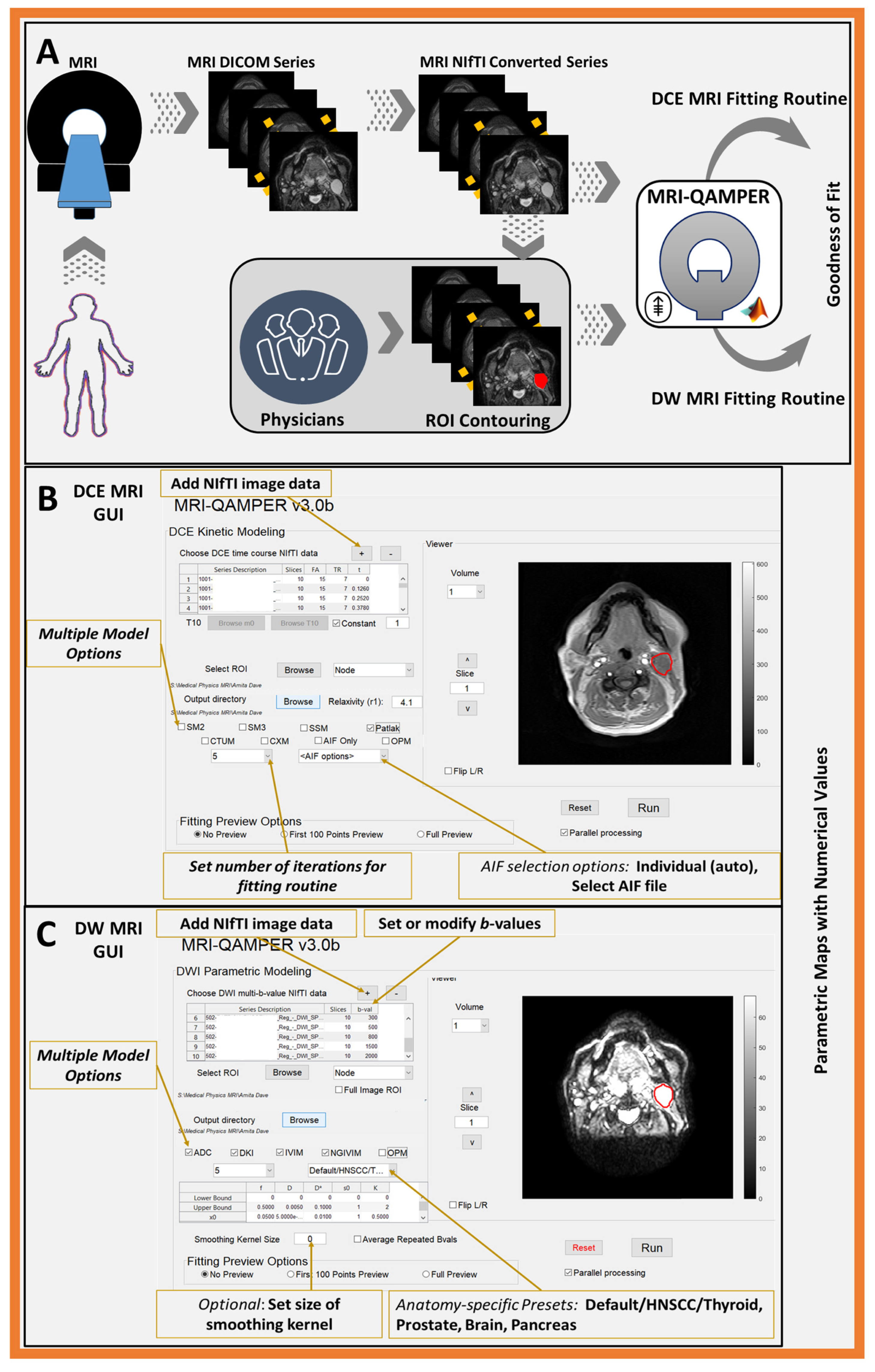
2.1. DCE MRI Pharmacokinetic Modeling
2.1.1. Extended Tofts Model (ETM)
2.1.2. Patlak Model (PM)
2.1.3. Fast Exchange Regime (FXR) or Shutter Speed Model (SSM)
2.1.4. Arterial Input Function (AIF) Selection
2.2. DW MRI Data Modeling
2.3. and Relaxometry
2.4. Optimal Model Mapping (OMM)
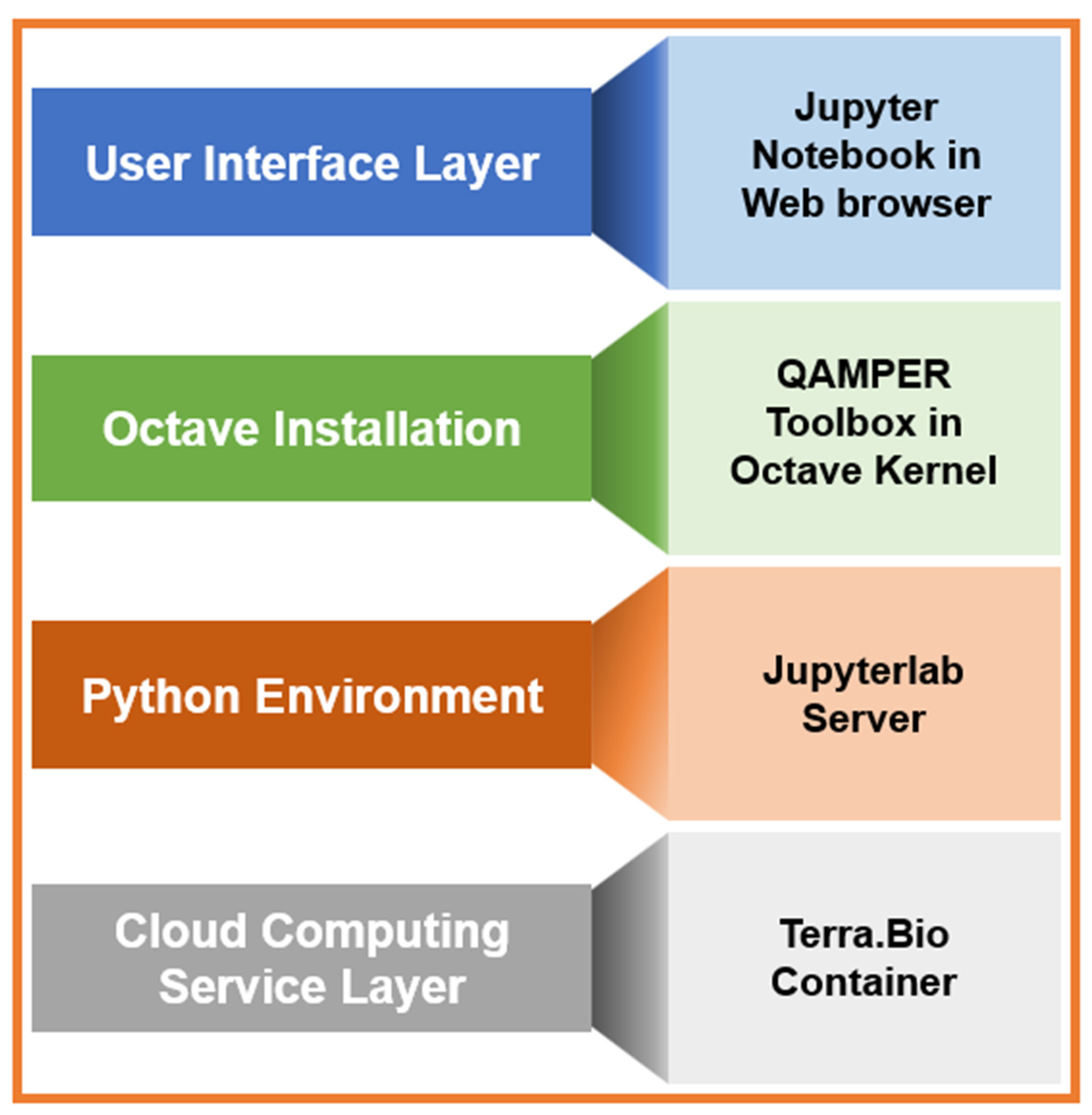
2.5. Imaging Formats and Conversion
3. Results
3.1. QAMPER QIN Software Validation: DROs and CCPs
3.2. DCE MRI DRO (RSNA)
3.3. DW MRI DRO (University of Michigan)
3.4. Collaborative Challenge Projects
3.4.1. DCE CCP (ISMRM, Open Science Initiative for Perfusion Imaging (OSIPI))
3.4.2. DW MRI CCP (QIN, MCW, Prostate)
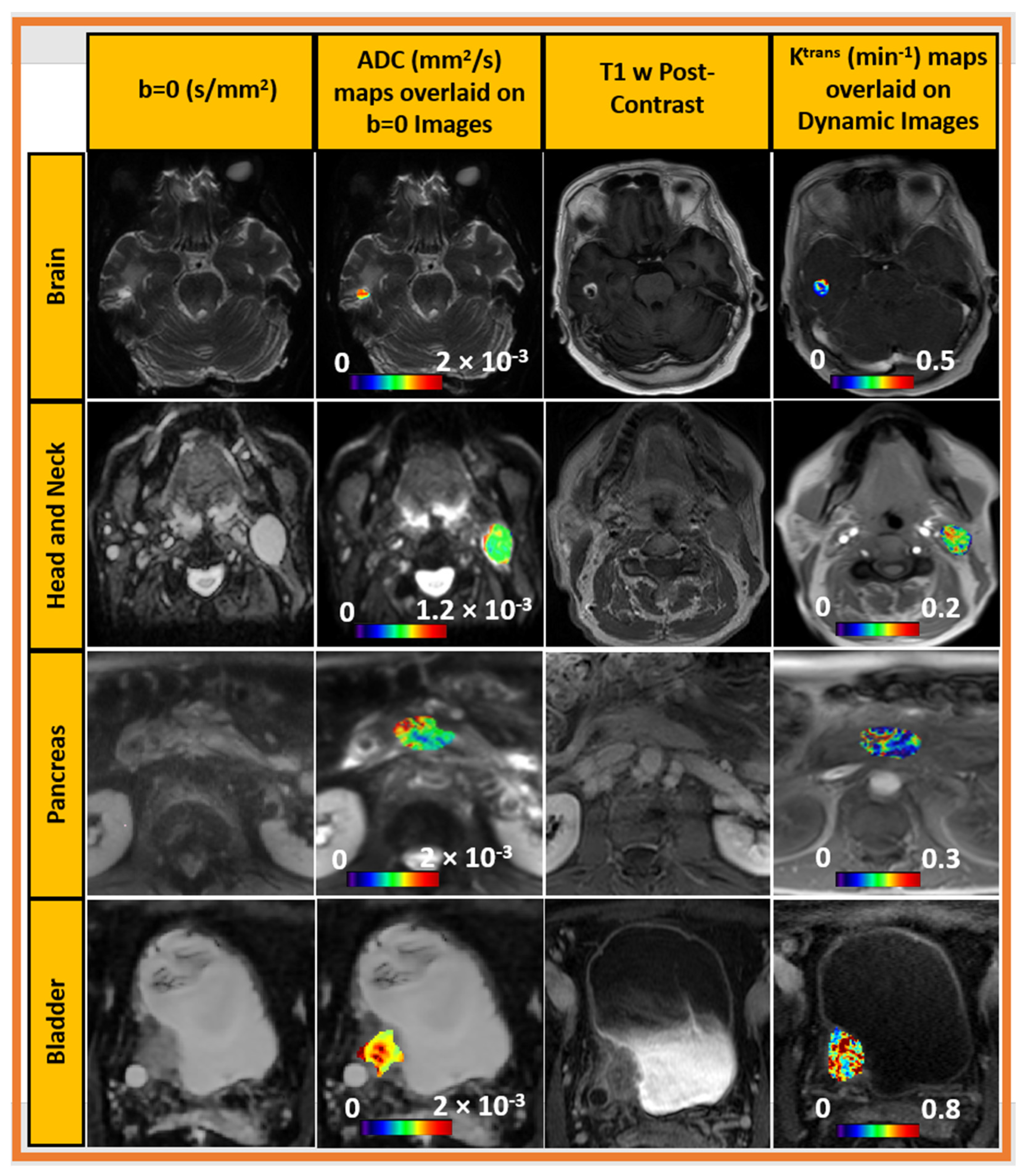
3.5. DCE and DW MRI in Clinical Trial (Oropharyngeal Cancer)
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
Appendix A.1. Precontrast Fitting
Appendix A.2. Signal-to-CA Concentration Calculation
Appendix A.3. Compartmental Tissue Uptake Model (CTUM)
Appendix A.4. Two-Compartment Exchange Model (2CXM)
References
- Grover, V.P.; Tognarelli, J.M.; Crossey, M.M.; Cox, I.J.; Taylor-Robinson, S.D.; McPhail, M.J. Magnetic Resonance Imaging: Principles and Techniques: Lessons for Clinicians. J. Clin. Exp. Hepatol. 2015, 5, 246–255. [Google Scholar] [CrossRef] [PubMed]
- Kabasawa, H. MR Imaging in the 21st Century: Technical Innovation over the First Two Decades. Magn. Reson. Med. Sci. 2022, 21, 71–82. [Google Scholar] [CrossRef] [PubMed]
- Chalavi, S.; Simmons, A.; Dijkstra, H.; Barker, G.J.; Reinders, A.A.T.S. Quantitative and qualitative assessment of structural magnetic resonance imaging data in a two-center study. BMC Med. Imaging 2012, 12, 27. [Google Scholar] [CrossRef] [PubMed]
- Konar, A.S.; Paudyal, R.; Shah, A.D.; Fung, M.; Banerjee, S.; Dave, A.; Lee, N.; Hatzoglou, V.; Shukla-Dave, A. Qualitative and quantitative performance of magnetic resonance image compilation (MAGiC) method: An exploratory analysis for head and neck imaging. Cancers 2022, 14, 3624. [Google Scholar] [CrossRef] [PubMed]
- Sourbron, S.P.; Buckley, D.L. On the scope and interpretation of the Tofts models for DCE-MRI. Magn. Reson. Med. 2011, 66, 735–745. [Google Scholar] [CrossRef]
- Chawla, S.; Kim, S.; Wang, S.; Poptani, H. Diffusion-weighted imaging in head and neck cancers. Future Oncol. 2009, 5, 959–975. [Google Scholar] [CrossRef] [PubMed]
- Keenan, K.E.; Biller, J.R.; Delfino, J.G.; Boss, M.A.; Does, M.D.; Evelhoch, J.L.; Griswold, M.A.; Gunter, J.L.; Hinks, R.S.; Hoffman, S.W.; et al. Recommendations towards standards for quantitative MRI (qMRI) and outstanding needs. J. Magn. Reson. Imaging 2019, 49, e26–e39. [Google Scholar] [CrossRef]
- Shukla-Dave, A.; Obuchowski, N.A.; Chenevert, T.L.; Jambawalikar, S.; Schwartz, L.H.; Malyarenko, D.; Huang, W.; Noworolski, S.M.; Young, R.J.; Shiroishi, M.S.; et al. Quantitative imaging biomarkers alliance (QIBA) recommendations for improved precision of DWI and DCE-MRI derived biomarkers in multicenter oncology trials. J. Magn. Reson. Imaging 2019, 49, e101–e121. [Google Scholar] [CrossRef]
- Padhani, A.R.; Khan, A.A. Diffusion-weighted (DW) and dynamic contrast-enhanced (DCE) magnetic resonance imaging (MRI) for monitoring anticancer therapy. Target. Oncol. 2010, 5, 39–52. [Google Scholar] [CrossRef]
- Riaz, N.; Sherman, E.; Pei, X.; Schöder, H.; Grkovski, M.; Paudyal, R.; Katabi, N.; Selenica, P.; Yamaguchi, T.N.; Ma, D.; et al. Precision Radiotherapy: Reduction in Radiation for Oropharyngeal Cancer in the 30 ROC Trial. J. Natl. Cancer Inst. 2021, 113, 742–751. [Google Scholar] [CrossRef]
- Winfield, J.M.; Payne, G.S.; Weller, A.; deSouza, N.M. DCE-MRI, DW-MRI, and MRS in Cancer: Challenges and Advantages of Implementing Qualitative and Quantitative Multi-parametric Imaging in the Clinic. Top. Magn. Reson. Imaging 2016, 25, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Farahani, K.; Tata, D.; Nordstrom, R.J. QIN Benchmarks for Clinical Translation of Quantitative Imaging Tools. Tomography 2019, 5, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Obuchowski, N.A.; Reeves, A.P.; Huang, E.P.; Wang, X.F.; Buckler, A.J.; Kim, H.J.; Barnhart, H.X.; Jackson, E.F.; Giger, M.L.; Pennello, G.; et al. Quantitative imaging biomarkers: A review of statistical methods for computer algorithm comparisons. Stat. Methods Med. Res. 2015, 24, 68–106. [Google Scholar] [CrossRef] [PubMed]
- Barboriak, D. QIBA DRO for DCE MRI. 2018. [Google Scholar]
- Kurland, B.F.; Gerstner, E.R.; Mountz, J.M.; Schwartz, L.H.; Ryan, C.W.; Graham, M.M.; Buatti, J.M.; Fennessy, F.M.; Eikman, E.A.; Kumar, V.; et al. Promise and pitfalls of quantitative imaging in oncology clinical trials. Magn. Reson. Imaging 2012, 30, 1301–1312. [Google Scholar] [CrossRef] [PubMed]
- Shah, A.D.; Shridhar Konar, A.; Paudyal, R.; Oh, J.H.; LoCastro, E.; Nuñez, D.A.; Swinburne, N.; Vachha, B.; Ulaner, G.A.; Young, R.J.; et al. Diffusion and Perfusion MRI Predicts Response Preceding and Shortly After Radiosurgery to Brain Metastases: A Pilot Study. J. Neuroimaging 2021, 31, 317–323. [Google Scholar] [CrossRef] [PubMed]
- Paudyal, R.; Oh, J.H.; Riaz, N.; Venigalla, P.; Li, J.; Hatzoglou, V.; Leeman, J.; Nunez, D.A.; Lu, Y.; Deasy, J.O.; et al. Intravoxel incoherent motion diffusion-weighted MRI during chemoradiation therapy to characterize and monitor treatment response in human papillomavirus head and neck squamous cell carcinoma. J. Magn. Reson. Imaging 2017, 45, 1013–1023. [Google Scholar] [CrossRef] [PubMed]
- Nunez, D.A.; Lu, Y.; Paudyal, R.; Hatzoglou, V.; Moreira, A.L.; Oh, J.H.; Stambuk, H.E.; Mazaheri, Y.; Gonen, M.; Ghossein, R.A.; et al. Quantitative Non-Gaussian Intravoxel Incoherent Motion Diffusion-Weighted Imaging Metrics and Surgical Pathology for Stratifying Tumor Aggressiveness in Papillary Thyroid Carcinomas. Tomography 2019, 5, 26–35. [Google Scholar] [CrossRef]
- Paudyal, R.; Lu, Y.; Hatzoglou, V.; Moreira, A.; Stambuk, H.E.; Oh, J.H.; Cunanan, K.M.; Aramburu Nunez, D.; Mazaheri, Y.; Gonen, M.; et al. Dynamic contrast-enhanced MRI model selection for predicting tumor aggressiveness in papillary thyroid cancers. NMR Biomed. 2020, 33, e4166. [Google Scholar] [CrossRef]
- McGarry, S.D.; Brehler, M.; Bukowy, J.D.; Lowman, A.K.; Bobholz, S.A.; Duenweg, S.R.; Banerjee, A.; Hurrell, S.L.; Malyarenko, D.; Chenevert, T.L.; et al. Multi-Site Concordance of Diffusion-Weighted Imaging Quantification for Assessing Prostate Cancer Aggressiveness. J. Magn. Reson. Imaging 2022, 55, 1745–1758. [Google Scholar] [CrossRef]
- Kinh Do, R.; Reyngold, M.; Paudyal, R.; Oh, J.H.; Konar, A.S.; LoCastro, E.; Goodman, K.A.; Shukla-Dave, A. Diffusion-Weighted and Dynamic Contrast-Enhanced MRI Derived Imaging Metrics for Stereotactic Body Radiotherapy of Pancreatic Ductal Adenocarcinoma: Preliminary Findings. Tomography 2020, 6, 261–271. [Google Scholar] [CrossRef]
- Berks, M.; Parker, G.; Little, R.; Cheung, S. Madym: A C++ toolkit for quantitative DCE-MRI analysis. J. Open Source Softw. 2021, 6, 3523. [Google Scholar] [CrossRef]
- Barnes, S.R.; Ng, T.S.; Santa-Maria, N.; Montagne, A.; Zlokovic, B.V.; Jacobs, R.E. ROCKETSHIP: A flexible and modular software tool for the planning, processing and analysis of dynamic MRI studies. BMC Med. Imaging 2015, 15, 19. [Google Scholar] [CrossRef]
- Ewing, J.R.; Bagher-Ebadian, H. Model selection in measures of vascular parameters using dynamic contrast-enhanced MRI: Experimental and clinical applications. NMR Biomed. 2013, 26, 1028–1041. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Jansen, J.F.; Mazaheri, Y.; Stambuk, H.E.; Koutcher, J.A.; Shukla-Dave, A. Extension of the intravoxel incoherent motion model to non-gaussian diffusion in head and neck cancer. J. Magn. Reson. Imaging 2012, 36, 1088–1096. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Morgan, P.S.; Ashburner, J.; Smith, J.; Rorden, C. The first step for neuroimaging data analysis: DICOM to NIfTI conversion. J. Neurosci. Methods 2016, 264, 47–56. [Google Scholar] [CrossRef]
- Yushkevich, P.A.; Gao, Y.; Gerig, G. ITK-SNAP: An interactive tool for semi-automatic segmentation of multi-modality biomedical images. In Proceedings of the 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Orlando, FL, USA, 16–20 August 2016; pp. 3342–3345. [Google Scholar]
- Tofts, P.S.; Brix, G.; Buckley, D.L.; Evelhoch, J.L.; Henderson, E.; Knopp, M.V.; Larsson, H.B.; Lee, T.Y.; Mayr, N.A.; Parker, G.J.; et al. Estimating kinetic parameters from dynamic contrast-enhanced T(1)-weighted MRI of a diffusable tracer: Standardized quantities and symbols. J. Magn. Reson. Imaging 1999, 10, 223–232. [Google Scholar] [CrossRef]
- Le Bihan, D. Intravoxel incoherent motion imaging using steady-state free precession. Magn. Reson. Med. 1988, 7, 346–351. [Google Scholar] [CrossRef]
- Jensen, J.H.; Helpern, J.A.; Ramani, A.; Lu, H.; Kaczynski, K. Diffusional kurtosis imaging: The quantification of non-gaussian water diffusion by means of magnetic resonance imaging. Magn. Reson. Med. 2005, 53, 1432–1440. [Google Scholar] [CrossRef]
- Bagher-Ebadian, H.; Jain, R.; Nejad-Davarani, S.P.; Mikkelsen, T.; Lu, M.; Jiang, Q.; Scarpace, L.; Arbab, A.S.; Narang, J.; Soltanian-Zadeh, H.; et al. Model selection for DCE-T1 studies in glioblastoma. Magn. Reson. Med. 2012, 68, 241–251. [Google Scholar] [CrossRef]
- Deoni, S.C.; Peters, T.M.; Rutt, B.K. High-resolution T1 and T2 mapping of the brain in a clinically acceptable time with DESPOT1 and DESPOT2. Magn. Reson. Med. 2005, 53, 237–241. [Google Scholar] [CrossRef]
- Patlak, C.S.; Blasberg, R.G.; Fenstermacher, J.D. Graphical evaluation of blood-to-brain transfer constants from multiple-time uptake data. J. Cereb. Blood Flow. Metab. 1983, 3, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Landis, C.S.; Li, X.; Telang, F.W.; Coderre, J.A.; Micca, P.L.; Rooney, W.D.; Latour, L.L.; Vétek, G.; Pályka, I.; Springer, C.S., Jr. Determination of the MRI contrast agent concentration time course in vivo following bolus injection: Effect of equilibrium transcytolemmal water exchange. Magn. Reson. Med. 2000, 44, 563–574. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Rooney, W.D.; Springer, C.S., Jr. A unified magnetic resonance imaging pharmacokinetic theory: Intravascular and extracellular contrast reagents. Magn. Reson. Med. 2005, 54, 1351–1359. [Google Scholar] [CrossRef]
- Paudyal, R.; Poptani, H.; Cai, K.; Zhou, R.; Glickson, J.D. Impact of transvascular and cellular-interstitial water exchange on dynamic contrast-enhanced magnetic resonance imaging estimates of blood to tissue transfer constant and blood plasma volume. J. Magn. Reson. Imaging 2013, 37, 435–444. [Google Scholar] [CrossRef]
- Shukla-Dave, A.; Lee, N.; Stambuk, H.; Wang, Y.; Huang, W.; Thaler, H.T.; Patel, S.G.; Shah, J.P.; Koutcher, J.A. Average arterial input function for quantitative dynamic contrast enhanced magnetic resonance imaging of neck nodal metastases. BMC Med. Phys. 2009, 9, 4. [Google Scholar] [CrossRef]
- Li, X.; Welch, E.B.; Chakravarthy, A.B.; Mayer, I.; Kelley, M.; Meszoely, I.; Means-Powell, J.; Gore, J.C.; Yankeelov, T.E. A semi-automated method for obtaining the arterial input function in dynamic breast data. Proc. Intl. Soc. Mag. Reson. Med. (ISMRM) 2010, 18, 2729. [Google Scholar]
- Chenevert, T.L.; McKeever, P.E.; Ross, B.D. Monitoring early response of experimental brain tumors to therapy using diffusion magnetic resonance imaging. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 1997, 3, 1457–1466. [Google Scholar]
- Cheng, H.L.; Stikov, N.; Ghugre, N.R.; Wright, G.A. Practical medical applications of quantitative MR relaxometry. J. Magn. Reson. Imaging 2012, 36, 805–824. [Google Scholar] [CrossRef]
- Wansapura, J.P.; Holland, S.K.; Dunn, R.S.; Ball, W.S., Jr. NMR relaxation times in the human brain at 3.0 tesla. J. Magn. Reson. Imaging 1999, 9, 531–538. [Google Scholar] [CrossRef]
- Gelman, N.; Gorell, J.M.; Barker, P.B.; Savage, R.M.; Spickler, E.M.; Windham, J.P.; Knight, R.A. MR imaging of human brain at 3.0 T: Preliminary report on transverse relaxation rates and relation to estimated iron content. Radiology 1999, 210, 759–767. [Google Scholar] [CrossRef]
- Gelman, N.; Ewing, J.R.; Gorell, J.M.; Spickler, E.M.; Solomon, E.G. Interregional variation of longitudinal relaxation rates in human brain at 3.0 T: Relation to estimated iron and water contents. Magn. Reson. Med. 2001, 45, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Fatemi, Y.; Danyali, H.; Helfroush, M.S.; Amiri, H. Fast T(2) mapping using multi-echo spin-echo MRI: A linear order approach. Magn. Reson. Med. 2020, 84, 2815–2830. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.P.; Urayama, S.I.; Isa, T.; Fukuyama, H. Optimal Model Mapping for Intravoxel Incoherent Motion MRI. Front. Hum. Neurosci. 2021, 15, 617152. [Google Scholar] [CrossRef] [PubMed]
- Ewing, J.R.; Brown, S.L.; Lu, M.; Panda, S.; Ding, G.; Knight, R.A.; Cao, Y.; Jiang, Q.; Nagaraja, T.N.; Churchman, J.L.; et al. Model selection in magnetic resonance imaging measurements of vascular permeability: Gadomer in a 9L model of rat cerebral tumor. J. Cereb. Blood Flow. Metab. 2006, 26, 310–320. [Google Scholar] [CrossRef]
- Kim, S.; Quon, H.; Loevner, L.A.; Rosen, M.A.; Dougherty, L.; Kilger, A.M.; Glickson, J.D.; Poptani, H. Transcytolemmal water exchange in pharmacokinetic analysis of dynamic contrast-enhanced MRI data in squamous cell carcinoma of the head and neck. J. Magn. Reson. Imaging Off. J. Int. Soc. Magn. Reson. Med. 2007, 26, 1607–1617. [Google Scholar] [CrossRef]
- Akaike, H. A new look at the statistical model identification. IEEE Trans. Autom. Control 1974, 19, 716–723. [Google Scholar] [CrossRef]
- Banerji, A.; Naish, J.H.; Watson, Y.; Jayson, G.C.; Buonaccorsi, G.A.; Parker, G.J. DCE-MRI model selection for investigating disruption of microvascular function in livers with metastatic disease. J. Magn. Reson. Imaging 2012, 35, 196–203. [Google Scholar] [CrossRef]
- Luypaert, R.; Ingrisch, M.; Sourbron, S.; De Mey, J. The Akaike information criterion in DCE-MRI: Does it improve the haemodynamic parameter estimates? Phys. Med. Biol. 2012, 57, 3609. [Google Scholar] [CrossRef]
- Schwarz, G. Estimating the dimension of a model. Ann. Stat. 1978, 6, 461–464. [Google Scholar] [CrossRef]
- Ger, R.B.; Mohamed, A.S.R.; Awan, M.J.; Ding, Y.; Li, K.; Fave, X.J.; Beers, A.L.; Driscoll, B.; Elhalawani, H.; Hormuth, D.A.; et al. A Multi-Institutional Comparison of Dynamic Contrast-Enhanced Magnetic Resonance Imaging Parameter Calculations. Sci. Rep. 2017, 7, 11185. [Google Scholar] [CrossRef]
- Malyarenko, D.; Pang, Y.; Amouzandeh, G.; Chenevert, T. Numerical DWI Phantoms to Optimize Accuracy and Precision of Quantitative Parametric Maps for Non-Gaussian Diffusion; SPIE: Bellingham, DC, USA, 2020; Volume 11313. [Google Scholar]
- Harrington, K.A.; Shukla-Dave, A.; Paudyal, R.; Do, R.K.G. MRI of the Pancreas. J. Magn. Reson. Imaging 2021, 53, 347–359. [Google Scholar] [CrossRef] [PubMed]
- Debus, C.; Floca, R.; Ingrisch, M.; Kompan, I.; Maier-Hein, K.; Abdollahi, A.; Nolden, M. MITK-ModelFit: A generic open-source framework for model fits and their exploration in medical imaging–design, implementation and application on the example of DCE-MRI. BMC Bioinform. 2019, 20, 31. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Shao, L.; Zhou, C.; Yan, Z.; Han, Y.; Song, Y. DCE-Net: A Dynamic Context Encoder Network for Liver Tumor Segmentation. 2023. Available online: https://www.researchsquare.com/article/rs-2272616/v1 (accessed on 10 October 2023).
- Shridhar Konar, A.; Qian, E.; Geethanath, S.; Buonincontri, G.; Obuchowski, N.A.; Fung, M.; Gomez, P.; Schulte, R.; Cencini, M.; Tosetti, M.; et al. Quantitative imaging metrics derived from magnetic resonance fingerprinting using ISMRM/NIST MRI system phantom: An international multicenter repeatability and reproducibility study. Med. Phys. 2021, 48, 2438–2447. [Google Scholar] [CrossRef] [PubMed]
- Konar, A.S.; Shah, A.D.; Paudyal, R.; Fung, M.; Banerjee, S.; Dave, A.; Hatzoglou, V.; Shukla-Dave, A. Quantitative Relaxometry Metrics for Brain Metastases Compared to Normal Tissues: A Pilot MR Fingerprinting Study. Cancers 2022, 14, 5606. [Google Scholar] [CrossRef] [PubMed]
- Apte, A.P.; LoCastro, E.; Iyer, A.; Jiang, J.; Oh, J.H.; Veeraraghavan, H.; Shukla-Dave, A.; Deasy, J.O. Technical Note: STRATIS: A Cloud-enabled Software Toolbox for Radiotherapy and Imaging Analysis. bioRxiv 2022. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
LoCastro, E.; Paudyal, R.; Konar, A.S.; LaViolette, P.S.; Akin, O.; Hatzoglou, V.; Goh, A.C.; Bochner, B.H.; Rosenberg, J.; Wong, R.J.; et al. A Quantitative Multiparametric MRI Analysis Platform for Estimation of Robust Imaging Biomarkers in Clinical Oncology. Tomography 2023, 9, 2052-2066. https://doi.org/10.3390/tomography9060161
LoCastro E, Paudyal R, Konar AS, LaViolette PS, Akin O, Hatzoglou V, Goh AC, Bochner BH, Rosenberg J, Wong RJ, et al. A Quantitative Multiparametric MRI Analysis Platform for Estimation of Robust Imaging Biomarkers in Clinical Oncology. Tomography. 2023; 9(6):2052-2066. https://doi.org/10.3390/tomography9060161
Chicago/Turabian StyleLoCastro, Eve, Ramesh Paudyal, Amaresha Shridhar Konar, Peter S. LaViolette, Oguz Akin, Vaios Hatzoglou, Alvin C. Goh, Bernard H. Bochner, Jonathan Rosenberg, Richard J. Wong, and et al. 2023. "A Quantitative Multiparametric MRI Analysis Platform for Estimation of Robust Imaging Biomarkers in Clinical Oncology" Tomography 9, no. 6: 2052-2066. https://doi.org/10.3390/tomography9060161
APA StyleLoCastro, E., Paudyal, R., Konar, A. S., LaViolette, P. S., Akin, O., Hatzoglou, V., Goh, A. C., Bochner, B. H., Rosenberg, J., Wong, R. J., Lee, N. Y., Schwartz, L. H., & Shukla-Dave, A. (2023). A Quantitative Multiparametric MRI Analysis Platform for Estimation of Robust Imaging Biomarkers in Clinical Oncology. Tomography, 9(6), 2052-2066. https://doi.org/10.3390/tomography9060161