A Novel Multi-Dynamic Coupled Neural Mass Model of SSVEP
Abstract
1. Introduction
- Develop a bidirectionally coupled neural mass model of SSVEP, for the first time, to simulate cross-regional brain interactions. The developed multi-dynamic coupled NMM helps to elucidate the generation mechanisms of SSVEP signals.
- Design parallel linear transfer functions and weight coefficient matrices for multiple stimulation frequencies. The entailed key parameters are further identified with the optimization using particle swarm algorithms.
- Implement various data augmentation methods (signal translation, amplitude distortion, temporal masking, scale transformation, and noise addition). Utilize augmented real datasets to train high-performance SSVEP classifiers that can be used with advanced deep learning methods.
- The application of advanced FPF-net classification algorithms for simulated signal recognition, with the further successful implementation on vehicle control.
2. Multi-Dynamic Coupled Neural Mass Model
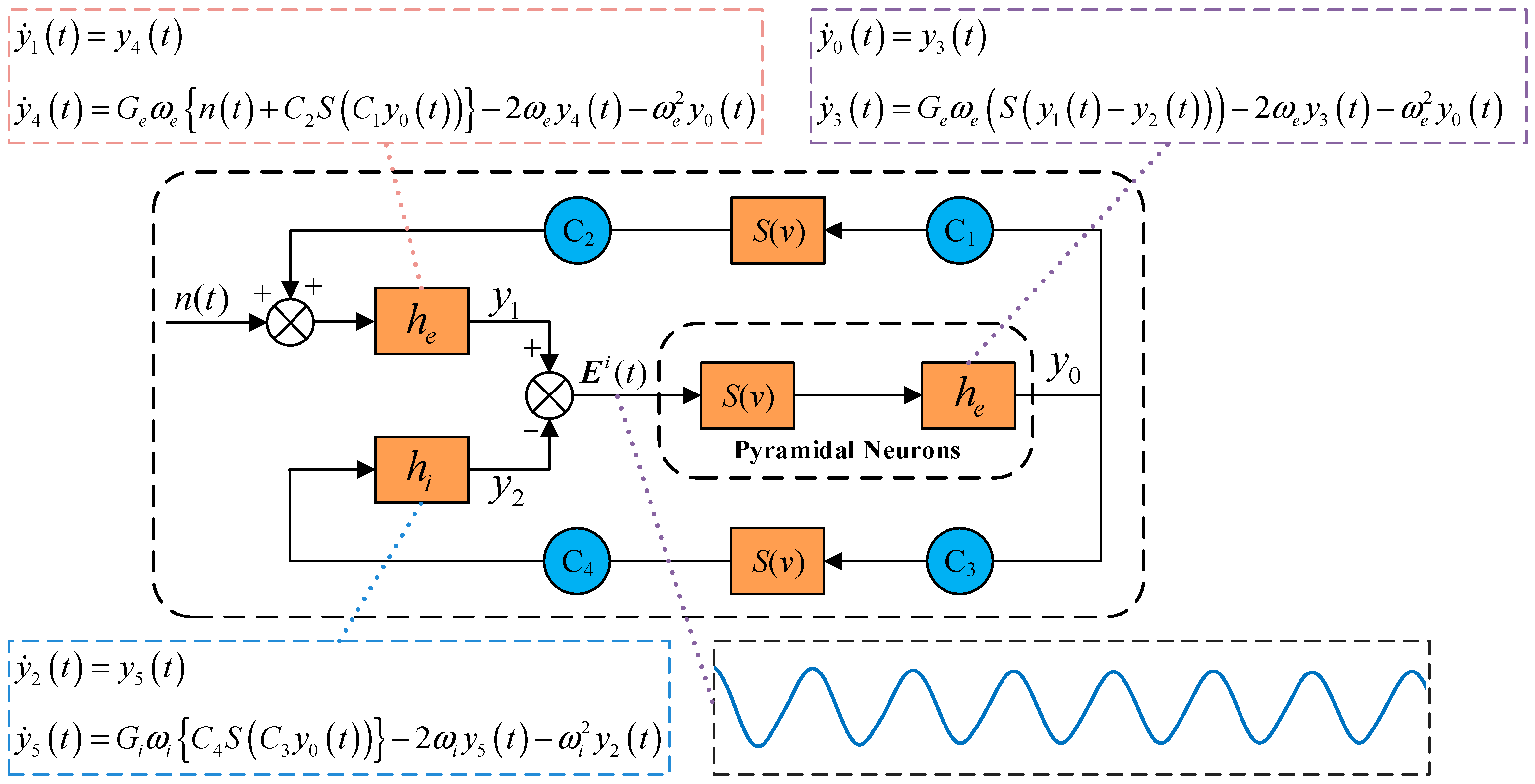
2.1. Traditional Neural Mass Model
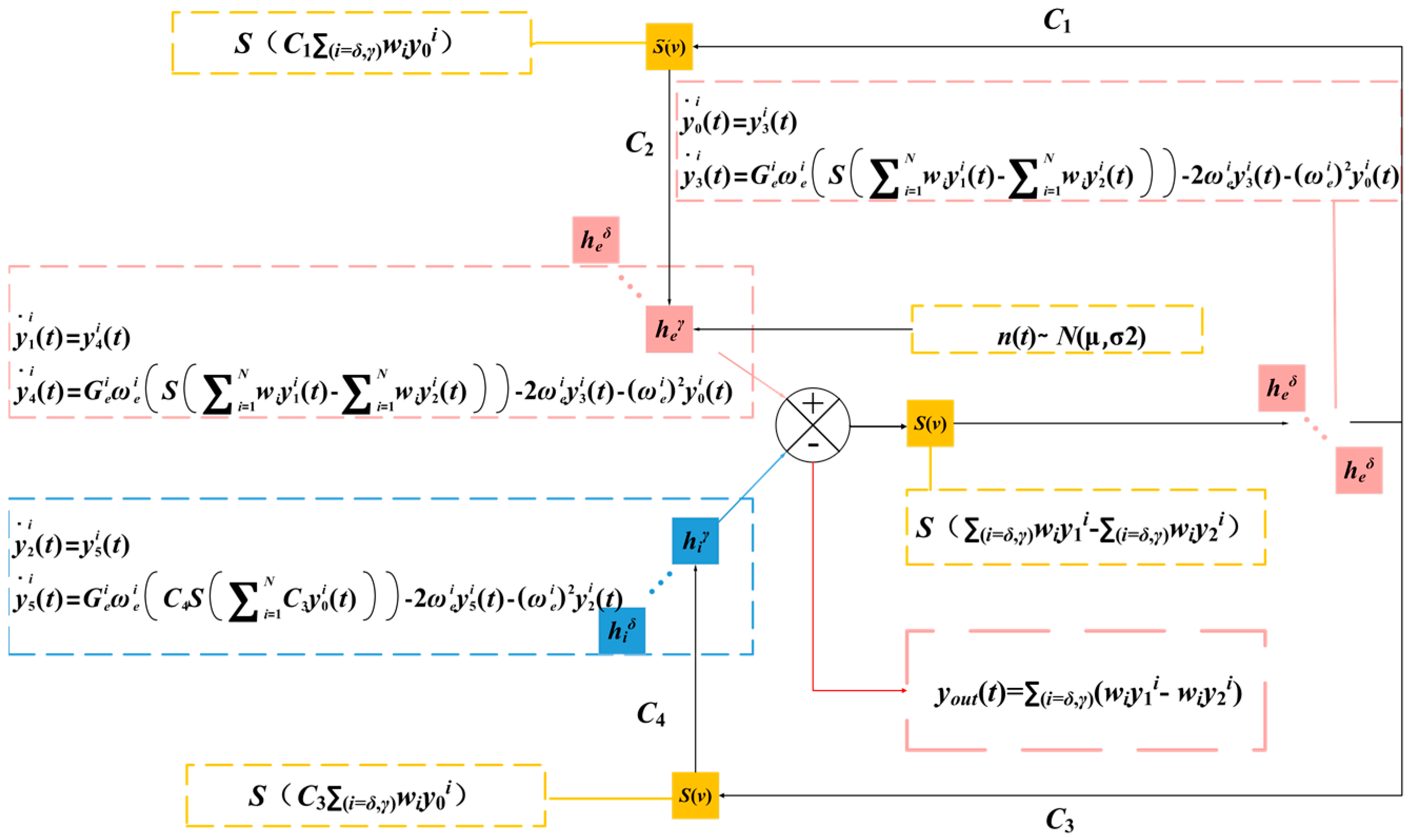
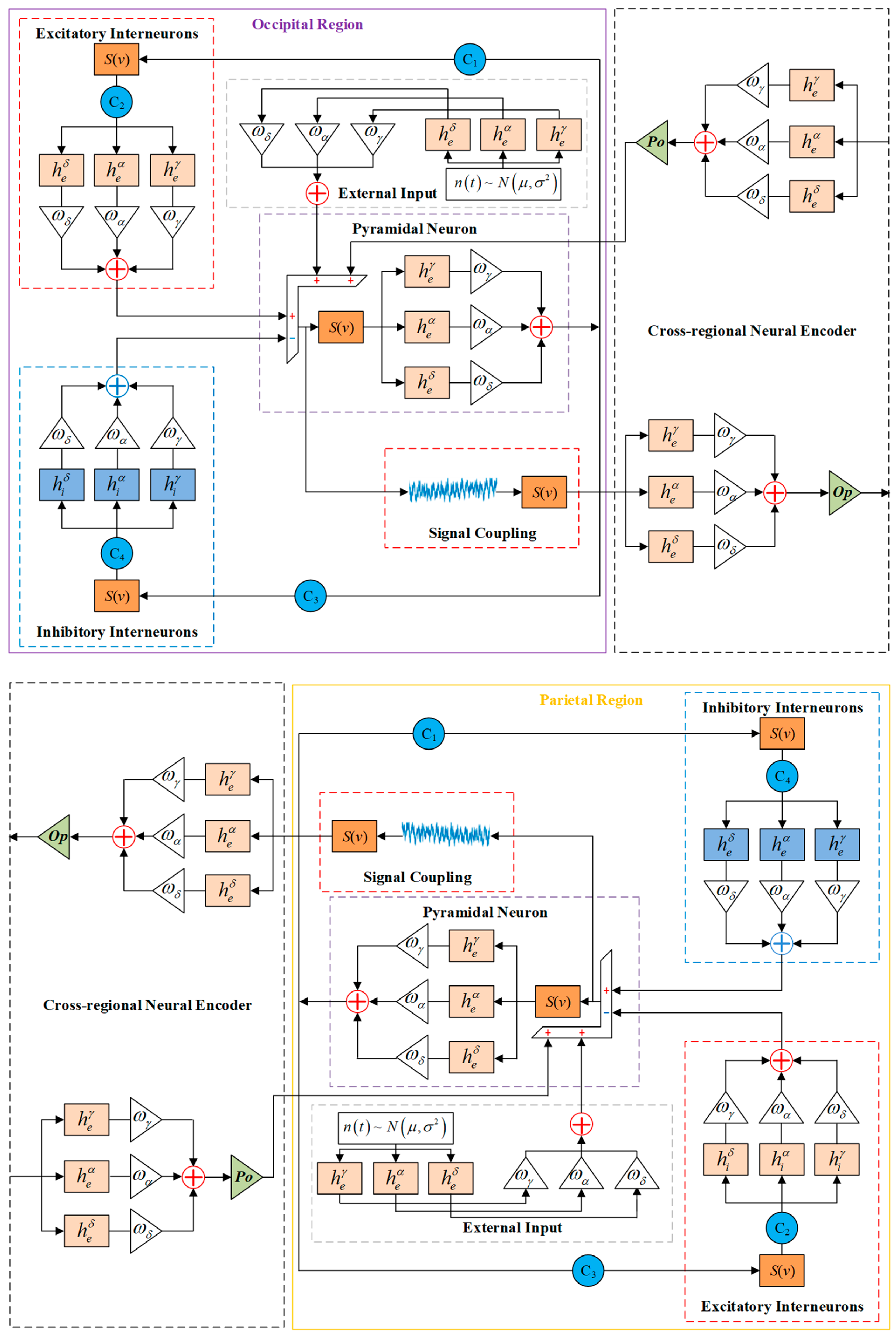
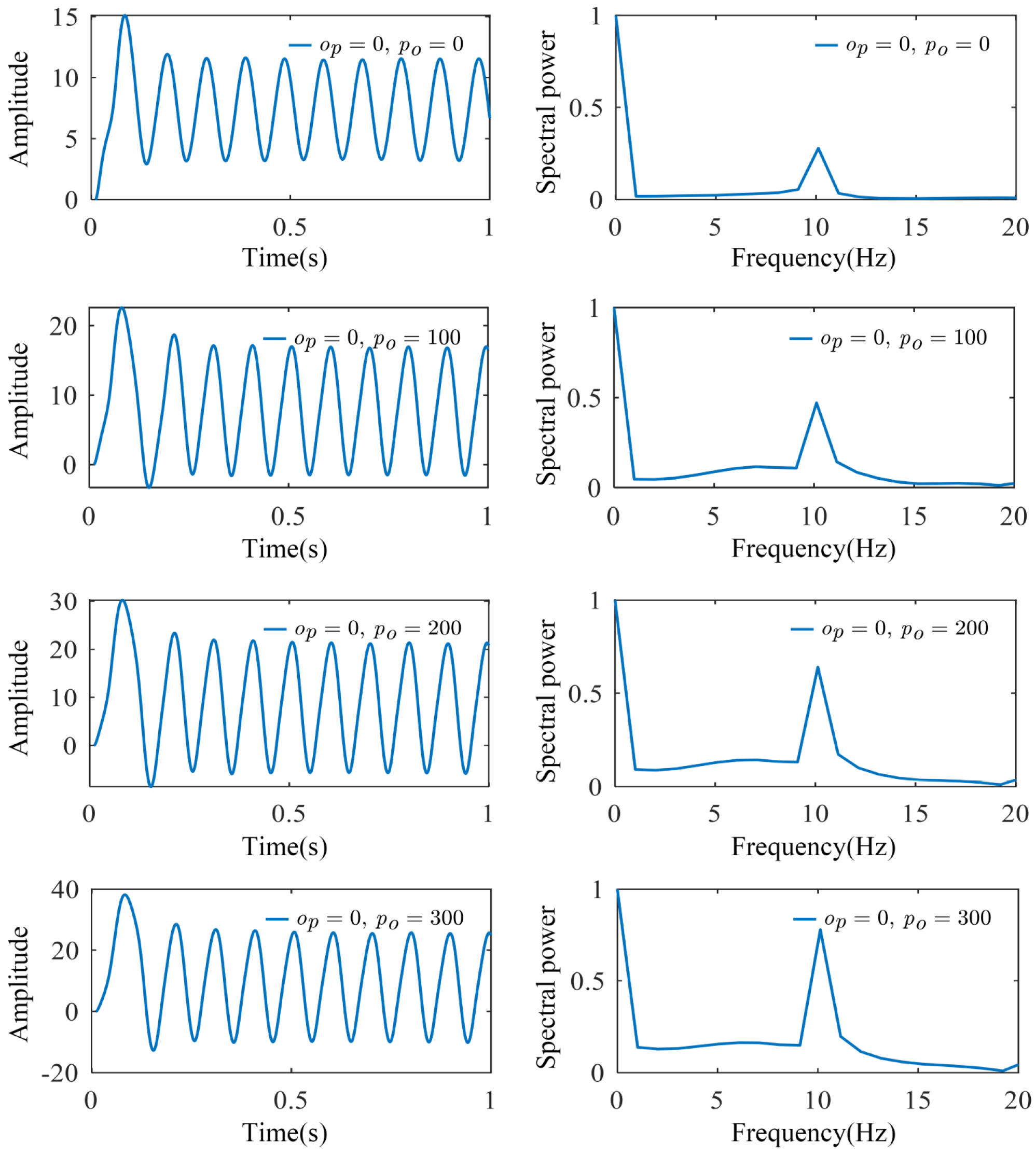
2.2. Multi-Dynamic Coupled NMM for SSVEP-BCI
- δ wave (0–4 Hz): characterizing low-frequency response properties;
- α wave (8–16 Hz): corresponding to the peak SSVEP response interval;
- γ wave (32–64 Hz): capturing high-frequency dynamics.
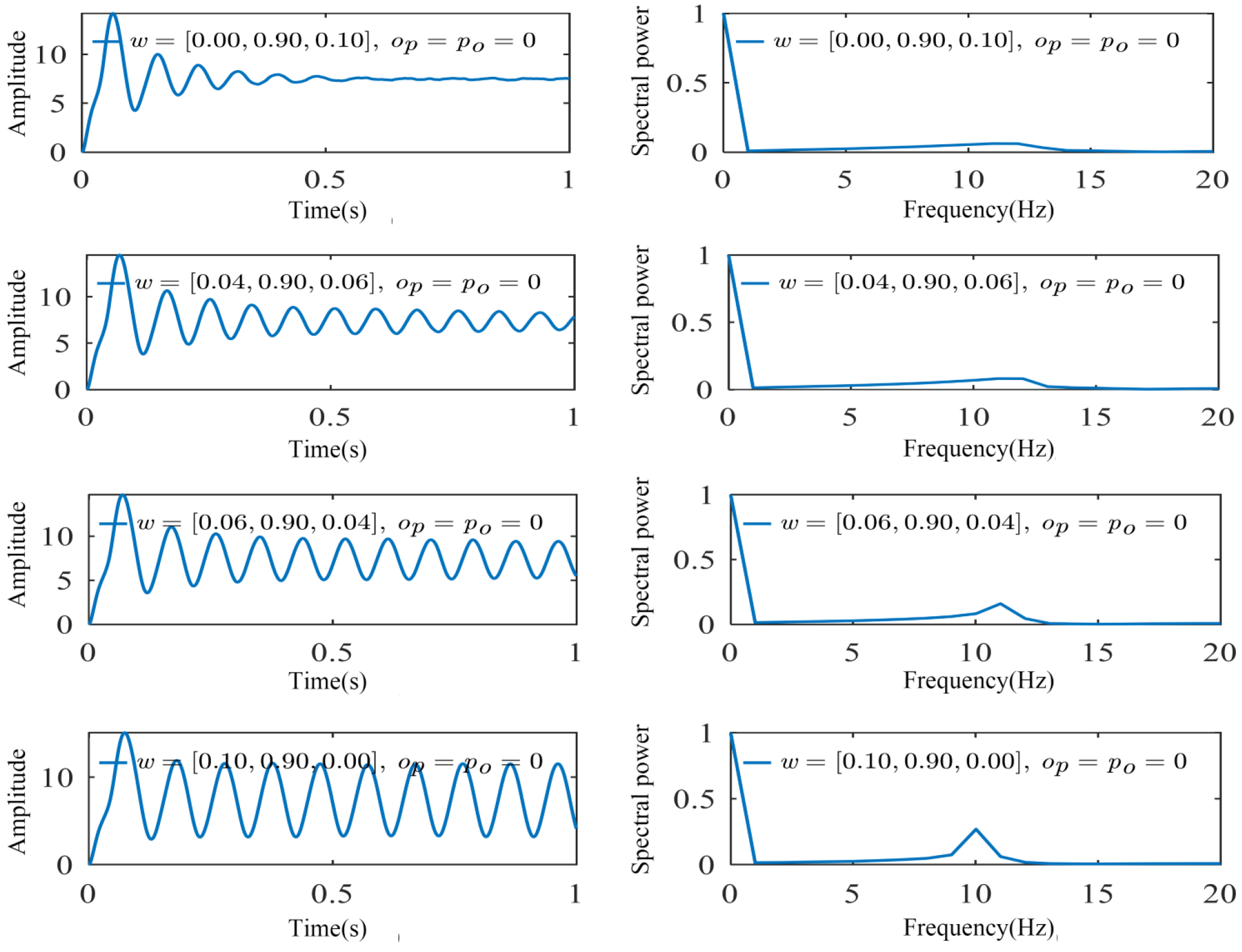
2.3. Weight Parameter Identification for Oscillators
2.4. Particle Swarm Optimization-Based Optimization for Identified Parameters
- Particle Representation and Initialization: Each particle is represented by a position vector in the solution space, and its velocity vector defines the direction and rate of search. For the i-th particle, its position is represented as Xi = (xi1, xi2, …, xin), and its velocity as Vi = (vi1, vi2, …, vin).
- Objective Function Definition: The Mean Squared Error (MSE) is used as the objective function to measure the difference between the model’s predicted frequency spectrum and the real collected frequency spectrum:
- 3.
- Particle Velocity and Position Update: Each particle updates its velocity and position according to the following formulas:
- 4.
- Update of Individual and Global Best Positions: Each particle dynamically updates its personal best (pbest) by retaining the parameter combination (position) that yields the lowest MSE value encountered during its search history. The swarm collaboratively identifies the global best (gbest)—i.e., the single position across all 50 particles that achieves the minimum MSE throughout the optimization process.
- 5.
- Termination Condition: The PSO terminates when the preset number of iterations (t = 70) is reached.
3. Results
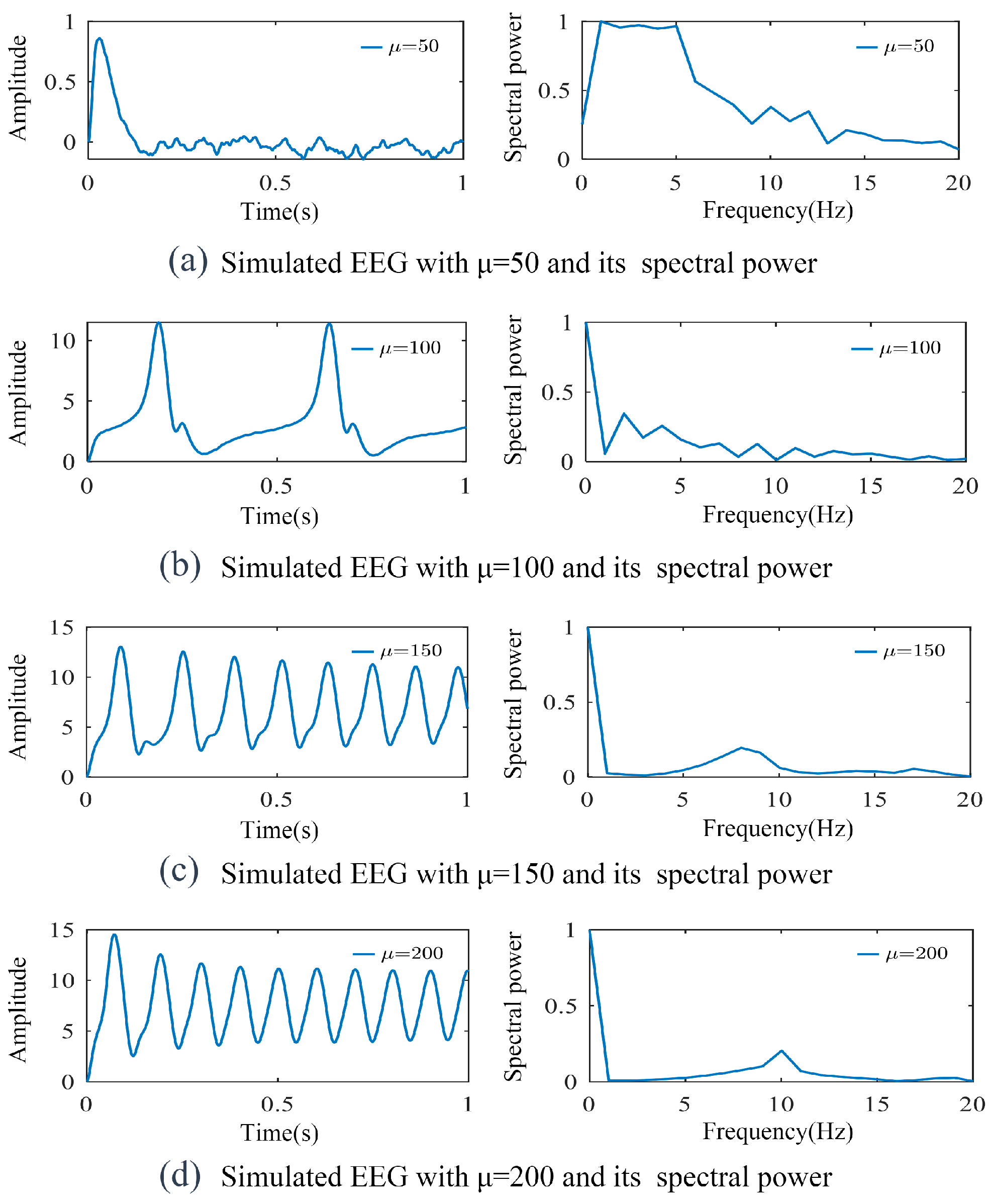
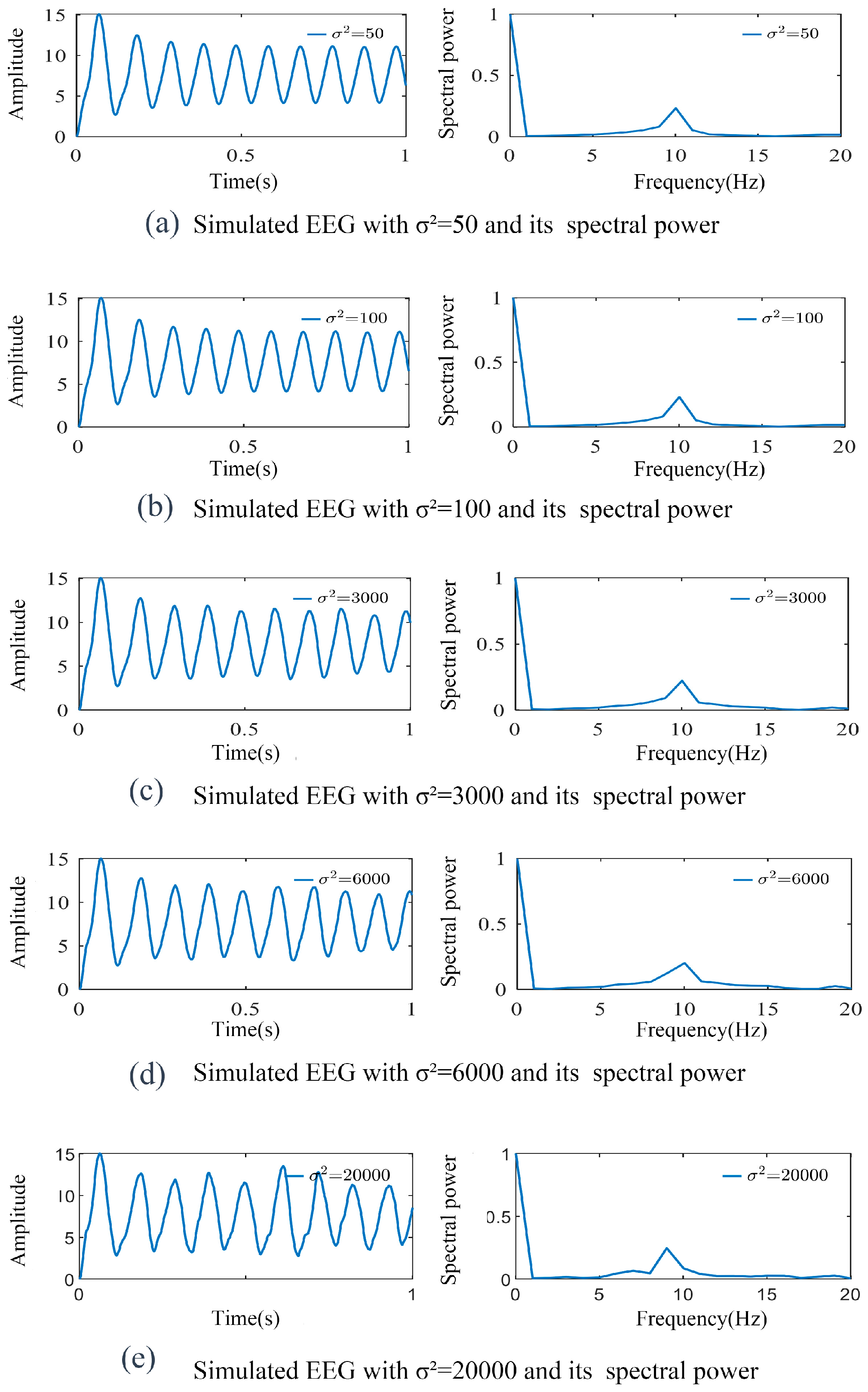
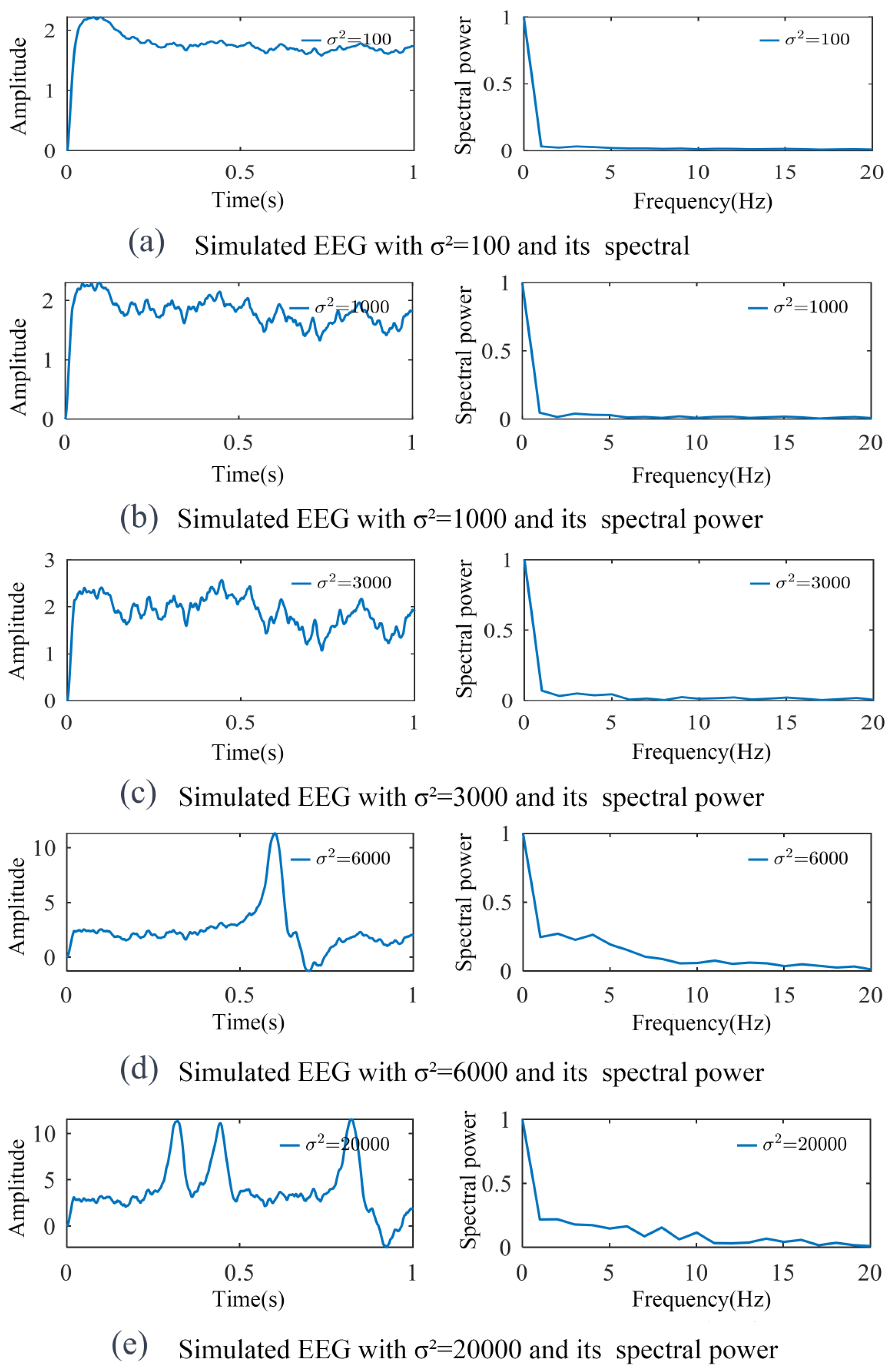
3.1. Simulation of SSVEP-BCI Multi-Dynamic Coupled Neural Mass Model
3.2. Parameter Identification and Model Validation
3.3. Simulated SSVEP Signal Processing and Classification
4. Discussion
- Incorporating additional brain regions, such as the prefrontal and temporal areas, into the model. This would involve developing a systematic framework based on the functional connectivity of the cerebral cortex network, thereby enhancing the physiological realism of the model. However, this may also lead to an increase in computational complexity; therefore, future research will need to strike a balance between simulation fidelity and computational efficiency.
- Investigating the integration of advanced techniques, such as adaptive chaotic PSO or deep reinforcement learning, to further improve the model’s adaptability and robustness in dynamic and complex environments. Additionally, there is also the need to further examine the effects of individual neural system variations on SSVEP signals. This could lead to the development of more refined adaptation strategies, ultimately enhancing the model’s universality and effectiveness for diverse users.
- Validating the overall performance of the SSVEP-integrated BCI model. To ensure that the generated signal can be used by real SSVEP-BCIs, more performance comparisons need to be conducted to verify if the simulated SSVEP signal is suitable for current algorithms oriented towards real SSVEP signal processing. Whether there are significant differences in performance comparisons under machine learning and deep learning methods in the processing steps, such as preprocessing, feature extraction, and feature classification, needs to be further verified.
- Extending the model to other non-visual BCI modalities, such as motor imagery- and auditory-based paradigms. However, these signals have different frequency characteristics and dynamic patterns, requiring specific adjustments to the model’s structure, dynamic coupling, and optimized parameters.
- Extending the model’s applications to a range of devices, including brain-controlled drones, wheelchairs, and smart home systems. This will involve optimizing the mechanisms for controlling multiple devices simultaneously, ensuring precise signal mapping. Moreover, conducting performance validation in dynamic real-world environments also helps to assess the model’s reliability, stability, and practical applicability.
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nam, C.S.; Choi, I.; Wadeson, A.; Whang, M. Brain Computer Interface. An Emerging Interaction Technology. In Brain Computer Interfaces Handbook: Technological and Theoretical Advances; Pergamon Press: Oxford, UK, 2018; pp. 11–52. [Google Scholar]
- Herath, H.M.K.K.M.B.; de Mel, W.R. Controlling an Anatomical Robot Hand Using the Brain-Computer Interface Based on Motor Imagery. Adv. Hum.-Comput. Interact. 2021, 2021, 5515759. [Google Scholar] [CrossRef]
- Malki, A.; Yang, C.; Wang, N.; Li, Z. Mind Guided Motion Control of Robot Manipulator Using EEG Signals. In Proceedings of the International Conference on Information Science and Technology, Sanya, China, 14–15 November 2015. [Google Scholar]
- Sandesh, R.S.; Venkatesan, N. Steady State VEP-Based BCI to Control a Five-Digit Robotic Hand Using LabVIEW. Int. J. Biomed. Eng. Technol. 2022, 38, 109–127. [Google Scholar] [CrossRef]
- Li, H.; Bi, L.; Li, X. Robust Predictive Control for EEG-Based Brain-Robot Teleoperation. IEEE Trans. Intell. Transp. Syst. 2024, 25, 9130–9140. [Google Scholar] [CrossRef]
- Li, H.; Bi, L.; Yi, J. Sliding-Mode Nonlinear Predictive Control of Brain-Controlled Mobile Robots. IEEE Trans. Cybern. 2022, 52, 5419–5431. [Google Scholar] [CrossRef] [PubMed]
- Müller-Putz, G.R.; Pfurtscheller, G. Control of an Electrical Prosthesis with an SSVEP-Based BCI. IEEE Trans. Biomed. Eng. 2008, 55, 361–364. [Google Scholar] [CrossRef]
- Li, H.; Li, X.; Millán, J.D.R. Noninvasive EEG-Based Intelligent Mobile Robots: A Systematic Review. IEEE Trans. Autom. Sci. Eng. 2024. [Google Scholar] [CrossRef]
- Jansen, B.H.; Zouridakis, G.; Brandt, M.E. A Neurophysiologically-Based Mathematical Model of Flash Visual Evoked Potentials. Biol. Cybern. 1993, 68, 275–283. [Google Scholar] [CrossRef]
- Jansen, B.H.; Rit, V.G. Electroencephalogram and Visual Evoked Potential Generation in a Mathematical Model of Coupled Cortical Columns. Biol. Cybern. 1995, 73, 357–366. [Google Scholar] [CrossRef]
- Wendling, F.; Bellanger, J.J.; Bartolomei, F. Relevance of Nonlinear Lumped-Parameter Models in the Analysis of Depth-EEG Epileptic Signals. Biol. Cybern. 2000, 83, 367–378. [Google Scholar] [CrossRef]
- Wendling, F. Neurocomputational Models in the Study of Epileptic Phenomena. J. Clin. Neurophysiol. 2005, 22, 285–287. [Google Scholar]
- Wendling, F.; Bartolomei, F.; Bellanger, J.J. Epileptic Fast Activity Can Be Explained by a Model of Impaired GABAergic Dendritic Inhibition. Eur. J. Neurosci. 2002, 15, 1499–1508. [Google Scholar] [CrossRef] [PubMed]
- David, O.; Friston, K.J. A Neural Mass Model for MEG/EEG: Coupling and Neuronal Dynamics. NeuroImage 2003, 20, 1743–1755. [Google Scholar] [CrossRef]
- Cui, D.; Li, X.L.; Ji, X.Q. Multichannel Neural Mass Modelling and Analyzing. Sci. China Ser. F Inf. Sci. 2011, 54, 1283–1292. [Google Scholar] [CrossRef]
- Eeckman, F.H.; Freeman, W.J. Asymmetric Sigmoid Non-Linearity in the Rat Olfactory System. Brain Res. 1991, 557, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Robinson, P.A.; Rennie, C.J.; Wright, J.J. Prediction of Electroencephalographic Spectra from Neurophysiology. Phys. Rev. E 2001, 63, 021903. [Google Scholar] [CrossRef]
- Wu, L.; Liu, A.; Ward, R.K. Signal Processing for Brain-Computer Interfaces: A Review and Current Perspectives. IEEE Signal Process. Mag. 2023, 40, 80–91. [Google Scholar] [CrossRef]
- Fang, J.; Liu, W.; Chen, L. A Survey of Algorithms, Applications and Trends for Particle Swarm Optimization. Int. J. Netw. Dyn. Intell. 2023, 2, 24–50. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, X.; Gao, X. A Benchmark Dataset for SSVEP-Based Brain-Computer Interfaces. IEEE Trans. Neural Syst. Rehabil. Eng. 2017, 25, 1746–1752. [Google Scholar] [CrossRef]
- Liu, B.; Huang, X.; Wang, Y. BETA: A Large Benchmark Database Toward SSVEP-BCI Application. Front. Neurosci. 2020, 14, 00627. [Google Scholar] [CrossRef]
- Gao, S.; Yang, J.; Shen, T.; Jiang, W. A Parallel Feature Fusion Network Combining GRU and CNN for Motor Imagery EEG Decoding. Brain Sci. 2022, 12, 1233. [Google Scholar] [CrossRef]
- Kobayashi, N.; Ishizuka, K. LSTM-Based Classification of Multiflicker-SSVEP in Single Channel Dry-EEG for Low-Power/High-Accuracy Quadcopter-BMI System. In Proceedings of the IEEE International Conference on Systems, Man and Cybernetics, Bari, Italy, 6–9 October 2019. [Google Scholar]
- Aznan, N.K.N.; Bonner, S.; Connolly, J.; Al Moubayed, N.; Breckon, T. On the Classification of SSVEP-Based Dry-EEG Signals via Convolutional Neural Networks. In Proceedings of the IEEE International Conference on Systems, Man, and Cybernetics, Miyazaki, Japan, 7–10 October 2018. [Google Scholar]
- Eldele, E.; Chen, Z.; Liu, C. An Attention-Based Deep Learning Approach for Sleep Stage Classification with Single-Channel EEG. IEEE Trans. Neural Syst. Rehabil. Eng. 2021, 29, 809–817. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Lu, C.; Yang, M. A Hybrid Method for Heartbeat Classification via Convolutional Neural Networks, Multilayer Perceptrons and Focal Loss. PeerJ Comput. Sci. 2020, 6, 324. [Google Scholar] [CrossRef] [PubMed]
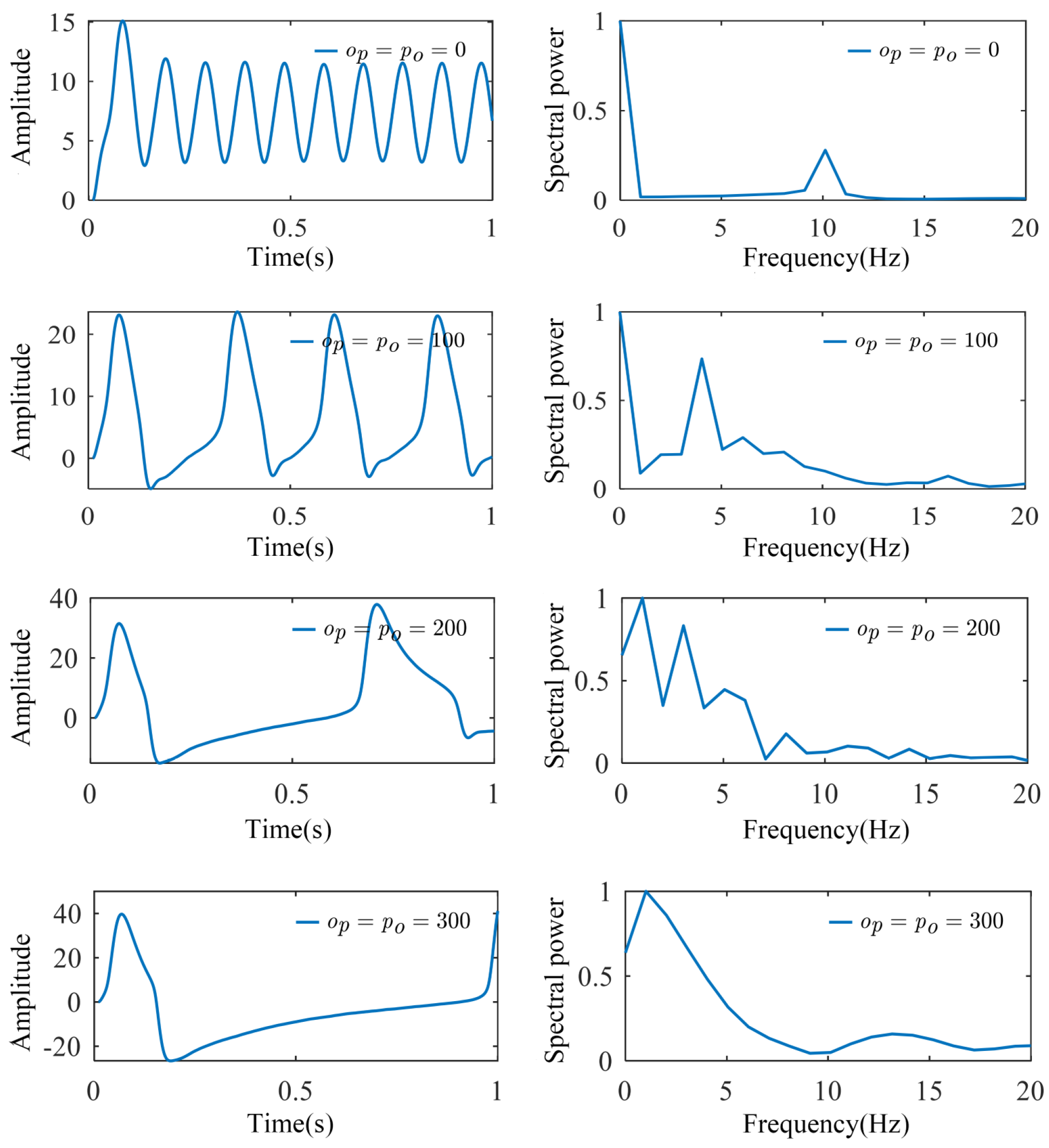
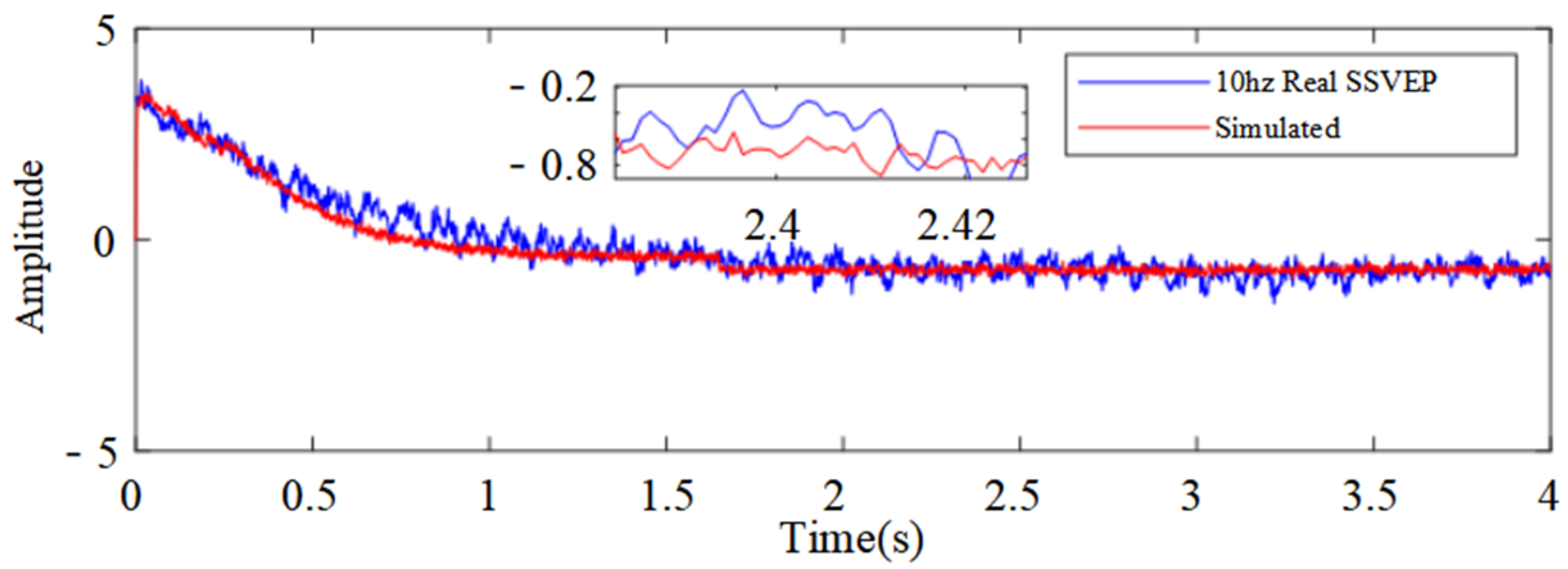
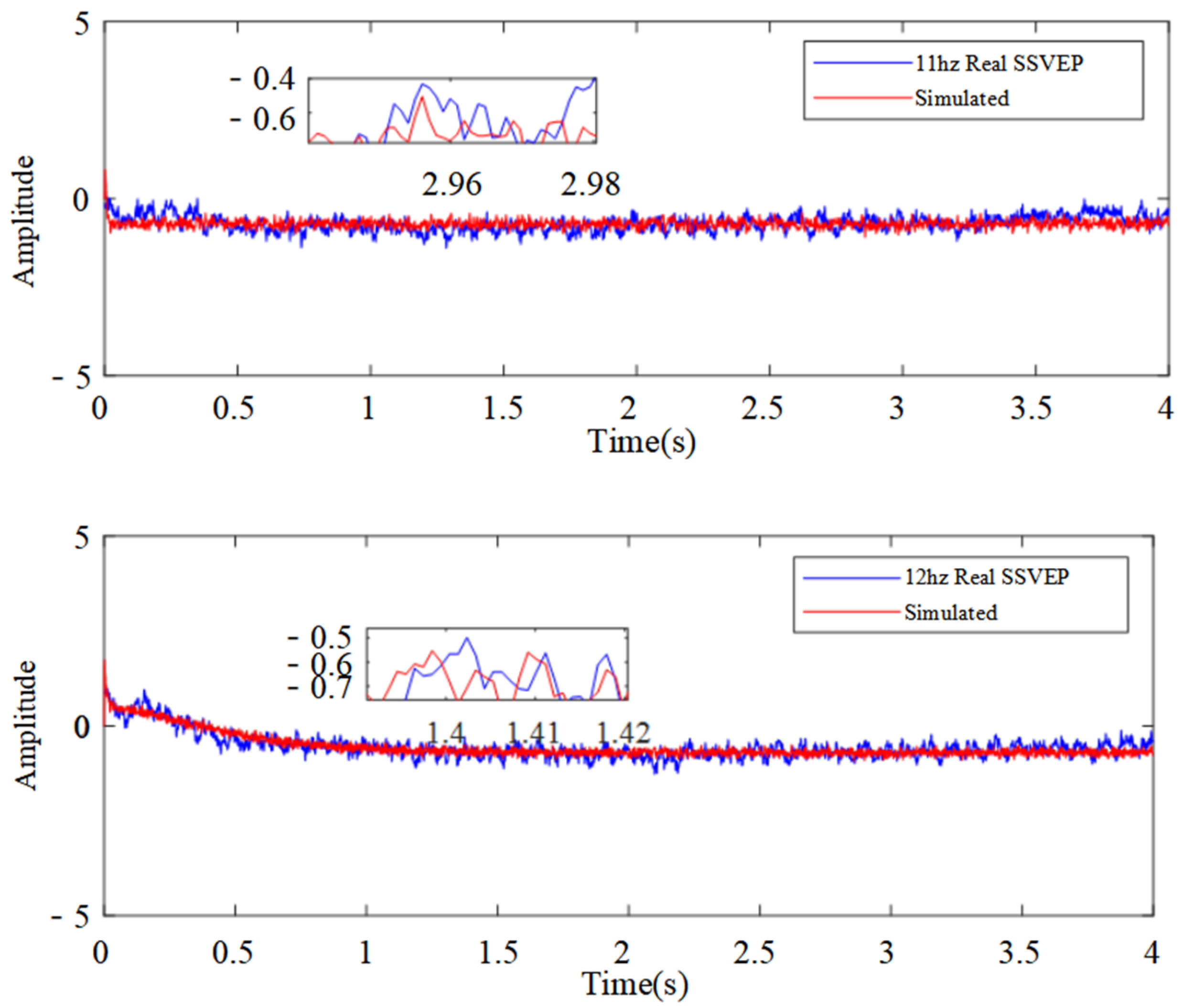
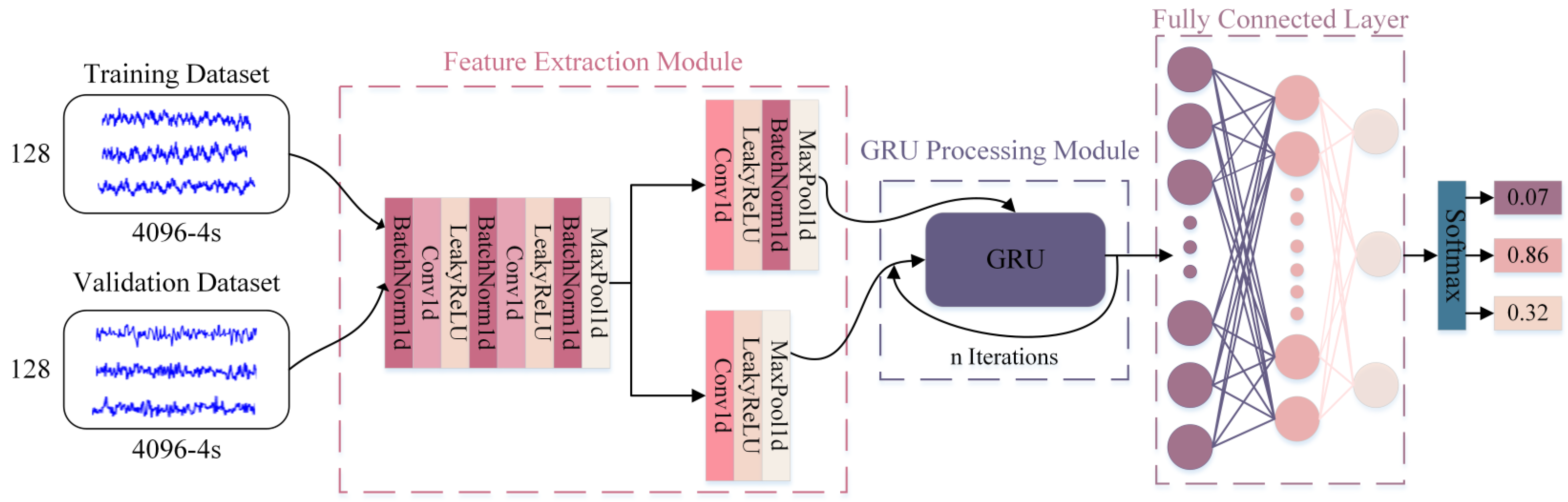
| Visual Stimulus Frequency | Energy Ratio of Rhythmic Wave Bands | ||
|---|---|---|---|
| 0–4 Hz (δ) | 8–16 Hz (α) | 32–64 Hz (γ) | |
| 10 Hz | 0.0459 | 0.1239 | 0.8303 |
| 11 Hz | 0.0400 | 0.1331 | 0.8269 |
| 12 Hz | 0.0384 | 0.1280 | 0.8336 |
| Visual Stimulus Frequency | The Coupling Coefficient and Gaussian White Noise Optimization Parameters | |||||
|---|---|---|---|---|---|---|
| op | po | μo | σ2o | μp | σ2p | |
| 10 Hz | 1234.18 | 17.78 | 1628.27 | 1709.47 | 1600.13 | 7345.33 |
| 11 Hz | 359.44 | 64.11 | 1354.51 | 17,978.49 | 1436.16 | 2261.88 |
| 12 Hz | 1566.06 | 370.92 | 1741.10 | 4611.35 | 899.50 | 7798.72 |
| Visual Stimulus Frequency | Performance Metrics | ||
|---|---|---|---|
| MAE | RMSE | ME | |
| 10 Hz | 0.3226 | 0.4182 | 2.2861 |
| 11 Hz | 0.1953 | 0.2458 | 1.1009 |
| 12 Hz | 0.1520 | 0.1906 | 0.8430 |
| SVM | LSTM | AttnSleep | CMFnet | FPF-Net | |
|---|---|---|---|---|---|
| Training set | 0.512 | 0.731 | 0.729 | 0.756 | 0.821 |
| Test set | 0.475 | 0.726 | 0.714 | 0.744 | 0.813 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, H.; Wang, Y.; Fu, P. A Novel Multi-Dynamic Coupled Neural Mass Model of SSVEP. Biomimetics 2025, 10, 171. https://doi.org/10.3390/biomimetics10030171
Li H, Wang Y, Fu P. A Novel Multi-Dynamic Coupled Neural Mass Model of SSVEP. Biomimetics. 2025; 10(3):171. https://doi.org/10.3390/biomimetics10030171
Chicago/Turabian StyleLi, Hongqi, Yujuan Wang, and Peirong Fu. 2025. "A Novel Multi-Dynamic Coupled Neural Mass Model of SSVEP" Biomimetics 10, no. 3: 171. https://doi.org/10.3390/biomimetics10030171
APA StyleLi, H., Wang, Y., & Fu, P. (2025). A Novel Multi-Dynamic Coupled Neural Mass Model of SSVEP. Biomimetics, 10(3), 171. https://doi.org/10.3390/biomimetics10030171






