An Overview of Fruit Allergens: Structural, Functional, Phylogenetical, and Clinical Aspects
Abstract
:1. Introduction
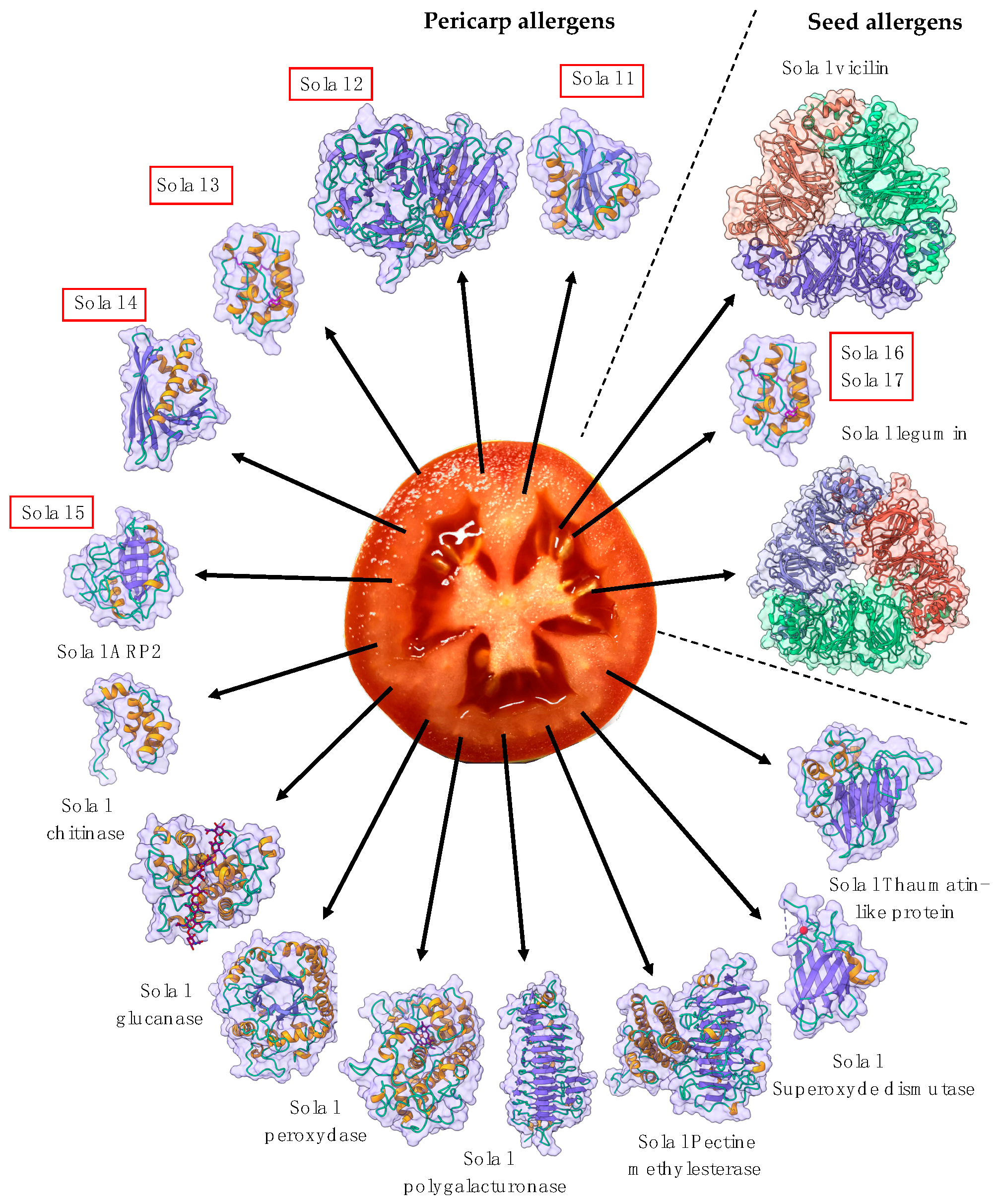
2. Repertoire of the Main Fruit Allergen Families
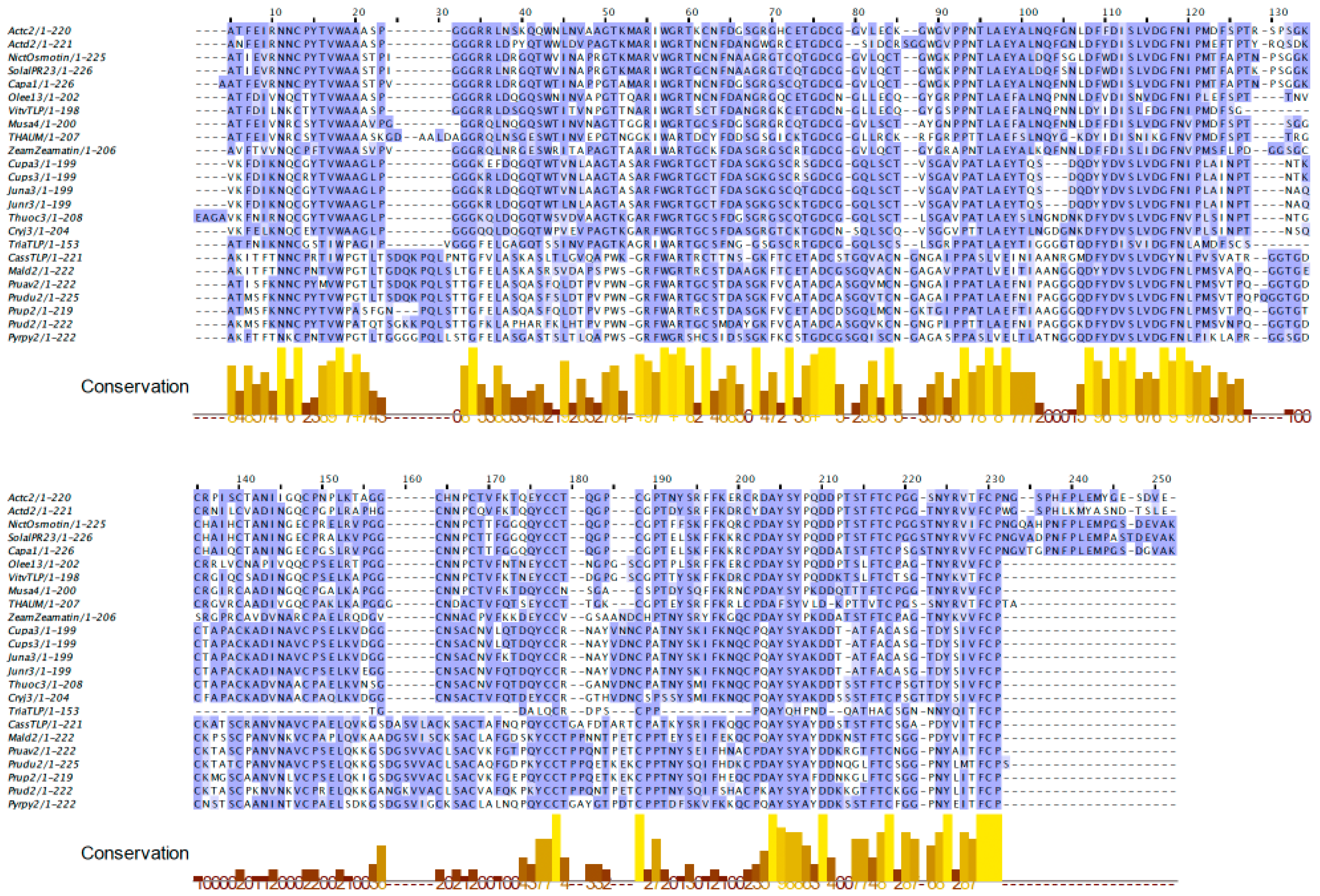
2.1. PR-5 Thaumatin-like Proteins (TLP)
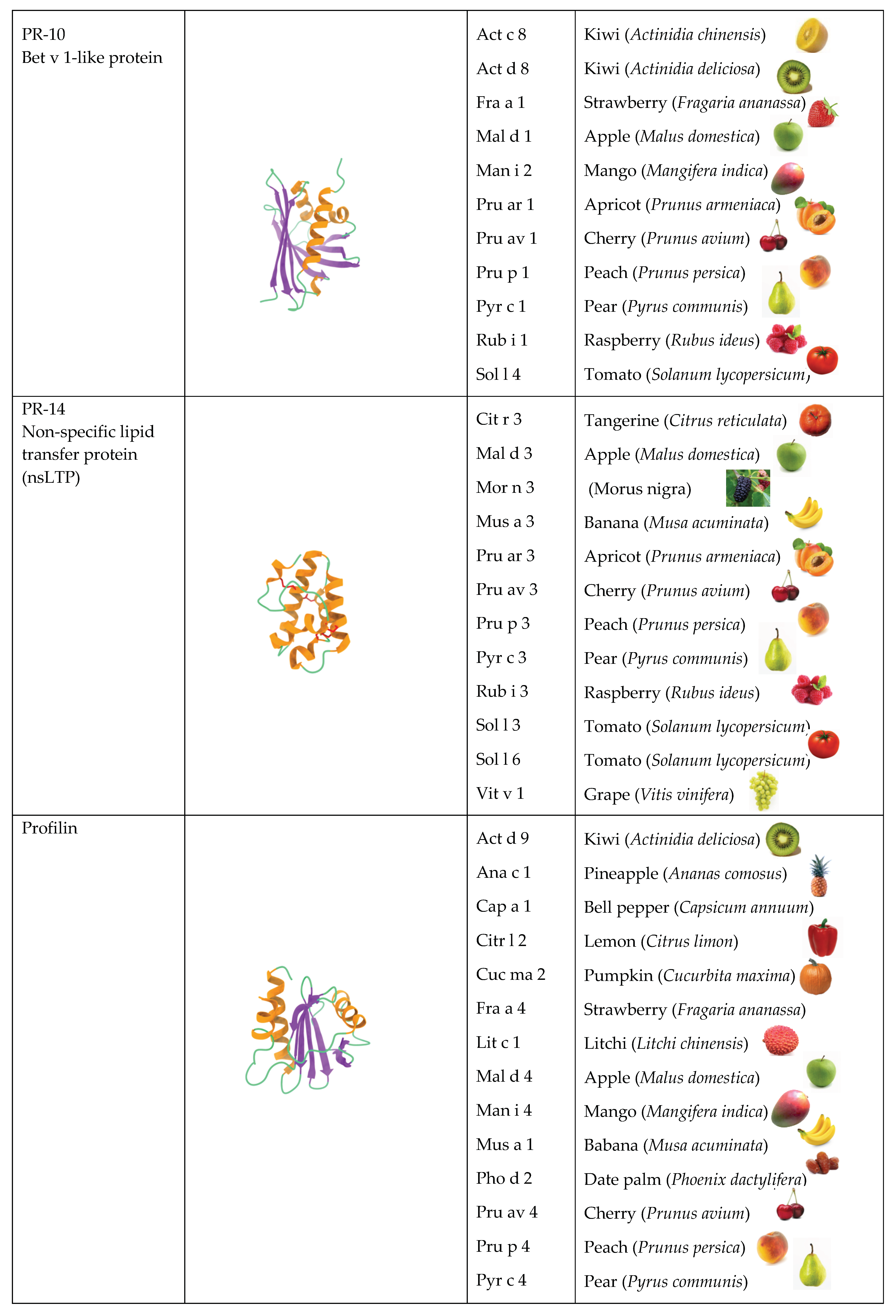
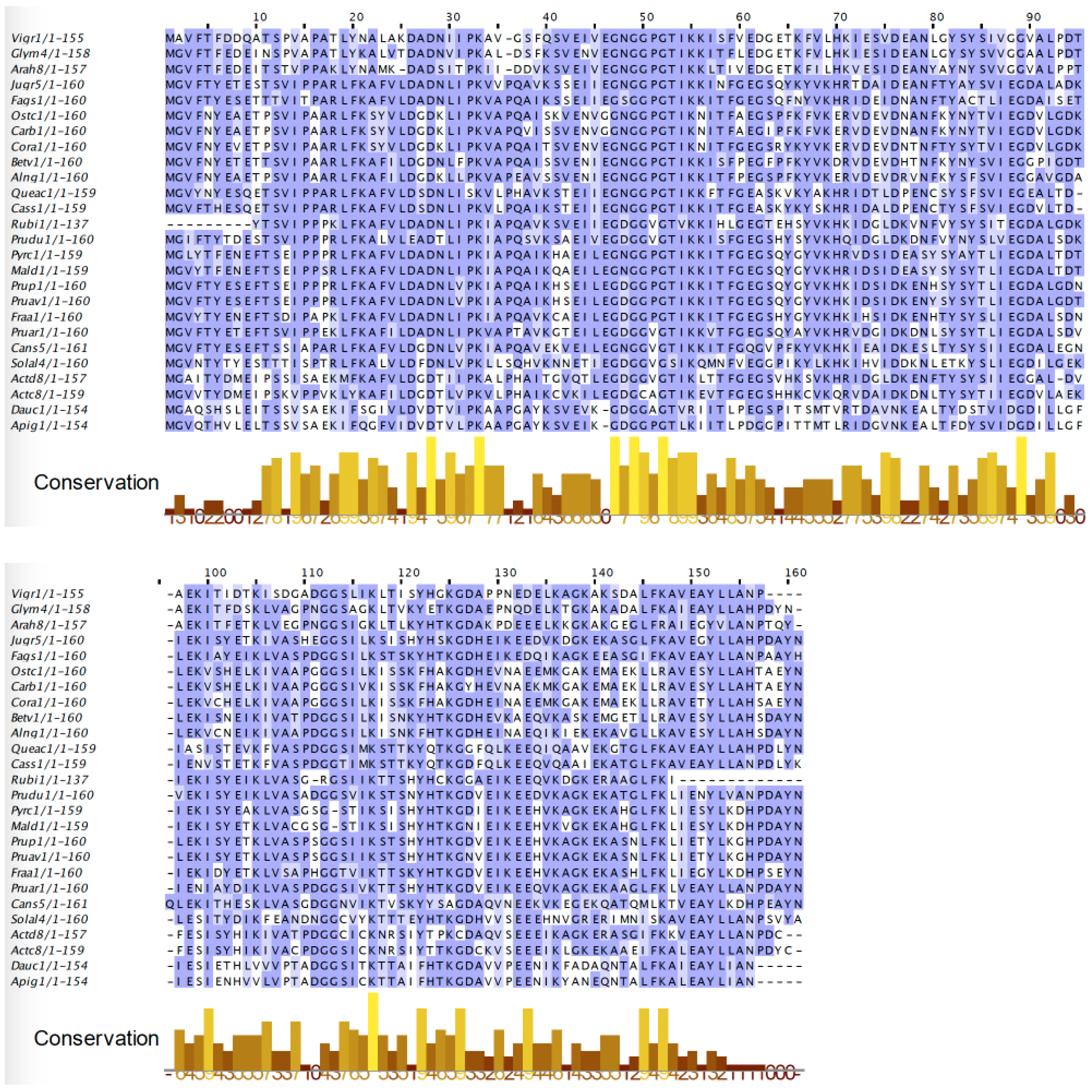
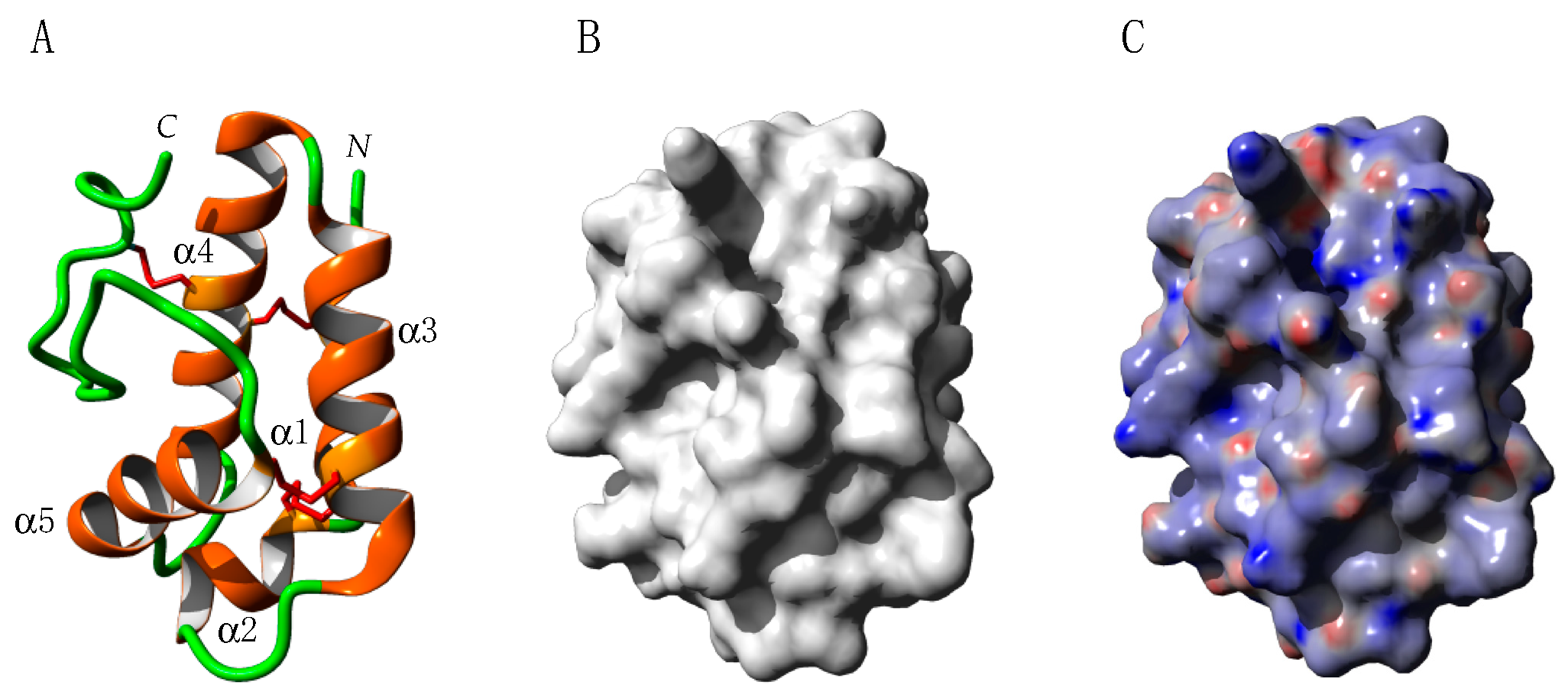
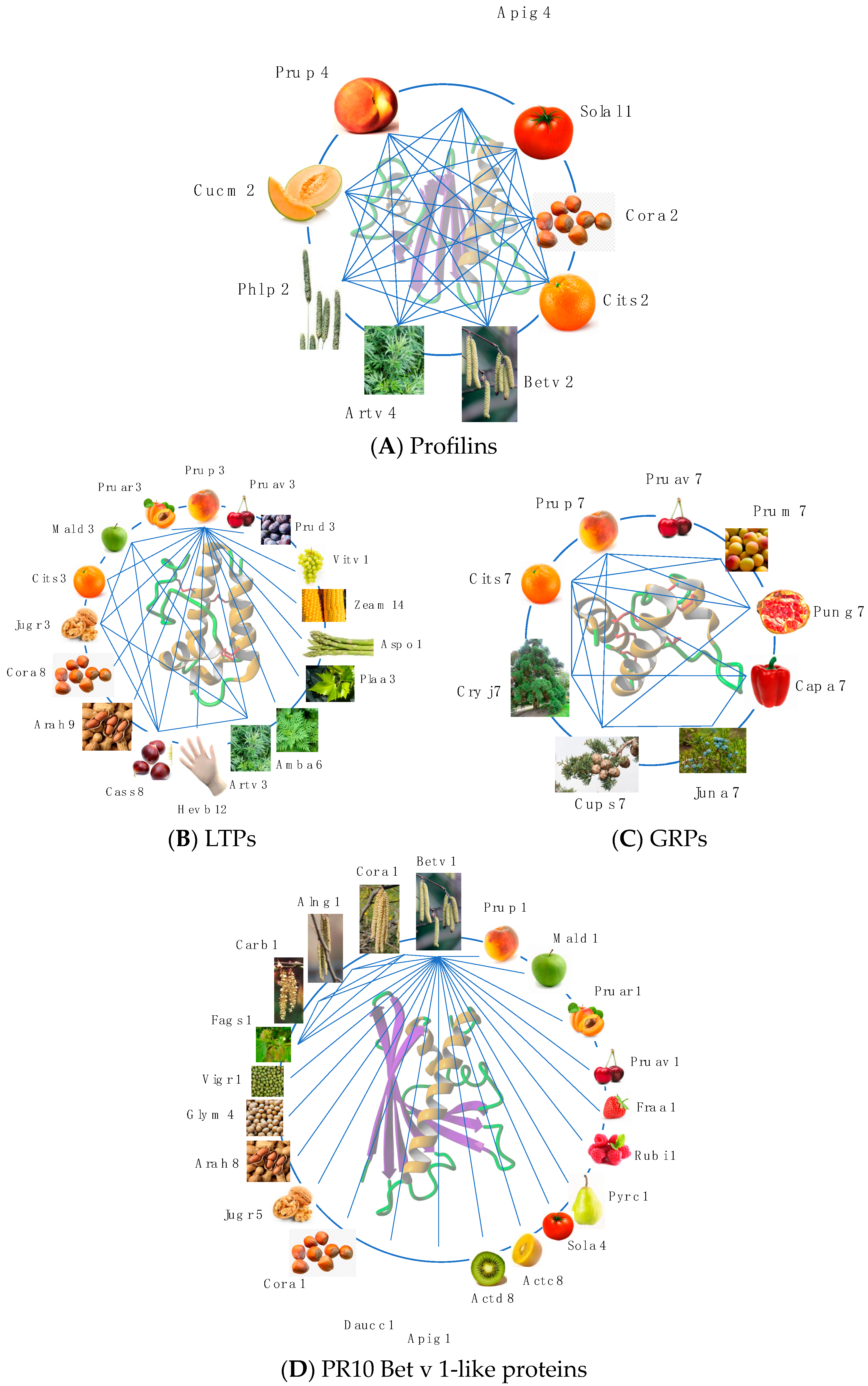
2.2. PR-10 Bet v 1-like Proteins
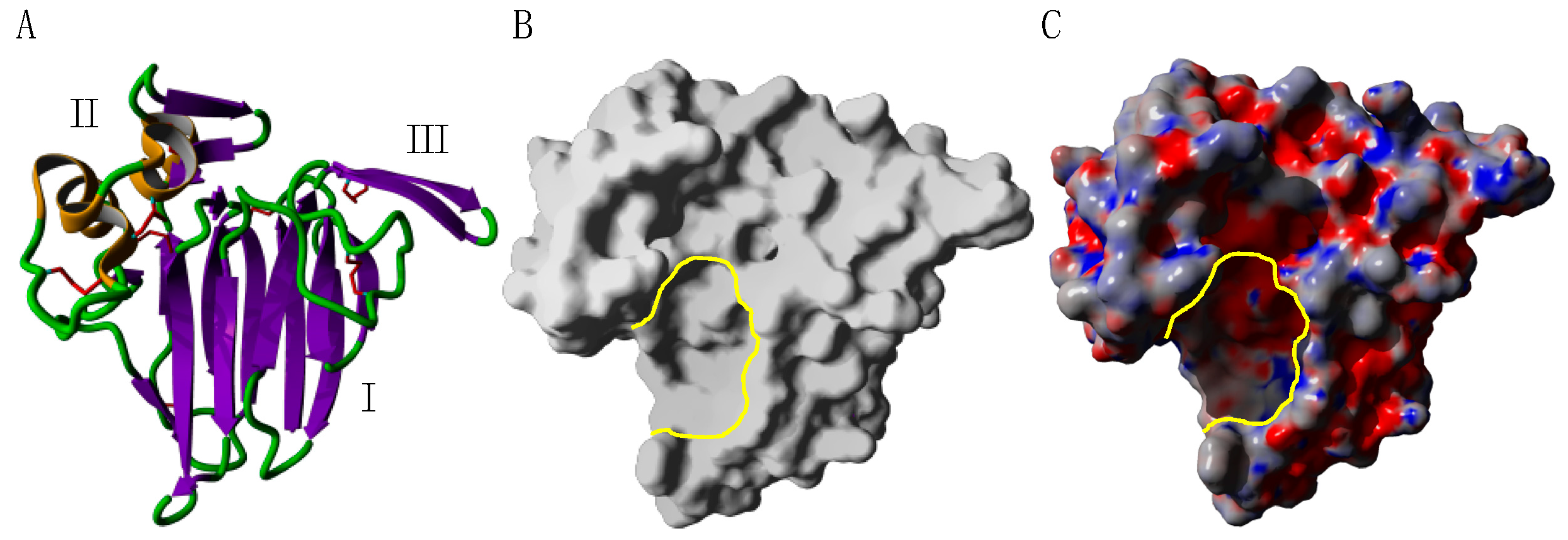
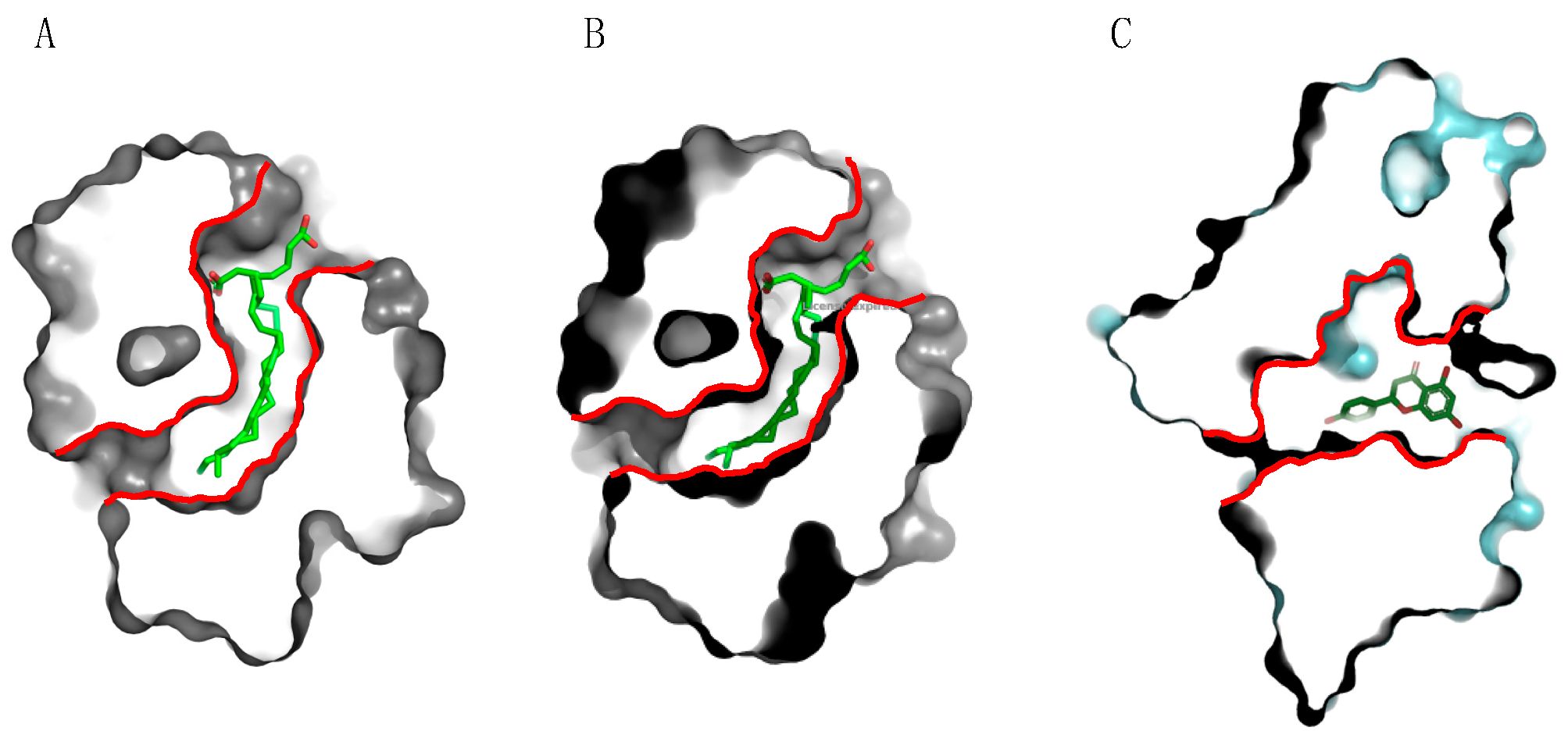
2.3. PR14 Non-Specific Lipid Transfer Proteins (nsLTP)
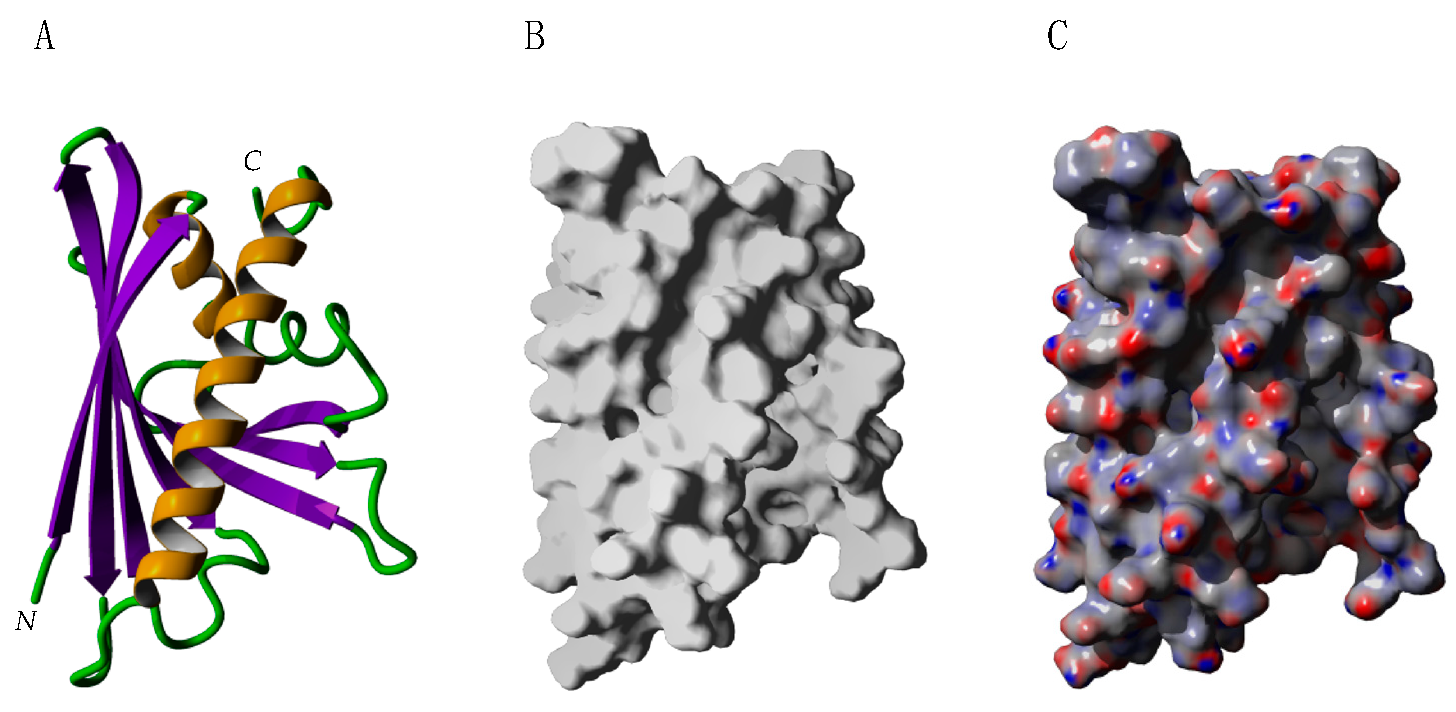
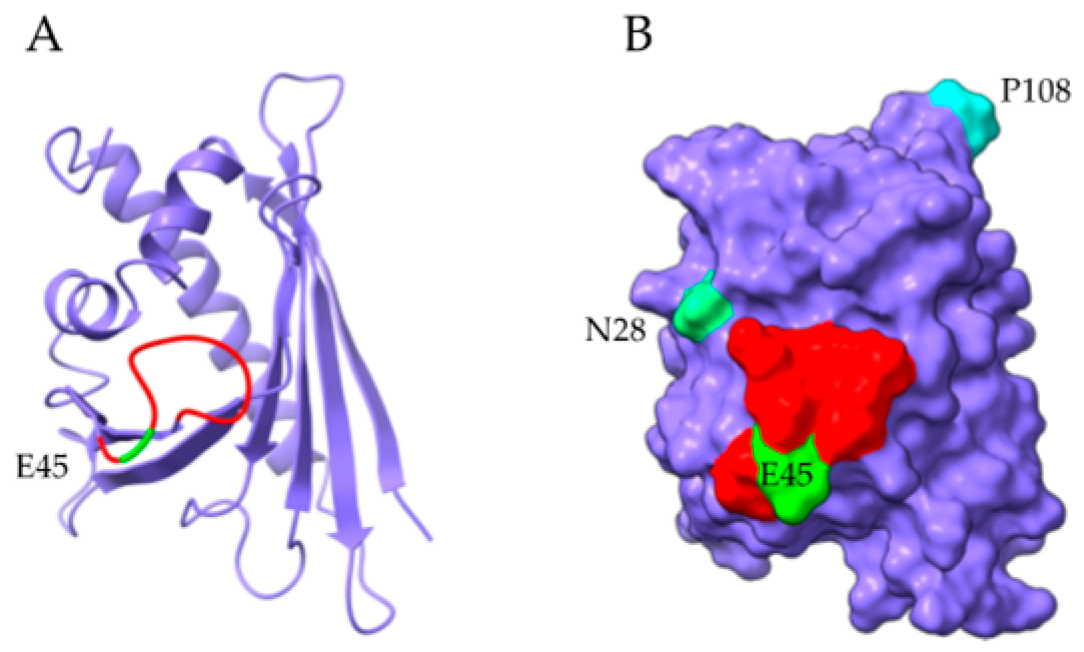
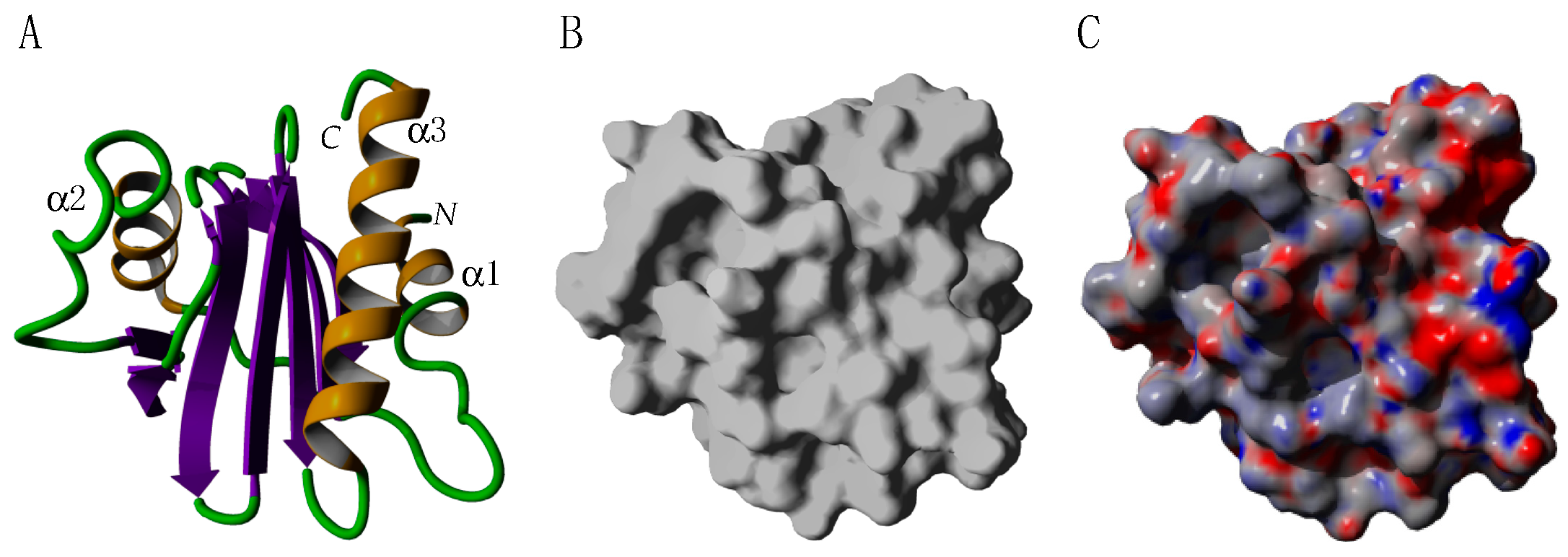
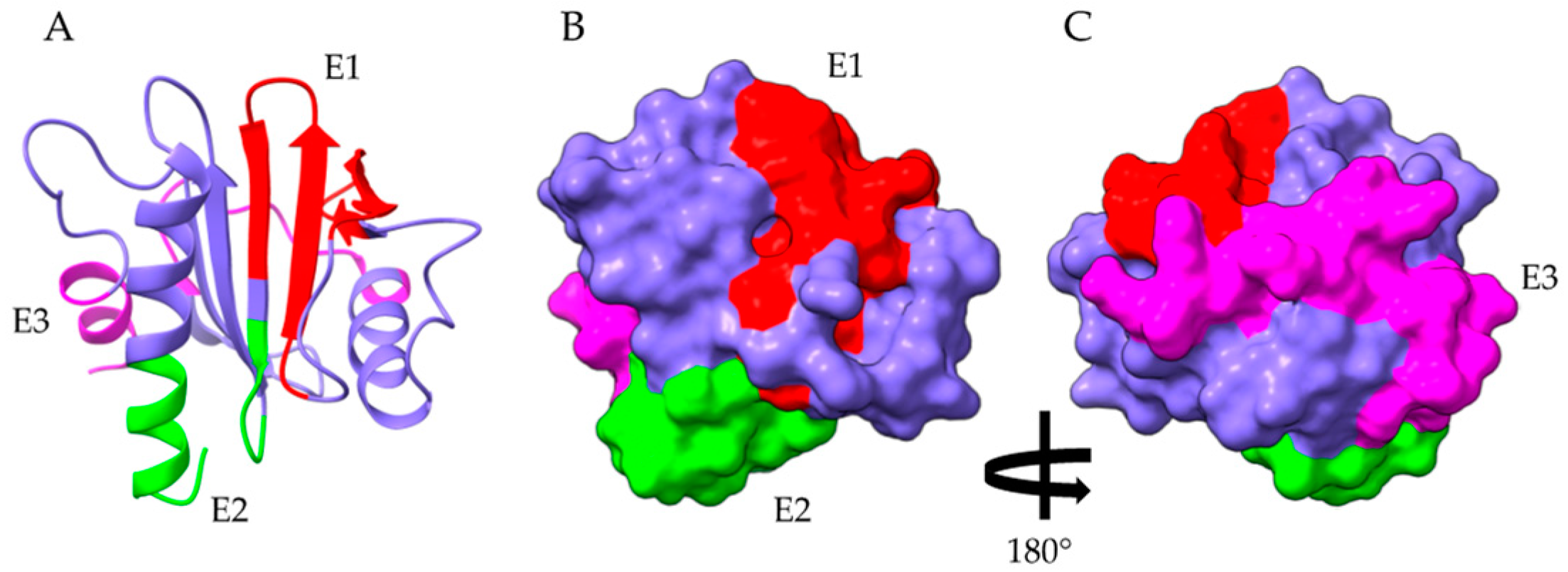
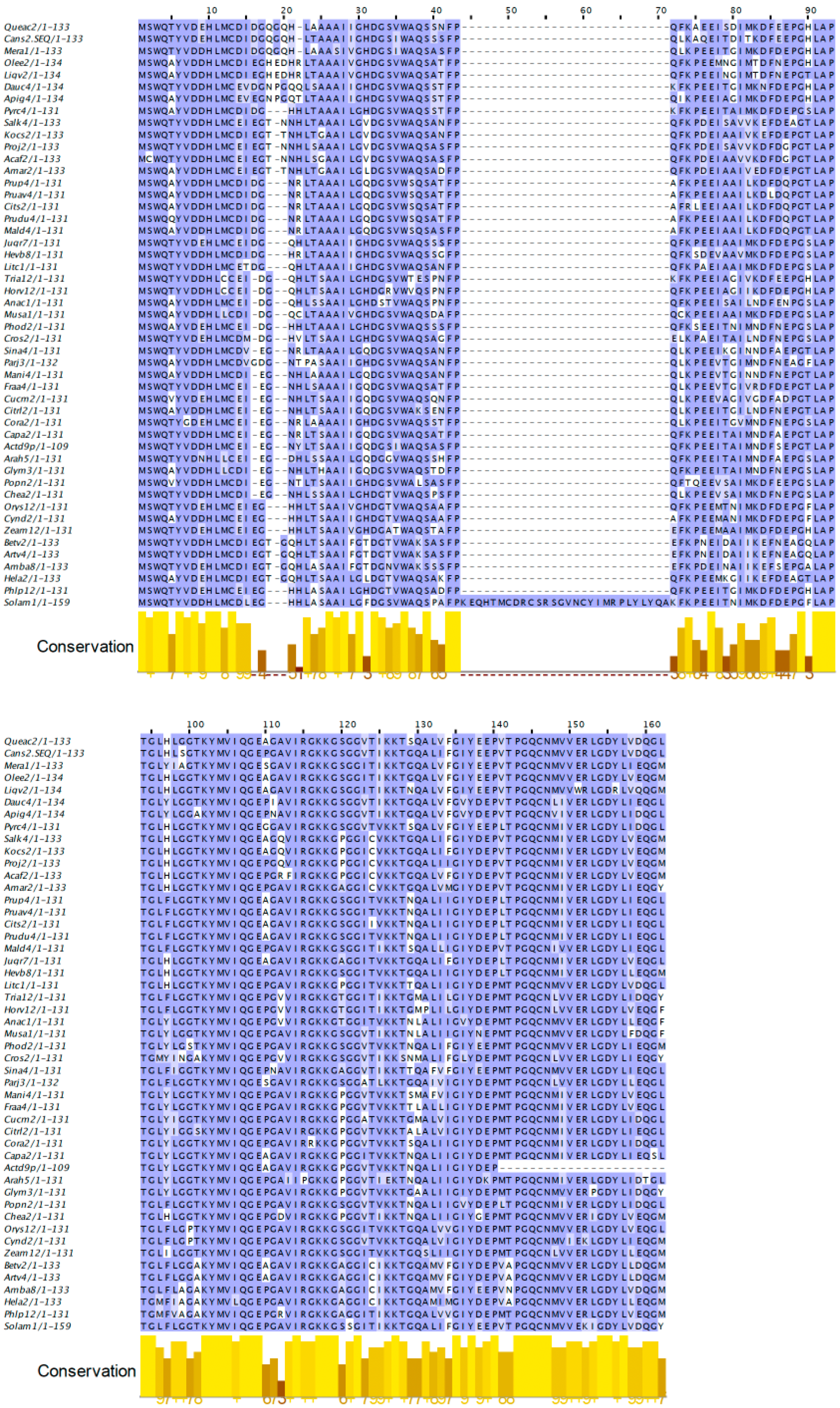
2.4. Profilins
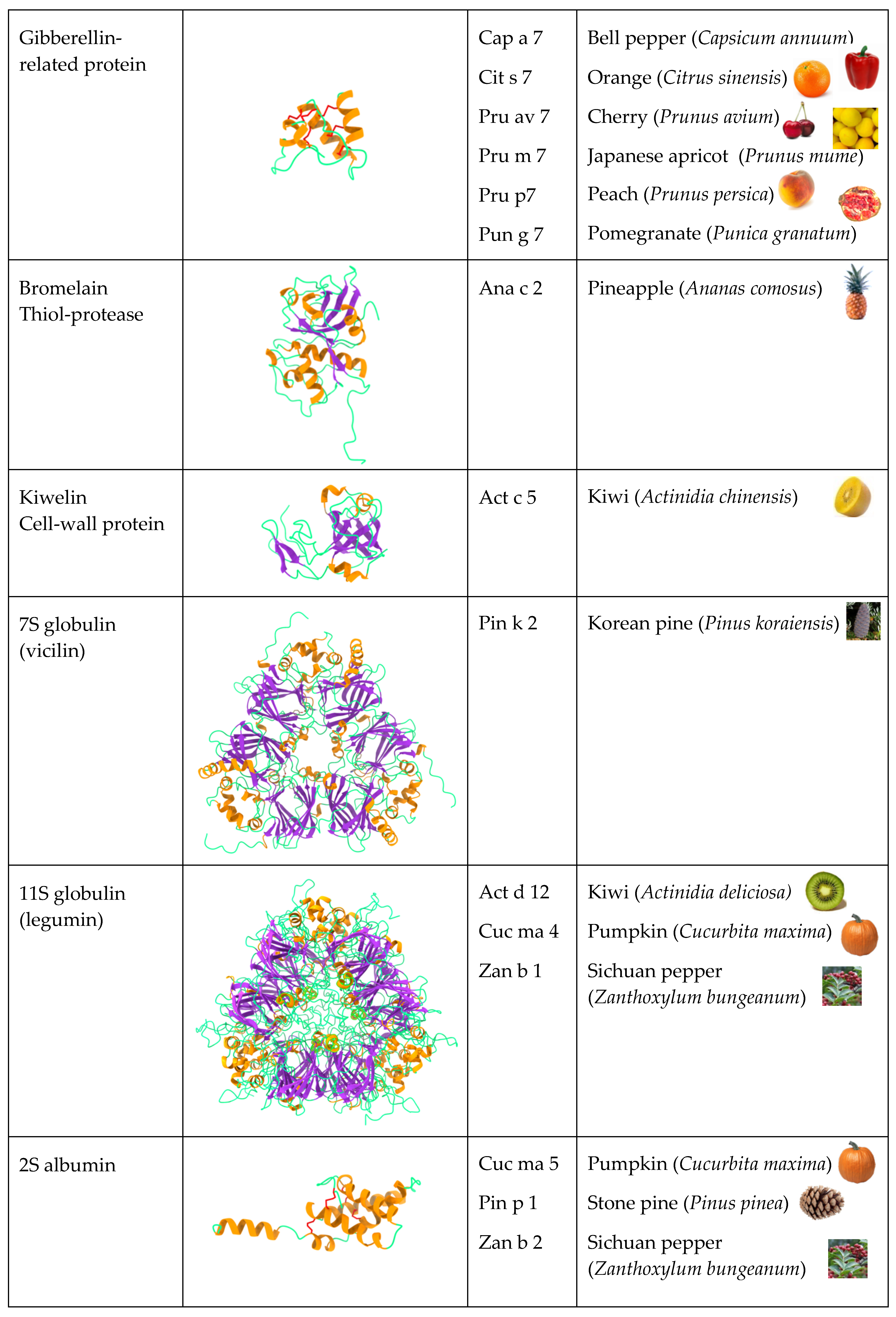
2.5. Gibberellin-Regulated Proteins (GRPs)
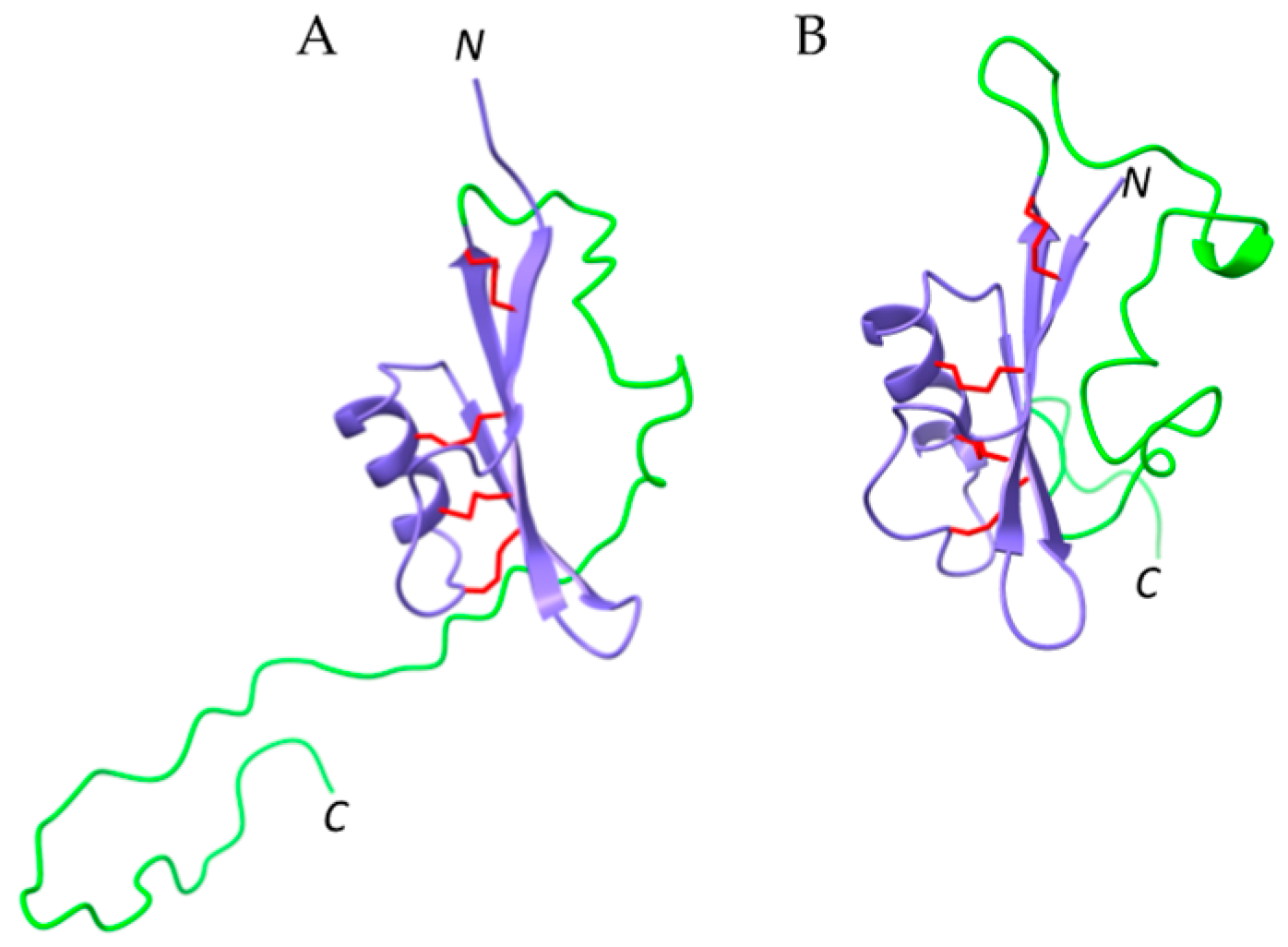
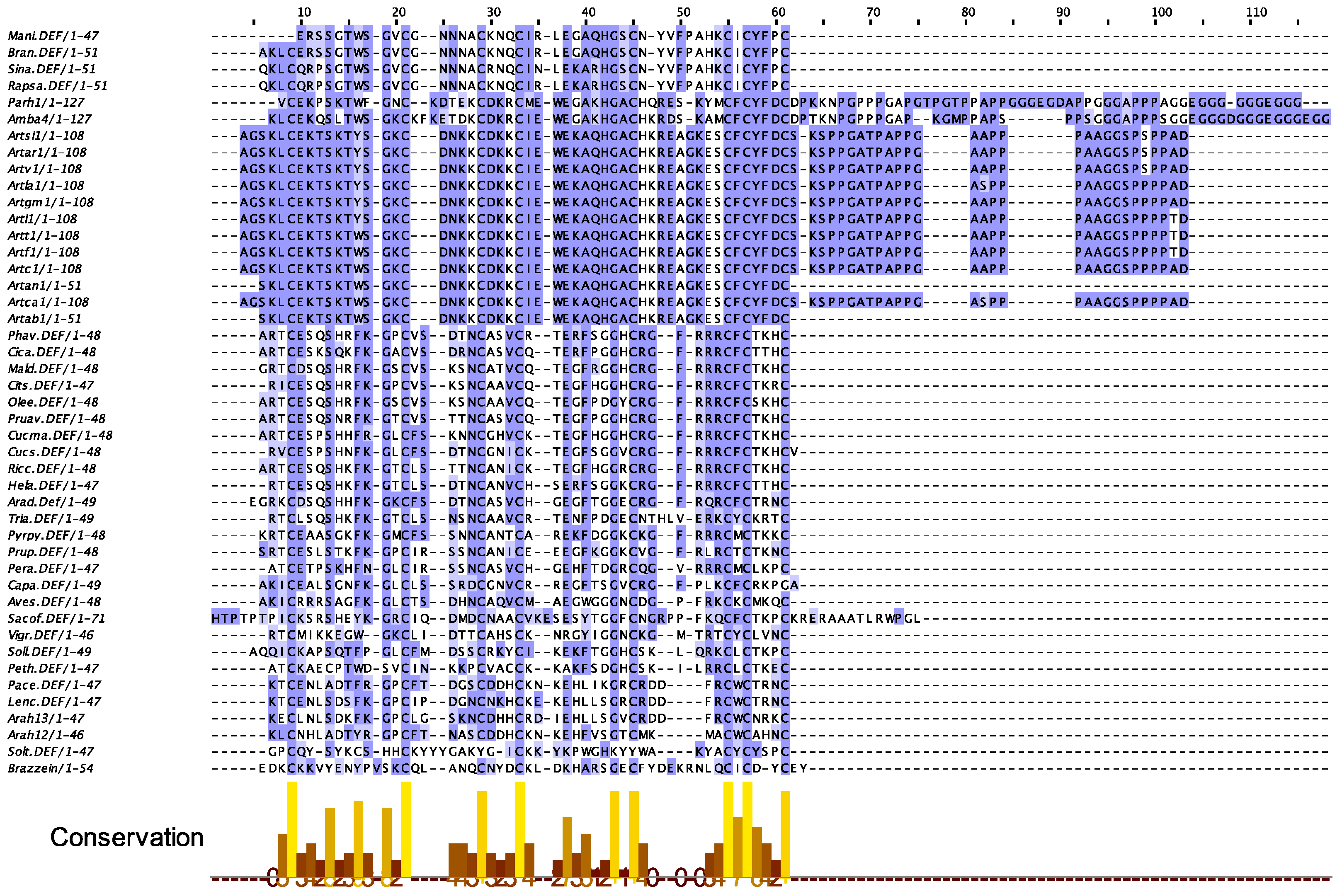
2.6. Defensins
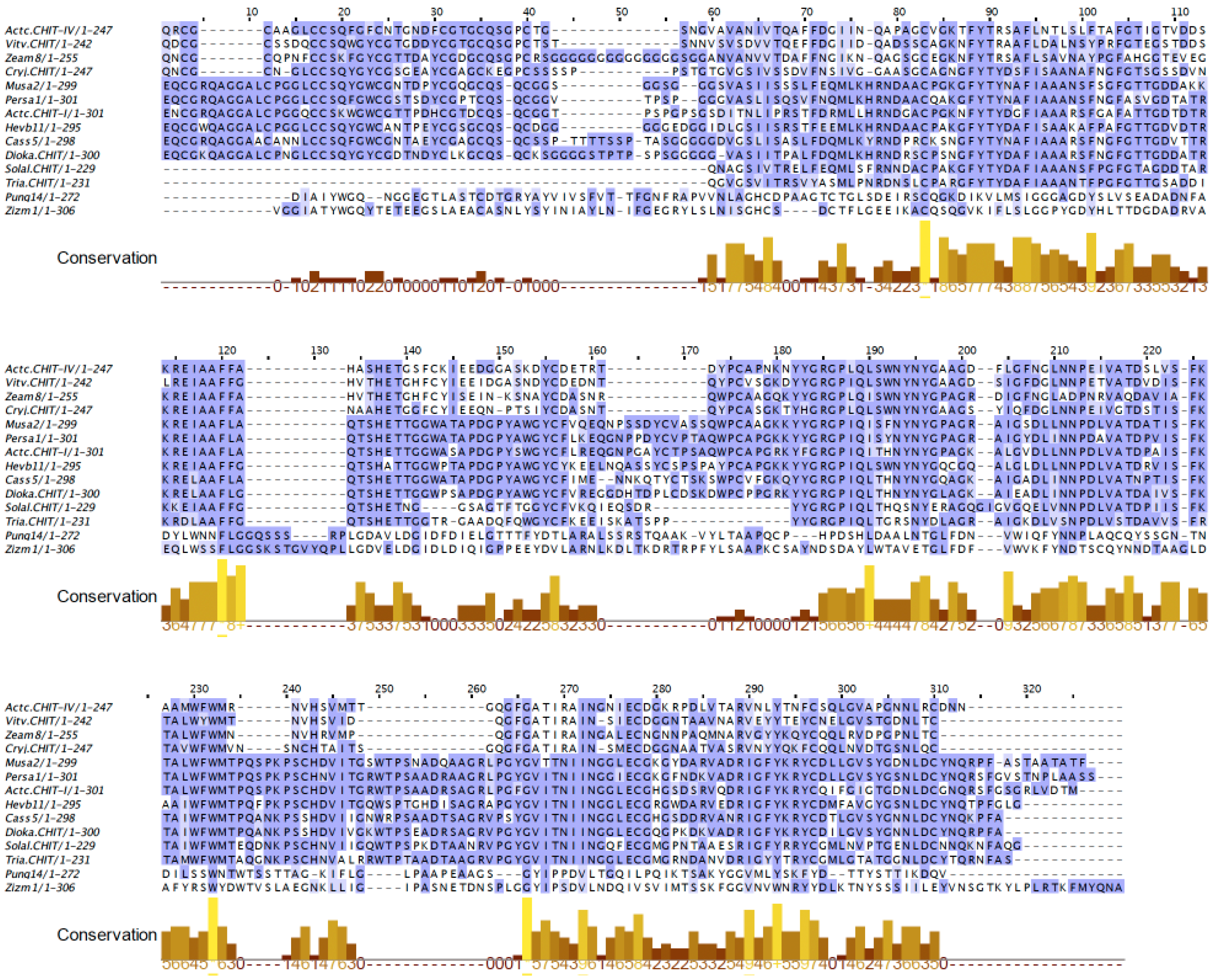
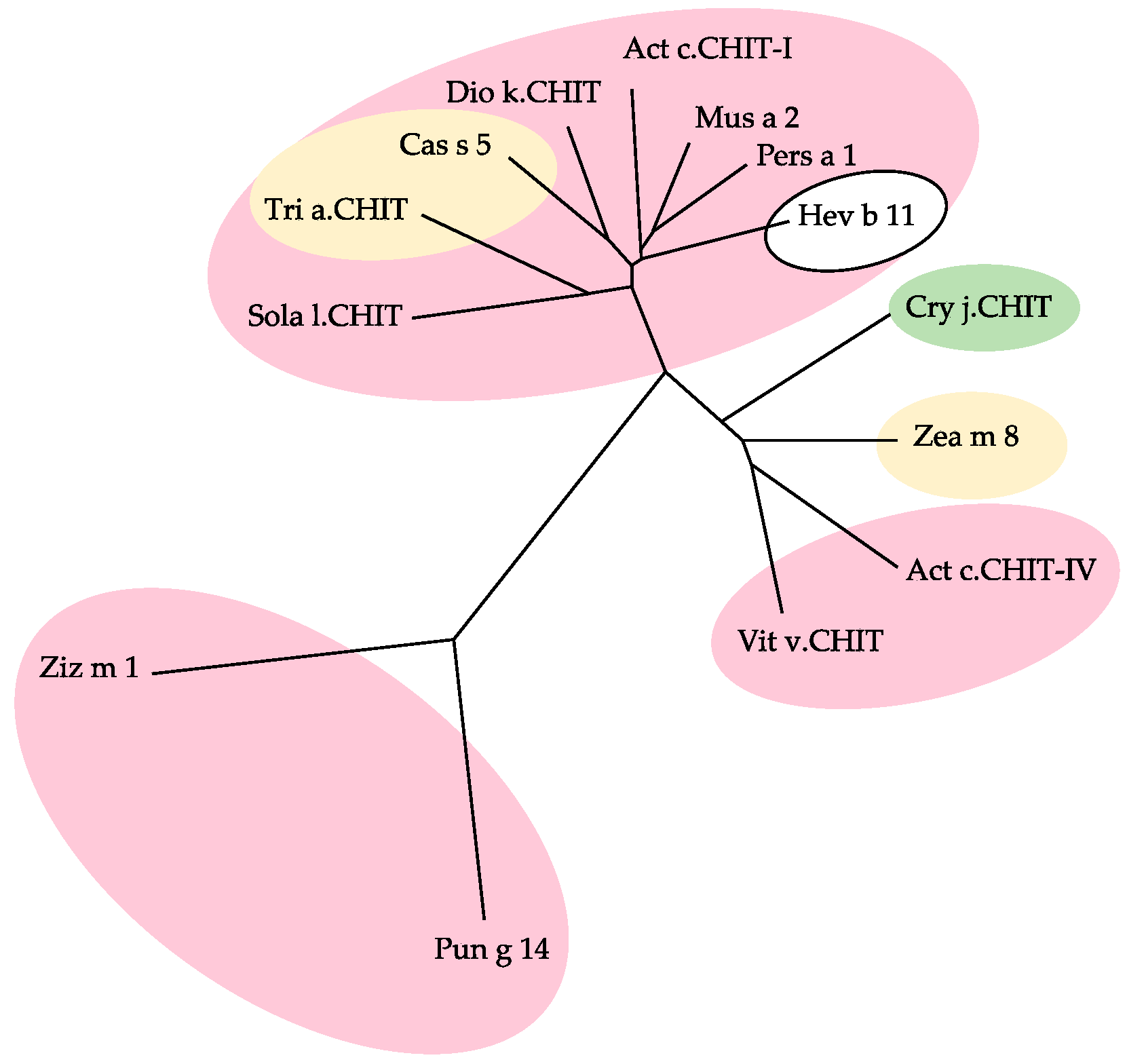
2.7. PR3 Chitinases
- -
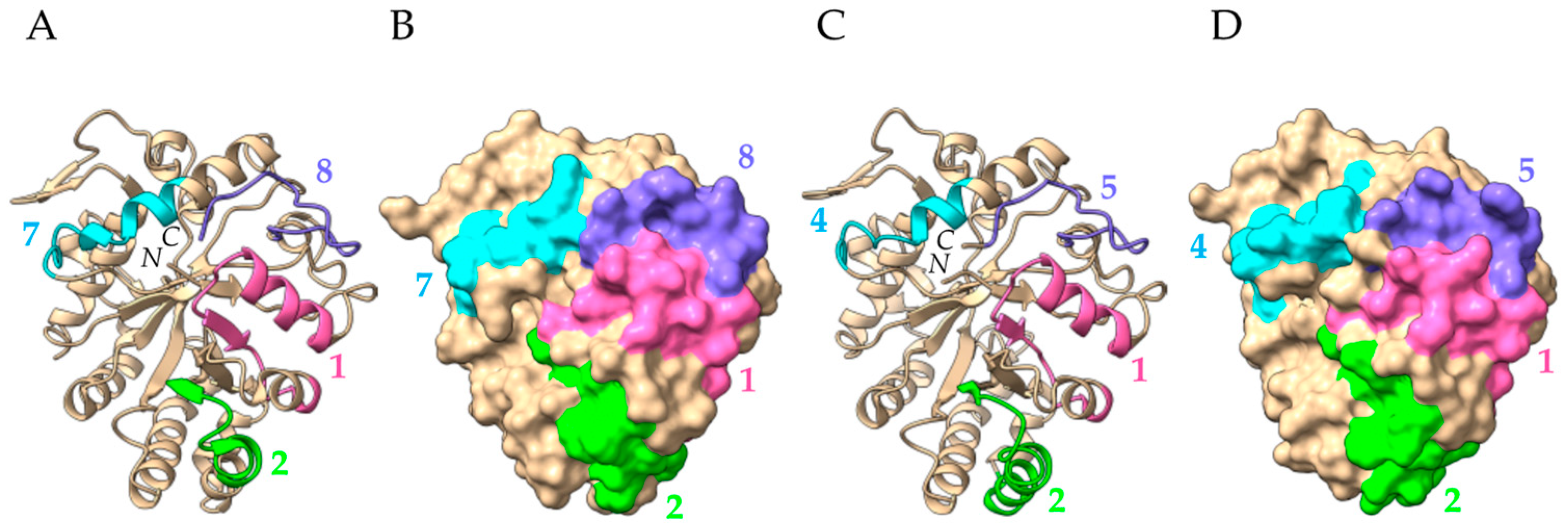
- Class-I and class-IV chitinases exhibit a structural organization in two domains, comprising a N-terminal chitin-binding domain made of a cysteine-rich polypeptide, similarly folded as the hevein domain of the latex from rubber tree Hevea brasiliensis (Hev b 6), linked by an extended linker loop to a C-terminal α-helical folded domain with a chitin-cleavage activity (Figure 29a). Class-I and class-IV chitinases use an inverting mechanism for cleaving the β1,4-GlcNAc linkage of chitin to generate shorter fragments of chitobiose (GlcNAc)2, chitotriose (GlcNAc)3 and chitotetraose (GlcNAc)4. The chitinase allergens from banana (Mus a 2), avocado (Pers a 1), and Hev b 11 from the latex of rubber tree, belong to this group of chitinases.
- -
- Class-III chitinases readily differ from class-I chitinases by a different α8β8 β-barrel organization made of a central crown of eight β-strands, linked to a peripheral crown of eight α-helix by interconnecting loops (Figure 29b) [56]. In addition, class-III chitinases use a different retaining mechanism for the cleavage of chitin chains in shorter fragments. The chitinase allergens from pomegranate (Pun g 14) and jujuba fruit (Ziz m 1), belong to the class-III chitinase group.
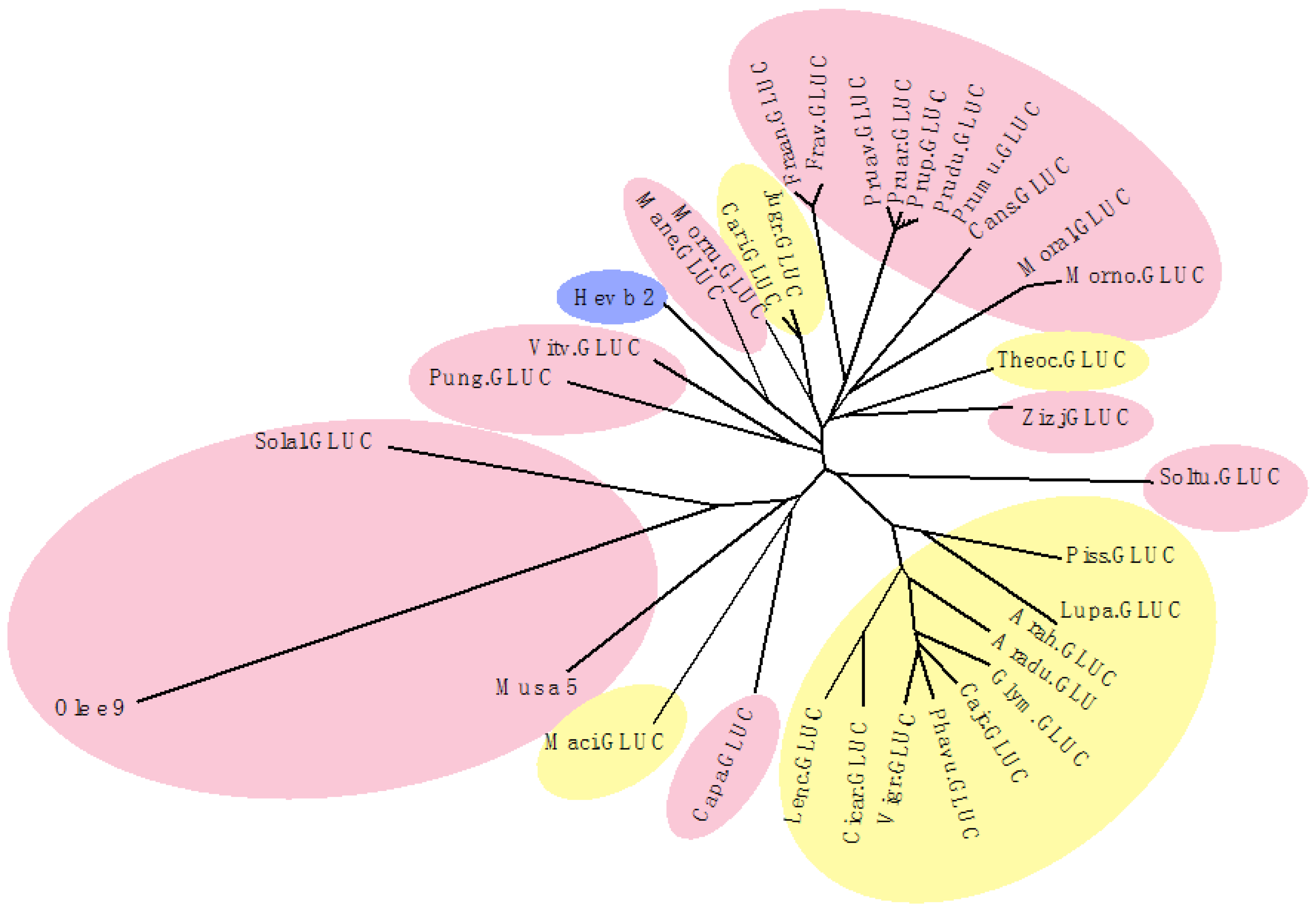
2.8. PR2 Glucanases
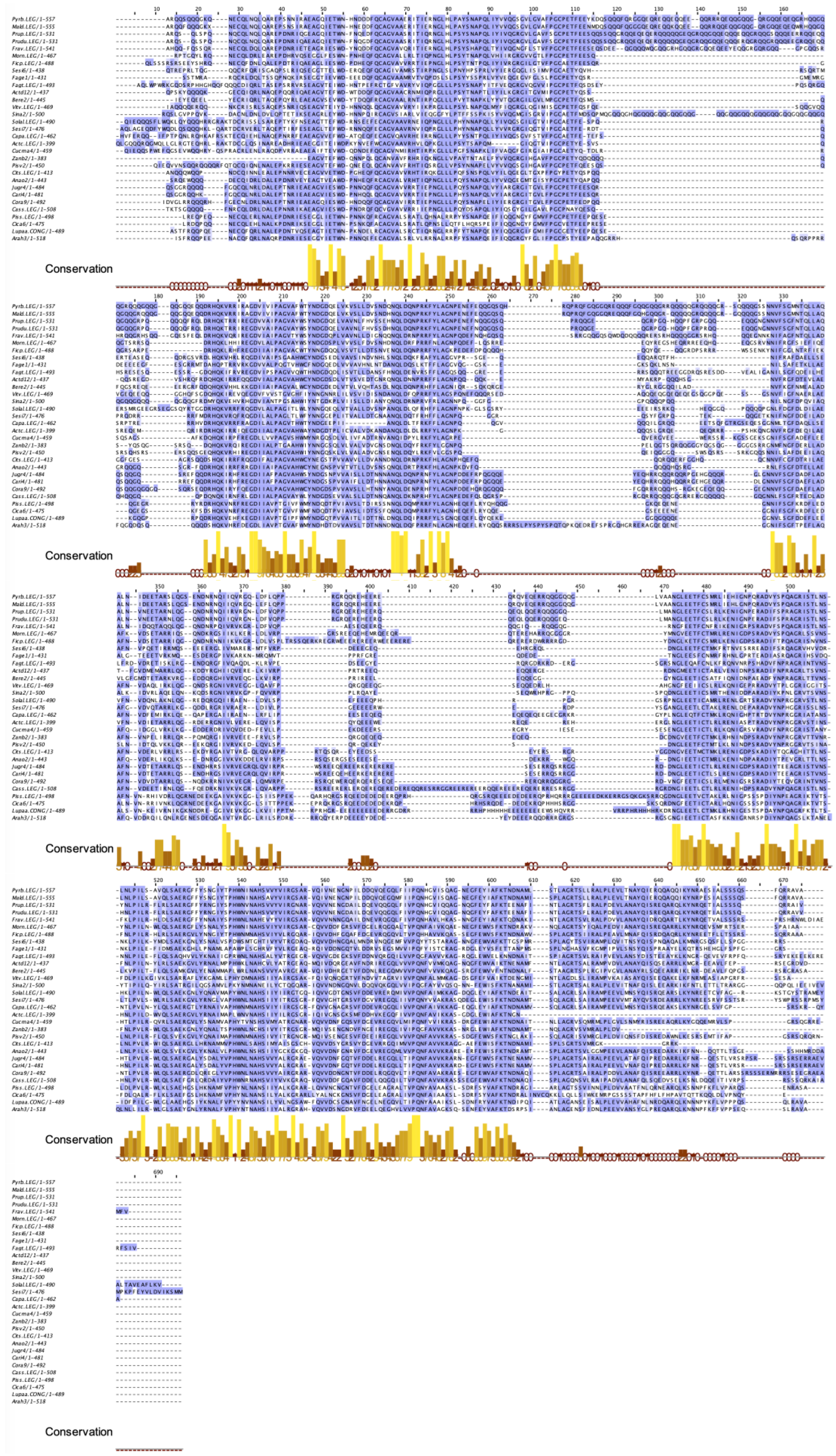
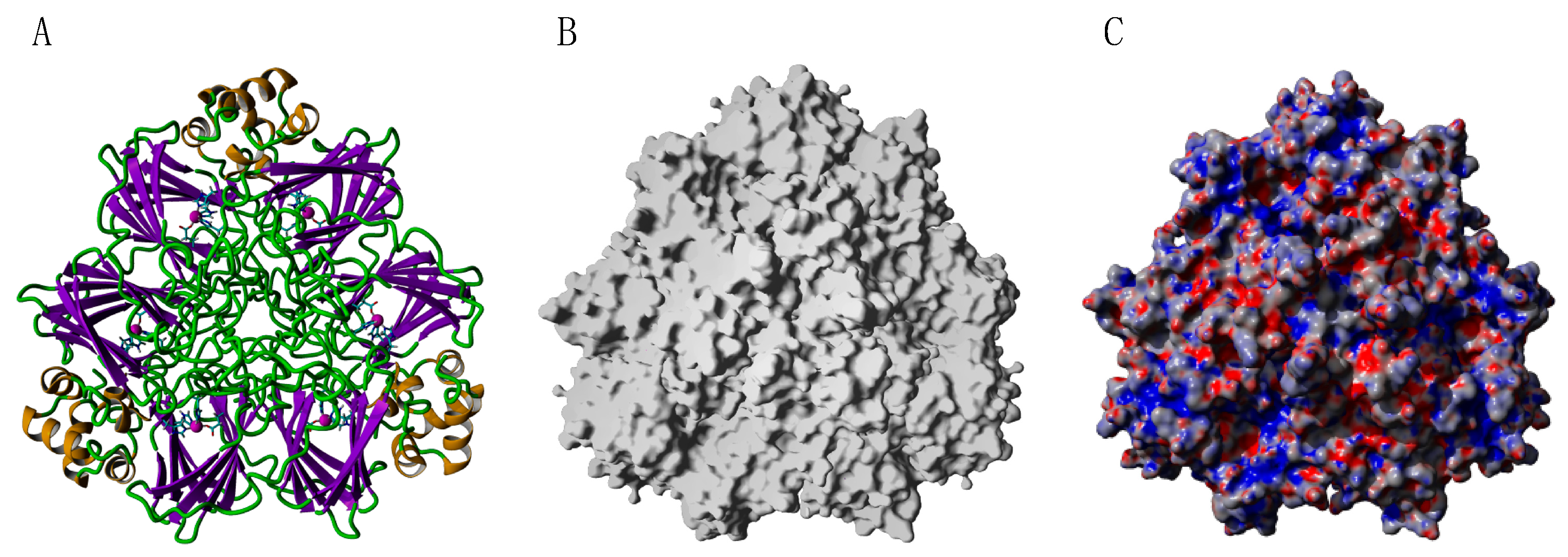
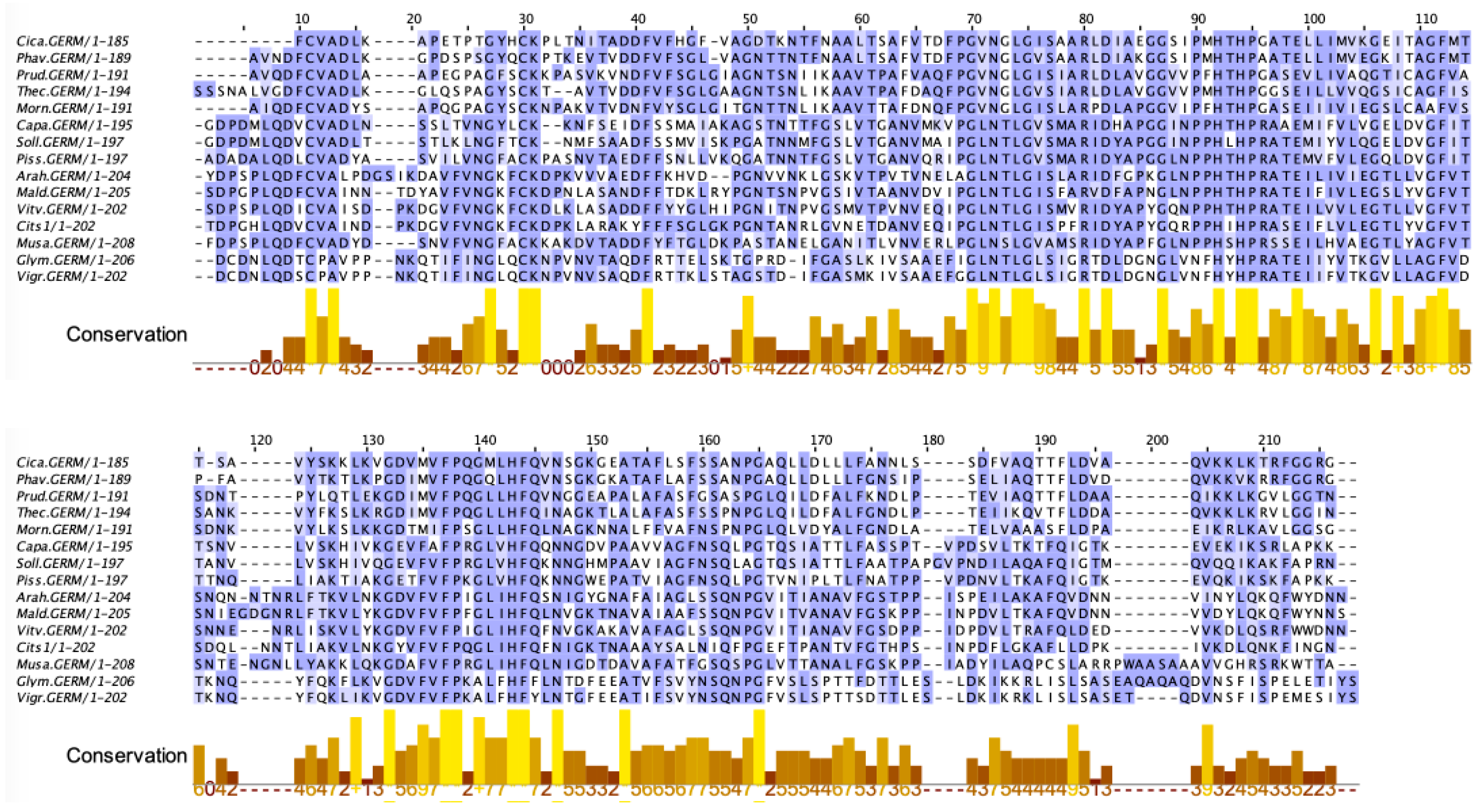
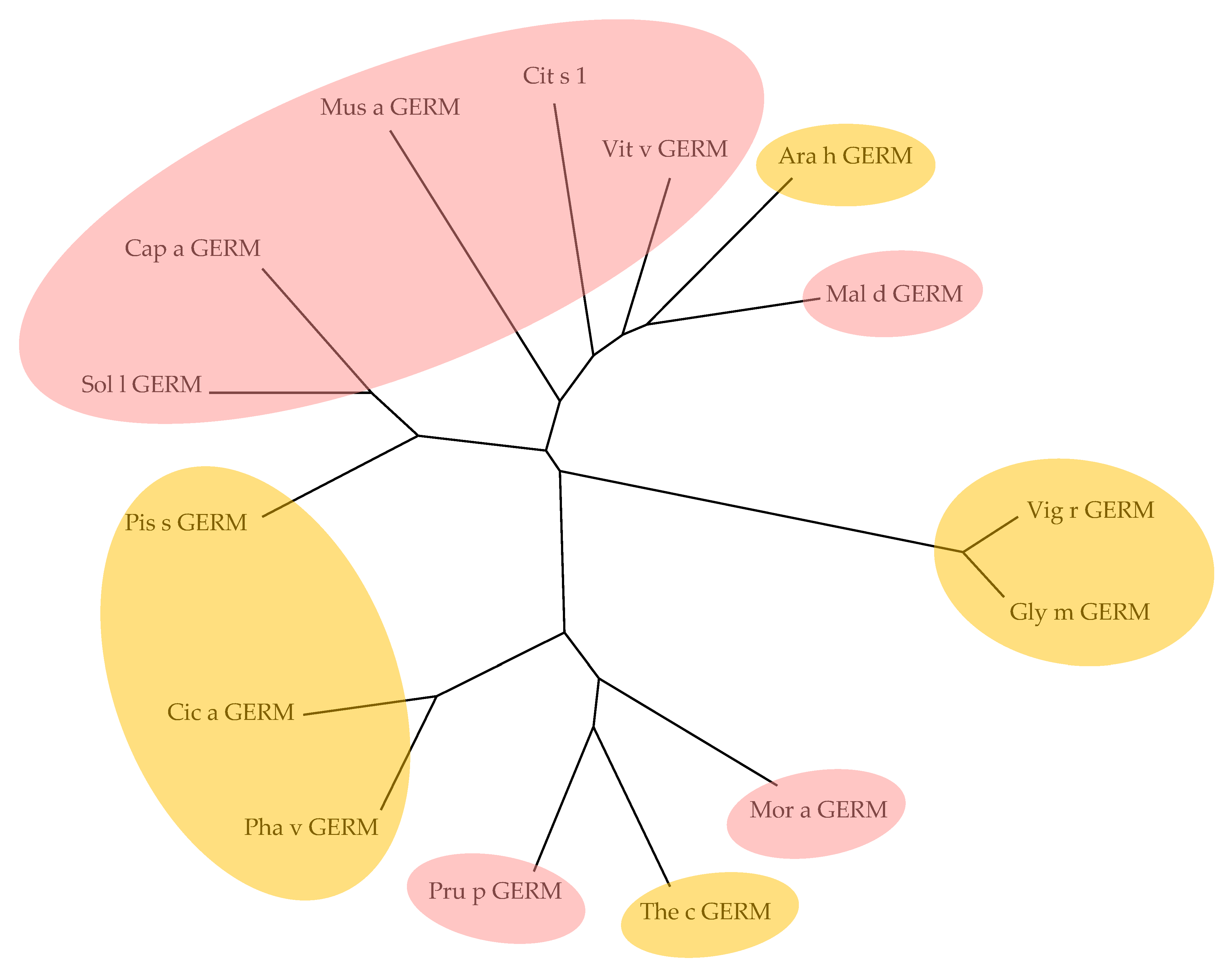
2.9. Seed Storage Proteins from Fruit Kernels
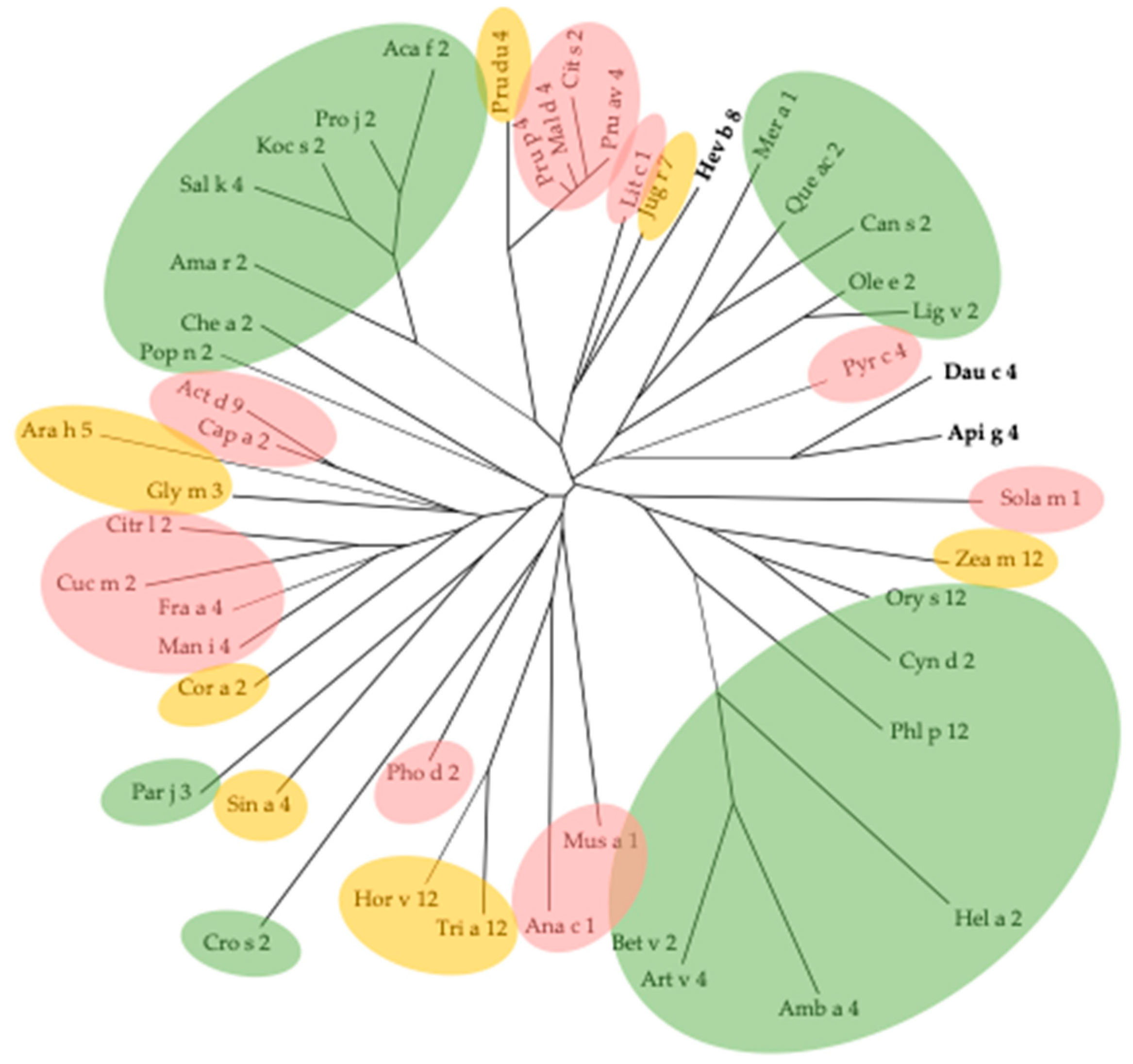
3. Cross-Reactivity of Fruit Allergens
4. Brief Clinical Aspects
- -
- The structural characteristics of the allergens (allergens exhibiting tightly packed conformation vs. allergens loosely structured) and some structure-associated properties, like the resistance to digestive proteases and heat (cooking) denaturation [36]. In this respect, fruit nsLTPs, like Pru p 3 and Mal d 3, possess a tightly packed three-dimensional conformation stabilized by four disulfide bridges, the so-called “saxophone-like conformation”, that contributes to the extreme resistance of these protein to the degradation by digestive proteases and to heat denaturation [22]. Similarly, the tightly packed GRPs offer an enhanced resistance to protease and heat denaturation [44]. Conversely, profilins, such as Pru p 4 and Mal d 4, are more loosely structured proteins and offer a weak resistance to both enzymatic digestion and heat denaturation [36];
- -
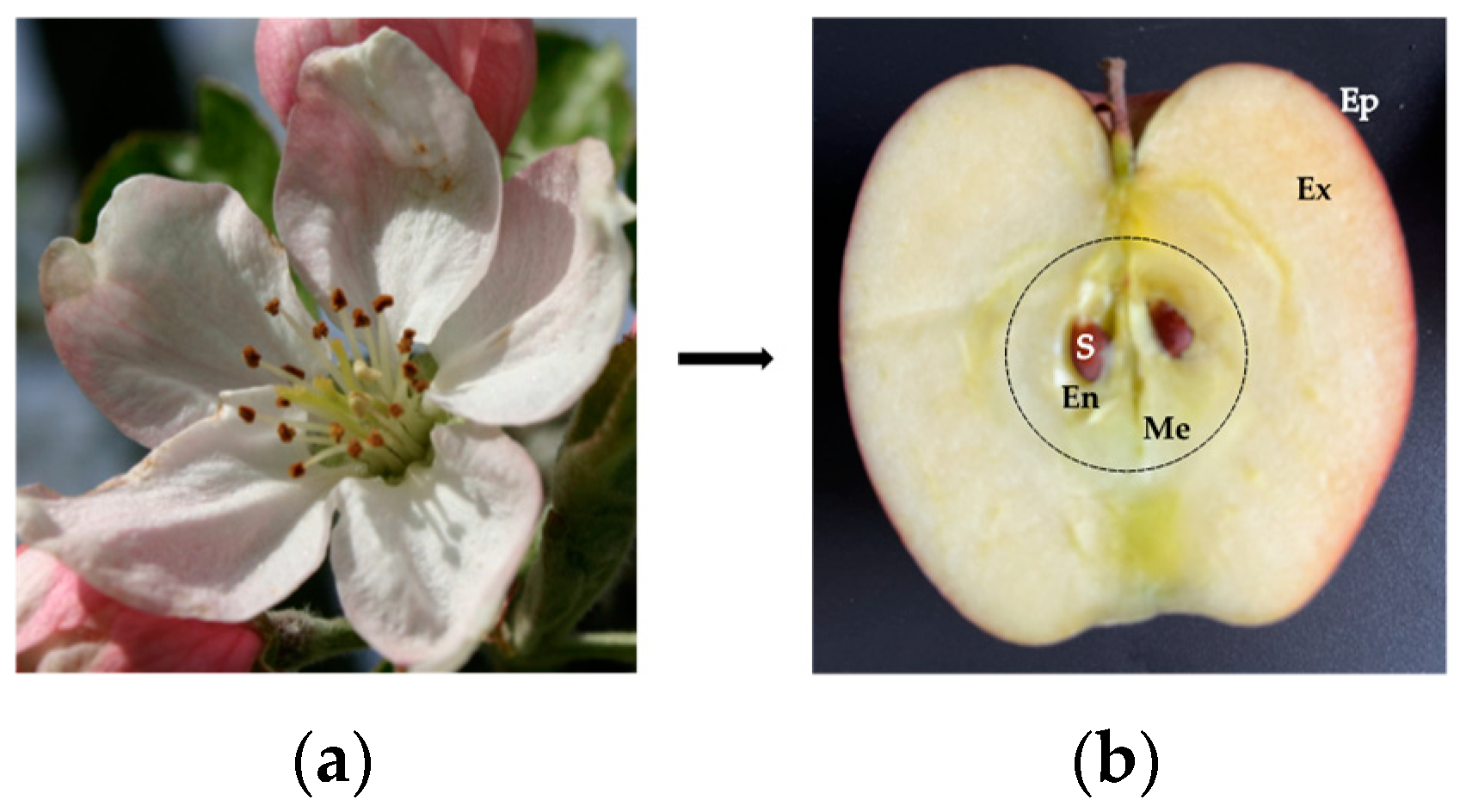
- The localization and accessibility of the allergens. In this regard, the surface localization of Pru p 3 in the fuzzy covering the peel of the peach [78], and the localization of Mal d 3 in the external cell layers forming the peel of the apple [24], favors the contact of the allergens with the body. Accordingly, removing the peel from peaches and apples is sufficient to eliminate most of the allergens from the fruits;
- -
- The amounts of allergens present in fruits and vegetables. This is an important point to consider, because many fruit allergens consist of PR proteins which interfere with the defense of plants against phytopathogenic fungi, bacteria, and viruses [79]. In addition, their synthesis can vary considerably, depending on the response of the plant to abiotic stresses, for example water stress or heat stress [80]. Moreover, large variations in the allergen content of fruits were measured in different varieties or cultivars of peach and apple [81,82,83]. Large variations were also measured in the allergen content of fruits, depending on the cultivation conditions [84,85], the degree of ripening of the harvested fruits, and the shelf life of the postharvest fruit storage [86,87,88,89,90]. Other factors can influence the sensitizing propensity of the allergens, such as the route of exposure [76] and the processing of fruits and fruit products before consumption [91].
5. Comparative Prevalence and Harmful Properties of Fruit Allergens
6. Conclusions
- Allergens from edible fruits, especially fleshy fruits, correspond to pan-allergens that are widely distributed in vegetables, seeds, and pollen from other apparently unrelated plants;
- Fruit allergens essentially belong to different PR-proteins, which play a role in the defense of plants against phytopathogenic fungi, bacteria, and viruses. Accordingly, their biosynthesis is largely influenced by the stress conditions to which the plants are subjected. The allergen content of fruits can also vary considerably, depending on the ripening stage and storage conditions of the fruits after harvest;
- The different families of allergenic PR-proteins exhibit highly conserved amino acid sequences and three-dimensional structures and display close phylogenetic relationships;
- Depending on their large distribution and their sequential, structural, and phylogenetical relationships, a high degree of cross-reactivity usually occurs between allergens from fruits and the counterparts from other sources like vegetables, seeds, or pollens. This cross-reactivity is at the origin of various clinical syndromes including, e.g., the apple-birch syndrome, the peach-cypress syndrome, and the peach-latex syndrome. However, many cross-sensitizations with pollen allergens are not clinically relevant;
- The allergenicity and toxicity vary largely among fruit allergens. Although all fruit allergens are potentially dangerous, some of them, like nsLTPs and GRPs, are responsible for food allergies, and provoke severe systemic reactions, especially in Mediterranean countries;
- In general, the consumption of allergen-containing fruits only results in mild oropharyngeal symptoms that corresponds to the so-called oral allergic syndrome, OAS. In some cases, however, more severe systemic reactions can develop, especially upon consumption of Rosaceae fruits or kiwi fruit.
7. Bioinformatics
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lefèvre, S.; Gregori, M.; Astier, C.; Rougé, P.; Kanny, G. Fruit allergies. Rev. Fr. Allergol. 2020, 60, 585–594. [Google Scholar] [CrossRef]
- Ebner, C.; Hoffmann-Sommergruber, K.; Breiteneder, H. Plant food allergens homologous to pathogenesis-related proteins. Allergy 2001, 56, 43–44. [Google Scholar] [CrossRef] [PubMed]
- Sinha, M.; Singh, R.P.; Singh Kushwaha, G.; Iqbal, N.; Singh, A.; Kaushik, S.; Kaur, P.; Sharma, S.; Singh, T.P. Current overview of allergens of plant pathogenesis related families. ScientificWorldJournal 2014, 2014, 543195. [Google Scholar] [CrossRef] [Green Version]
- Jain, A. Seed storage protein, functional diversity and association with allergy. Allergies 2023, 3, 25–38. [Google Scholar] [CrossRef]
- Barre, A.; Simplicien, M.; Benoist, H.; Rougé, P. Fruit allergies: Beware of the seed allergens! Rev. Fr. Allergol. 2018, 58, 308–317. [Google Scholar] [CrossRef]
- Mackenzie, A.; Pridham, J.B.; Saunders, N.A. Changes in the sweet proteins (thaumatins) in Thaumatoccocus daniellii fruit development. Phytochemistry 1985, 11, 2503–2506. [Google Scholar] [CrossRef]
- Leone, P.; Menu-Bouaouiche, L.; Peumans, W.J.; Payan, F.; Barre, A.; Roussel, A.; Van Damme, E.J.M.; Rougé, P. Resolution of the structure of the allergenic and antifungal banana fruit thaumatin-like protein at 1.7-Å. Biochimie 2006, 88, 45–52. [Google Scholar] [CrossRef]
- Smole, U.; Bublin, M.; Radauer, C.; Ebner, C.; Breiteneder, H. Mal d 2, the thaumatin-like allergen from apple, is highly resistant to gastrointestinal digestion and thermal processing. Int. Arch. Allergy Immunol. 2008, 147, 289–298. [Google Scholar] [CrossRef]
- Kumar, H.G.A.; Venkatesh, Y.P. In silico analyses of structural and allergenicity features of sapodilla (Manilkara zapota) acidic thaumatin-like protein in comparison with allergenic plant TLPs. Mol. Immunol. 2014, 57, 119–128. [Google Scholar] [CrossRef]
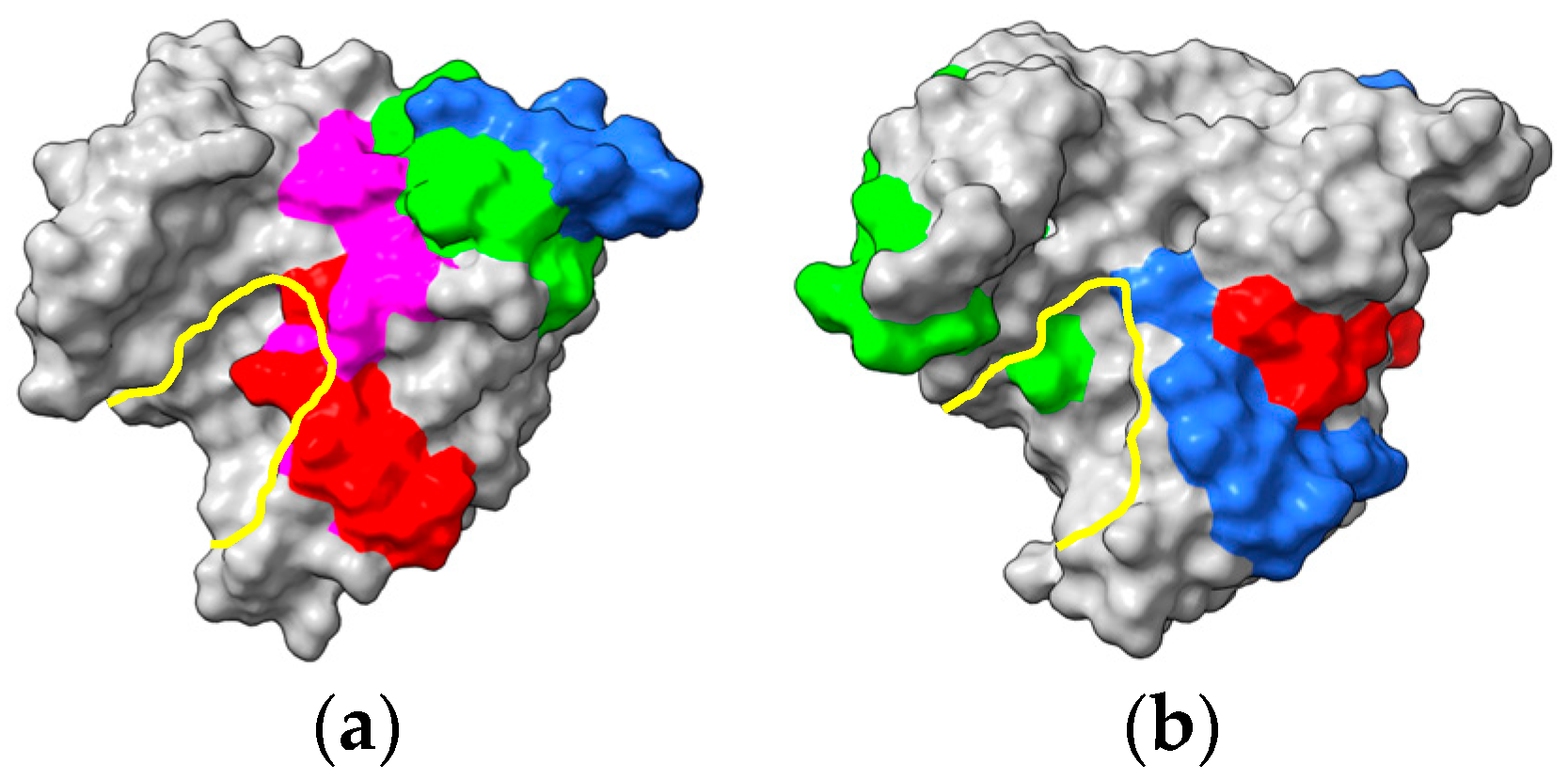
- Barre, A.; Sénéchal, H.; Nguyen, C.; Granier, C.; Poncet, P.; Rougé, P. Structural basis for the IgE-binding cross-reacting epitopic peptides of Cup s 3, a PR-5 thaumatin-like protein allergen from common cypress (Cupressus sempervirens) pollen. Allergies 2023, 3, 11–24. [Google Scholar] [CrossRef]
- Palacín, A.; Rivas, L.A.; Gómez-Casado, C.; Aguirre, J.; Tordesillas, L.; Bartra, J.; Blanco, C.; Carrillo, T.; Cuesta-Herranz, J.; Bonny, J.A.; et al. The involvement of thaumatin-like proteins in plant food cross-reactivity: A multicenter study using a specific protein microarray. PLoS ONE 2012, 7, e44088. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costa, J.; Mafra, I. Rosaceae food allergy: A review. Crit. Rev. Food Sci. Nutr. 2022, 1–38. [Google Scholar] [CrossRef] [PubMed]
- Breiteneder, H.; Hassfeld, W.; Pettenburger, K.; Jarolim, E.; Breitenbach, M.; Rumpold, D.; Kraft, D.; Scheiner, O. Isolation and characterization of messenger RNA from male inflorescences and pollen of the white birch (Betula verrucosa). Int. Arch. Allergy Appl. Immunol. 1988, 87, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Neudecker, P.; Schweimer, K.; Nerkamp, J.; Scheurer, S.; Vieths, S.; Sticht, H.; Rösch, P. Allergic cross-reactivity made visible: Solution structure of the major cherry allergen Pru av 1. J. Biol. Chem. 2001, 276, 22756–22763. [Google Scholar] [CrossRef] [Green Version]
- Skypala, I.J.; Asero, R.; Barber, D.; Cecchi, L.; Díaz-Perales, A.; Hoffmann-Sommergruber, K.; Pastorello, E.A.; Swoboda, I.; Bartra, J.; Ebo, D.G.; et al. Non-specific lipid-transfer proteins: Allergen structure and function, cross-reactivity, sensitization, and epidemiology. Clin. Transl. Allergy 2021, 11, e12010. [Google Scholar] [CrossRef]
- Neudecker, P.; Lehmann, K.; Nerkamp, J.; Haase, T.; Wangorsch, A.; Fötisch, K.; Hoffmann, S.; Rösch, P.; Vieths, S.; Scheurer, S. Mutational epitope analysis of Pru av 1 and Api g 1, the major allergens of cherry (Prunus avium) and celery (Apium graveolens): Correlating IgE reactivity with three-dimensional structure. Biochem. J. 2003, 376, 97–107. [Google Scholar] [CrossRef] [Green Version]
- Wiche, R.; Gubesch, M.; König, H.; Fötisch, K.; Hoffmann, A.; Wangorsch, A.; Scheurer, S.; Vieths, S. Molecular basis pf pollen-related food allergy: Identification of a second cross-reactive IgE epitope on Pru av 1, the major cherry (Prunus avium) allergen. Biochem. J. 2005, 385, 319–327. [Google Scholar] [CrossRef] [Green Version]
- Jahn-Schmid, B.; Radakovics, A.; Lüttkopf, D.; Scheurer, S.; Vieths, S.; Ebner, C.; Bohle, B. Bet v 1 142-156 is the dominant T-cell epitope of the major birch pollen allergen and important for cross-reactivity with Bet v 1-related food allergens. J. Allergy Clin. Immunol. 2005, 116, 213–219. [Google Scholar] [CrossRef]
- Breiteneder, H.; Aglas, L.; Kleine-Tebbe, J.; Hafner, C.; Dramburg, S.; Hilger, C.; Santos, A.F.; Vecillas, L.d.L.; Aalberse, R.C.; Acevedo, N.; et al. C02–PR-10-like allergens. EAACI Molecular Allergology User’s Guide 2.0. Pediatr. Allergy Immunol. 2023, 34, 272–280, e13854. [Google Scholar]
- Scala, E.; Maria Marra, A.; Asero, R.; Dramburg, S.; Hilger, C.; Santos, A.F.; Vecillas, L.d.L.; Aalberse, R.C.; Acevedo, N.; Aglas, L.; et al. B01—Tree pollen allergy. EAACI Molecular Allergology User’s Guide 2.0. Pediatr. Allergy Immunol. 2023, 34, 93–102, e13854. [Google Scholar]
- Pasquato, N.; Berni, R.; Folli, C.; Folloni, S.; Cianci, M.; Pantano, S.; Helliwell, J.R.; Zanotti, G. Crystal structure of peach Pru p 3, the prototypic member of the family of plant non-specific lipid transfer protein pan-allergens. J. Mol. Biol. 2005, 356, 684–694. [Google Scholar] [CrossRef] [PubMed]
- Toda, M.; Reese, G.; Gadermaier, G.; Schulten, V.; Lauer, I.; Egger, M.; Briza, P.; Randow, S.; Wolfheimer, S.; Kigongo, V.; et al. Protein unfolding strongly modulate the allergenicity and immunogenicity of Pru p 3, the major peach allergen. J. Allergy Clin. Immunol. 2011, 128, 1022–1030. [Google Scholar] [CrossRef] [PubMed]
- Akkerdaas, J.; Totis, M.; Barnett, B.; Bell, E.; Davis, T.; Edrington, T.; Glenn, K.; Graser, G.; Herman, R.; Knulst, A.; et al. Protease resistance of food proteins: A mixed picture for predicting alleregenicity but a useful tool for assessing exposure. Clin. Transl. Allergy 2018, 8, 30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borges, J.-P.; Jauneau, A.; Brulé, C.; Culerrier, R.; Barre, A.; Didier, A.; Rougé, P. The lipid transfer proteins (LTP) essentially concentrate in the skin of Rosaceae fruits as cell surface exposed allergens. Plant Physiol. Biochem. 2006, 44, 535–542. [Google Scholar] [CrossRef]
- García-Casado, G.; Pacios, L.F.; Díaz-Perales, A.; Sánchez-Monge, R.; Lombardero, M.; García-Selles, F.J.; Polo, F.; Barber, D.; Salcedo, G. Identification of IgE-binding epitopes of the major peach allergen Pru p 3. J. Allergy Clin. Immunol. 2003, 112, 599–605. [Google Scholar] [CrossRef]
- Borges, J.-P.; Barre, A.; Culerrier, R.; Granier, C.; Didier, A.; Rougé, P. Lipid transfer proteins from Rosaceae fruits share consensus epitopes responsible for vtheir IgE-binding cross-reactivity. Biochem. Biophys. Res. Commun. 2008, 365, 685–690. [Google Scholar] [CrossRef]
- Pastorello, E.A.; Gomez, F.; Barber, D.; Dramburg, S.; Hilger, C.; Santos, A.F.; Vecillas, L.d.L.; Aalberse, R.C.; Acevedo, N.; Aglas, L.; et al. C03—Nonspecific lipid transfer proteins (nsLTP). EAACI Molecular Allergology User’s Guide 2.0. Pediatr. Allergy Immunol. 2023, 34, 281–287, e13854. [Google Scholar]
- Scheurer, S.; Schulke, S. Interaction of non-specific lipid-transfer proteins with plant-derived lipids and its impact on allergic sensitization. Front. Immunol. 2018, 9, 1389. [Google Scholar] [CrossRef] [Green Version]
- Dubiela, P.; Aina, R.; Polak, D.; Geiselhart, S.; Humeniuk, P.; Bohle, B.; Alessandri, S.; Del Conte, R.; Cantini, F.; Borowski, T.; et al. Enhanced Pru p 3 IgE-binding activity by selected free fatty acid-interaction. J. Allergy Clin. Immunol. 2017, 140, 1728–1731. [Google Scholar] [CrossRef] [Green Version]
- Dubiela, P.; Del Conte, R.; Cantini, F.; Borowski, T.; Aina, R.; Radauer, C.; Bublin, M.; Hoffmann-Sommergruber, K.; Alessandri, S. Impact of lipid binding on the tertiary structure and allergenic potential of Jug r 3, the non-specific lipid transfer protein from walnut. Sci. Rep. 2019, 9, 2007. [Google Scholar] [CrossRef] [Green Version]
- Chruszcz, M.; Chew, F.T.; Hoffmann-Sommergruber, K.; Hurlburt, B.K.; Mueller, G.A.; Pomés, A.; Rouvinen, J.; Villalba, M.; Wöhrl, B.; Breiteneder, H. Allergens and their associated small molecule ligands—Their dual role in sensitization. Allergy 2021, 76, 2367–2382. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Klein, Z.; Pazos-Castro, D.; Hernandez-Ramirez, G.; Garrido-Aranda, M.; Díaz-Perales, A.; Tome-Amat, J. Lipid ligands and allergenic LTPs: Redefining the paradigm of the protein-centered vision in allergy. Front. Allergy 2022, 3, 864652. [Google Scholar] [CrossRef] [PubMed]
- Rougé, P.; Hilger, C.; Hoffmann-Sommergruber, K.; Traidl-Hoffmann, C.; Dramburg, S.; Hilger, C.; Santos, A.F.; Vecillas, L.d.L.; Aalberse, R.C.; Acevedo, N.; et al. A11—Small molecules as immunomodulators and allergen ligands. EAACI Molecular Allergology User’s Guide 2.0. Pediatr. Allergy Immunol. 2023, 34, 80–84, e13854. [Google Scholar]
- Brenda Kapingidza, A.; Pye, S.E.; Hyduke, N.; Dolamore, C.; Pote, S.; Schlachter, C.; Commins, S.P.; Kowal, K.; Chruszcz, M. Comparative structural and thermal stability studies of Cuc m 2.0101, Art v 4.0101 and other allergenic profilins. Mol. Immunol. 2019, 114, 19–29. [Google Scholar] [CrossRef]
- Asero, R.; Mistrello, G.; Roncarolo, D.; Amato, S.; Zanoni, D.; Barocci, F.; Caldironi, G. Detection of clinical markers of sensitization to profilin in patients allergic to plant-derived foods. J. Allergy Clin. Immunol. 2003, 112, 427–432. [Google Scholar] [CrossRef] [PubMed]
- Offermann, L.; Schlachter, C.R.; Perdue, M.L.; Majorek, K.A.; He, J.Z.; Booth, W.T.; Kowal, K.; Chruszcz, M. Structural, functional, and immunological characterization of profilin panallergens Amb a 4, Art v 4, and Bet v 2. J. Biol. Chem. 2016, 291, 15447–15459. [Google Scholar] [CrossRef] [Green Version]
- Hofmann, A.; Burks, A.W. Pollen food syndrome: Update on the allergens. Curr. Allergy Asthma Rep. 2008, 8, 413–417. [Google Scholar] [CrossRef]
- López-Torrejón, G.; Díaz-Perales, A.; Rodríguez, J.; Sánchez-Monge, R.; Crespo, J.F.; Salcedo, G.; Pacios, L.F. An experimental and modeling-based approach to locate IgE epitopes of plant profilin allergens. J. Allergy Clin. Immunol. 2007, 119, 1481–1488. [Google Scholar] [CrossRef]
- Tordesillas, L.; Pacios, L.F.; Palacín, A.; Cuesta-Herranz, J.; Madero, M.; Díaz-Perales, A. Characterization of IgE epitopes of Cuc m 2, the major melon allergen, and their role in cross-reactivity with pollen profilins. Clin. Exp. Allergy 2010, 40, 174–181. [Google Scholar] [CrossRef]
- Rihs, H.P.; Chen, Z.; Ruëff, F.; Petersen, A.; Rozynek, P.; Heimann, H.; Baur, X.D. IgE binding of the recombinant allergen soybean profilin (rGly m 3) is mediated by conformational epitopes. J. Allergy Clin. Immunol. 1999, 104, 1293–1301. [Google Scholar] [CrossRef]
- Sirvent, S.; Tordesillas, L.; Villalba, M.; Díaz-Perales, A.; Cuesta-Herranz, J.; Salcedo, G.; Rodríguez, R. Pollen and plant food profilin allergens show equivalent IgE reactivity. Ann. Allergy Asthma Immunol. 2011, 106, 429–435. [Google Scholar] [CrossRef] [PubMed]
- Terán, M.G.; García-Ramírez, B.; Mares-Mejía, I.; Ortega, E.; O’Malley, A.; Chruszcz, M.; Rodríguez-Romero, A. Molecular basis of plant profilins’ cross-reactivity. Biomolecules 2023, 13, 608. [Google Scholar] [CrossRef] [PubMed]
- Asero, R.; Barber, D.; Celi, G.; Dramburg, S.; Hilger, C.; Santos, A.F.; Vecillas, L.d.L.; Aalberse, R.C.; Acevedo, N.; Aglas, L.; et al. C01—Profilins. EAACI Molecular Allergology User’s Guide 2.0. Pediatr. Allergy Immunol. 2023, 34, 268–271, e13854. [Google Scholar]
- Tuppo, L.; Spadaccini, R.; Alessandri, C.; Wiek, H.; Boelens, R.; Giangrieco, I.; Tamburrini, M.; Mari, A.; Picone, D.; Ciardello, M.A. Structure, stability, and IgE binding of the peach allergen peamaclein (Pru p 7). Biopolymers 2014, 102, 416–425. [Google Scholar] [CrossRef] [PubMed]
- Iuzuka, T.; Barre, A.; Rougé, P.; Charpin, D.; Scala, E.; Baudin, B.; Aizawa, T.; Sénéchal, H.; Poncet, P. Gibberellin-regulated proteins: Emergent allergens. Front. Allergy 2022, 3, 877553. [Google Scholar] [CrossRef]
- Poncet, P.; Iizuka, T.; Sénéchal, H.; Scala, E.; Dramburg, S.; Hilger, C.; Santos, A.F.; Vecillas, L.d.L.; Aalberse, R.C.; Acevedo, N.; et al. C09–Gibberellin-regulated proteins. EAACI Molecular Allergology User’s Guide 2.0. Pediatr. Allergy Immunol. 2023, 34, 322–327, e13854. [Google Scholar]
- Poncet, P.; Sénéchal, H.; Charpin, D. Update on pollen-food allergy syndrome. Expert Rev. Clin. Immunol. 2020, 16, 561–578. [Google Scholar] [CrossRef]
- Takei, M.; Nin, C.; Izuka, T.; Pawlikowski, M.; Selva, M.A.; Chantran, Y.; Nakajima, Y.; Zheng, J.; Aizawa, T.; Ebisawa, M.; et al. Capsicum allergy: Involvement of Cap a 7, a new clinically relevant gibberellin-regulated protein cross-reactive with Cry j 7, the gibberellin-regulated protein from Japanese cedar pollen. Allergy asthma Immunol. Res. 2022, 14, 328–338. [Google Scholar] [CrossRef]
- Razzera, G.; Gadermaier, G.; de Paula, V.; Almeida, M.S.; Egger, M.; Jahn-schmid, B.; Almeida, F.C.L.; Ferreira, F.; Valente, A.P. Mapping the interactions between a major pollen allergen and human IgE antibodies. Structure 2010, 18, 1011–1021. [Google Scholar] [CrossRef] [Green Version]
- Gao, G.; Dai, J.; Ding, M.; Hellekant, G.; Wang, J.; Wang, D. Studies on solution NMR structure of brazzein: Secondary structure and molecular scaffold. Sci. China C Life Sci. 1999, 42, 409–419. [Google Scholar] [CrossRef]
- Nagata, K.; Hongo, N.; Kameda, Y.; Yamamura, A.; Sasaki, H.; Lee, W.C.; Ishikawa, K.; Suzuki, E.; Tanokura, M. The structure of brazzein, a sweet-tasting protein from the wild African plant Pentadiplandra brazzeana. Acta Crystallogr. D Biol. Crystallogr. 2013, 69, 642–647. [Google Scholar] [CrossRef] [PubMed]
- Ming, D.; Hellekant, G. Brazzein, a new high-potency thermostable sweet protein from Pentadiplandra brazzeana B. FEBS Lett. 1994, 355, 106–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pablos, I.; Eichhorn, S.; Machado, Y.; Briza, P.; Neunkirchner, A.; Jahn-Schmid, B.; Wildner, S.; Soh, W.T.; Ebner, C.; Park, J.-W.; et al. Distinct epitope structures of defensin-like proteins linked to prolin-rich regions give rise to differences in their allergenici activity. Allergy 2018, 73, 431–441. [Google Scholar] [CrossRef] [PubMed]
- Cosi, V.; Gadermaier, G. The role of defensins as pollen and food allergens. Curr. Allergy Asthma Rep. 2023, 23, 277–285. [Google Scholar] [CrossRef] [PubMed]
- Patil, R.S.; Ghormade, V.; Deshpande, M.V. Chitinolytic enzymes: An exploration. Enz. Mircrob. Technol. 2000, 26, 473–483. [Google Scholar] [CrossRef] [PubMed]
- Masuda, T.; Zhao, G.; Mikami, B. Crystal structure of class III chitinase from pomegranate provides the insight into its metal storage capacity. Biosci. Biotechnol. Biochem. 2015, 79, 45–50. [Google Scholar] [CrossRef] [Green Version]
- Reyes-López, C.; Hernández-Santoyo, A.; Pedraza-Escalona, M.; Mendoza, G.; Hernández-Arana, A.; Rodríguez-Romero, A. Insights into a conformational epitope of Hev b 6.02 (hevein). Biochem. Biophys. Res. Commun. 2004, 314, 123–130. [Google Scholar] [CrossRef]
- Blanco, C.; Carrillo, T.; Castillo, R.; Quiralte, J.; Cuevas, M. Latex allergy: Clinical features and cross-reactivity with fruits. Ann. Allergy 1994, 73, 309–314. [Google Scholar]
- Dall’Antonia, Y.; Pavlov, T.; Fuchs, H.; Breiteneder, H.; Keller, W. The high-resolution crtystal structure of an allergenic thaumatin-like protein, Pru av 2, isolated from ripe cherries. Pdb code 2AHN (to be published).
- Marangon, M.; Van Sluyter, S.C.; Waters, E.J.; Menz, R.I. Structure of haze forming proteins in white wines: Vitis vinifera thaumatin-like proteins. PLoS ONE 2014, 9, e113757. [Google Scholar] [CrossRef] [Green Version]
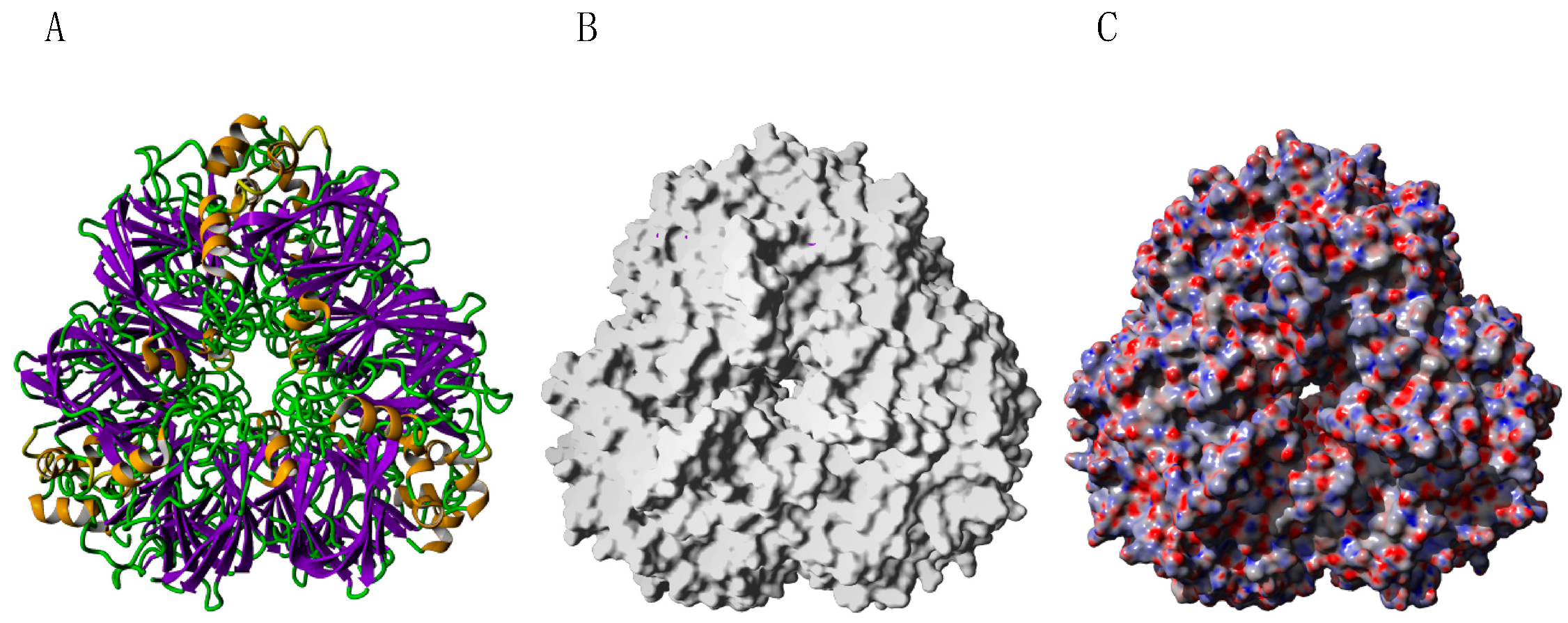
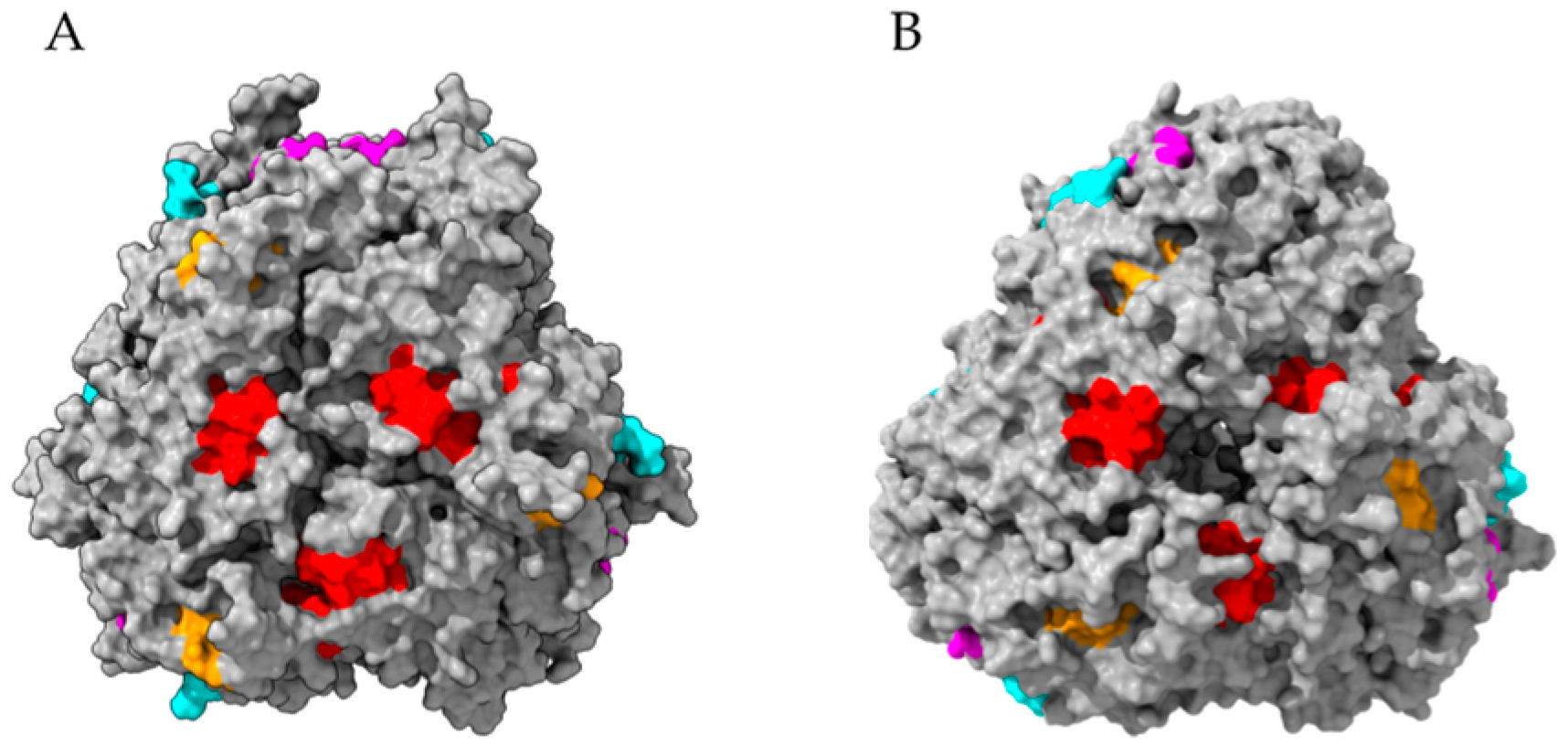
- Rodríguez-Romero, A.; Hernández-Santoyo, A.; Fuentes-Silva, D.; Palomares, L.A.; Muñoz-Cruz, S.; Yépez-Mulia, L.; Orozco-Martínez, S. Structural analysis of the endogenous glycoallergen Hev b 2 (endo-b-1,3-glucanase) from Hebea brasiliensis and its recognition by human basophils. Acta Crystallogr. D Biol. Crystallogr. 2014, 70, 329–341. [Google Scholar] [CrossRef] [Green Version]
- Barre, A.; Culerrier, R.; Granier, C.; Selman, L.; Peumans, W.J.; Van Damme, E.J.M.; Bienvenu, F.; Bienvenu, J.; Rougé, P. Mapping of IgE-binding epitopes on the major latex allergen Hev b 2 and the cross-reacting 1,3-β-glucanase fruit allergens as a molecular basis for the latex-fruit syndrome. Mol. Immunol. 2009, 46, 1595–1604. [Google Scholar] [CrossRef] [PubMed]
- Huecas, S.; Villalba, M.; Rodríguez, R. Ole e 9, a major olive pollen allergen is a 1,3-glucanase. Isolation, characterization, amino acid sequence, and tissue specificity. J. Biol. Chem. 2001, 276, 27959–27966. [Google Scholar] [CrossRef] [Green Version]
- Gallen, C.; Brunet, E.; Borges, J.P.; Rancé, F.; Barre, A.; Rougé, P. A case report of anaphylaxis to Physalis alkekengi fruit: The culprit is in the seeds! Rev. Fr. Allergol. 2018, 58, 527–529. [Google Scholar] [CrossRef]
- Shewry, P.R.; Napier, J.A.; Tatham, A.S. Seed storage proteins: Structures and biosynthesis. Plant Cell 1995, 7, 945–956. [Google Scholar]
- Jensen-Jarolim, E.; Schmid, B.; Bernier, F.; Berna, A.; Kinaciyan, T.; Focke, M.; Ebner, C.; Scheiner, O.; Boltz-Nitulescu, G. Allergologic exploration of germins and germin-like proteins, a new class of plant allergens. Allergy 2002, 57, 805–810. [Google Scholar] [CrossRef] [PubMed]
- Martín-Pedraza, L.; González, M.; Gómez, F.; Blanca-López, N.; Garrido-Arandia, M.; Rodríguez, R.; Torres, M.J.; Blanca, M.; Villalba, M.; Mayorga, C. Two non-specific lipid transfer proteins (nsLTPs) from tomato seeds are associated to severe symptoms of tomato-allergic patients. Mol. Nutr. Food Res. 2016, 60, 1172–1182. [Google Scholar] [CrossRef] [PubMed]
- Barre, A.; Nguyen, C.; Granier, C.; Benoist, H.; Rougé, P. IgE-binding epitopes of Pis v 1, Pis v 2 and Pis v 3, the pistachio (Pistacia vera) seed allergens. Allergies 2021, 1, 63–91. [Google Scholar] [CrossRef]
- Sirvent, S.; Cantó, B.; Cuesta-Herranz, J.; Gómez, F.; Blanca, N.; Canto, G.; Blanca, M.; Rodríguez, R.; Villalba, M.; Palomares, O. Act d 12 and Act d 13: Two novel, masked, relevant allergens in kiwifruit seeds. J. Allergy Clin. Immunol. 2014, 133, 1765–1767. [Google Scholar] [CrossRef] [Green Version]
- Barre, A.; Delplanque, A.; Simplicien, M.; Benoist, H.; Rougé, P. Molecular basis of cross-reactivity between Act c 12 and other seed 11S-globulin allergens. Rev. Fr. Allergol. 2017, 57, 58–66. [Google Scholar] [CrossRef]
- Crespo, J.F.; Retzek, M.; Foetisch, K.; Sierra-Maestro, E.; Cid-Sanchez, A.B.; Pascual, C.Y.; Conti, A.; Feliu, A.; Rodríguez, J.; Vieths, S.; et al. Germin-like protein Cit s 1 and profilin Cit s 2 are major allergens in orange (Citrus sinensis) fruits. Mol. Nutr. Food Res. 2006, 50, 282–290. [Google Scholar] [CrossRef]
- Ahrazem, O.; Ibañez, M.D.; López-Rorrejón, G.; Sánchez-Monge, R.; Sastre, J.; Lombardero, M.; Barber, D.; Salcedo, G. Orange germin-like glycoprotein Cit s 1: An equivocal allergen. Int. Arch. Allergy Immunol. 2006, 139, 96–103. [Google Scholar] [CrossRef] [PubMed]
- García, J.A.; Cuest-Herranz, J.; Perea Lam, N.; Díaz-Perales, A. Anaphylaxis mediated by thaumatin-like proteins. J. Investig. Allergol. Clin. Immunol. 2014, 24, 448–449. [Google Scholar]
- Bienvenu, F.; Barre, A.; Viel, S.; Garnier, L.; Guyon, C.; Favre-Metz, C.; Pauli, G.; Bienvenu, J.; Rougé, P. Are defensins important plant allergens ? Rev. Fr. Allergol. 2013, 53, 585–590. [Google Scholar] [CrossRef]
- Ballmer-Weber, B.; Hoffmann-Sommergruber, K.; Dramburg, S.; Hilger, C.; Santos, A.F.; Vecillas, L.d.L.; Aalberse, R.C.; Acevedo, N.; Aglas, L.; Altmann, F.; et al. B15—Fruit and vegetable allergy. EAACI Molecular Allergology User’s Guide 2.0. Pediatr. Allergy Immunol. 2023, 34, 200–208, e13854. [Google Scholar]
- van Ree, R.; Aalberse, R.C.; Dramburg, S.; Hilger, C.; Santos, A.F.; Vecillas, L.d.L.; Aalberse, R.C.; Acevedo, N.; Aglas, L.; Altmann, F.; et al. A02—Allergens and the allergenic composition of source materials. EAACI Molecular Allergology User’s Guide 2.0. Pediatr. Allergy Immunol. 2023, 34, 200–208, e13854. [Google Scholar]
- Pastorello, E.A.; Farioli, C.; Pravettoni, V.; Ispano, M.; Scibola, E.; Tranbaioli, C.; Giuffrida, M.G.; Ansaloni, R.; Godovac-Zimmermann, J.; Conti, A.; et al. The maize major allergen, which is responsible for food-induced allergic reactions, is a lipid transfer protein. J. Allergy Clin. Immunol. 2000, 106, 744–751. [Google Scholar] [CrossRef] [PubMed]
- Carnés, J.; Fernández-Caldas, E.; Gallego, M.T.; Ferrer, A.; Cuesta-Herranz, J. Pru p 3 (LTP) content in peach extracts. Allergy 2002, 57, 1071–1075. [Google Scholar] [CrossRef]
- Kaur, S.; Samota, M.K.; Choudhary, M.; Choudhary, M.; Pandey, A.K.; Sharma, A.; Thakur, J. How do plant defend themselves against pathogens—Biochemical mechanisms and genetic interventions. Physiol. Mol. Biol. Plants 2022, 28, 485–504. [Google Scholar] [CrossRef] [PubMed]
- Breen, S.; Williams, S.J.; Outram, M.; Kobe, B.; Solomon, P.S. Emerging insights into the functions of pathogenesis-related protein 1. Trends Plant Sci. 2017, 22, 871–879. [Google Scholar] [CrossRef]
- Ahrazem, O.; Jimeno, L.; López-Torrejón, G.; Herrero, M.; Espada, J.L.; Sánchez-Monge, R.; Duffort, O.; Barber, D.; Salcedo, G. Assessing allergen levels in peach and nectarine cultivars. Ann. Allergy Asthma Immunol. 2007, 99, 42–47. [Google Scholar] [CrossRef]
- Sancho, A.I.; van Ree, R.; van Leeuwen, A.; Meulenbroek, B.J.; van de Weg, E.W.; Gilissen, L.J.W.J.; Puehringer, H.; Laimer, M.; Martinelli, A.; Zaccharini, M.; et al. Measurement of lipid transfer protein in 88 apple cultivars. Int. Arch. Allergy Immunol. 2008, 146, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Gan, K.; Zhao, L.; Jia, H.; Zhu, Y.; Li, X.; Yang, Z.; Ye, Z.; Cao, K.; Wang, Z.; et al. Peach allergen Pru p 1 content is generally low in fruit but with large variations in different varieties. Clin. Transl. Allergy 2021, 11, e12034. [Google Scholar] [CrossRef]
- Matthes, A.; Schmitz-Eiberger, M. Apple (Malus domestica L. Borkh.) allergen Mad d 1: Effect of cultivar, cultivation system, and storage conditions. J. Agric. Food Chem. 2009, 57, 10548–10553. [Google Scholar] [CrossRef] [PubMed]
- Kurze, E.; Kock, V.; Scalzo, R.L.; Olbricht, K.; Schwab, W. Effect of strawberry genotype, cultivation and processing on the Fra a 1 allergen content. Nutrients 2018, 10, 857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sancho, A.I.; Foxall, R.; Browne, T.; Dey, R.; Zuidmeer, L.; Marzban, G.; Waldron, K.W.; van Ree, R.; Hoffmann-Sommergruber, K.; Laimer, M.; et al. Effect of postharvest storage on the expression of the apple allergen Mal d 1. J. Agric. Food Chem. 2006, 54, 5917–5923. [Google Scholar] [CrossRef] [PubMed]
- Schmitz-Eiberger, M.; Matthes, A. Effect of harvest maturity, duration of storage and shelf life of apples on the allergen Mal d 1, polyphenoloxidase activity and polyphenol content. Food Chem. 2011, 127, 1459–1464. [Google Scholar] [CrossRef]
- Kiewning, D.; Baab, G.; Schmitz-Eiberger, M. Impact of storage conditions of the apple allergen Mal d 1. Erwerbs-Obstbau 2012, 54, 177–183. [Google Scholar] [CrossRef]
- Hallmann, E.; Rozpara, E.; Slowianek, M.; Leszczynska, J. The effect of organic and conventional farm management on the allergenic potency and bioactive compounds status of apricots (Prunus armeniaca L.). Food Chem. 2019, 279, 171–178. [Google Scholar] [CrossRef]
- Song, H.; Asghari, M.; Zahedipour-Sheshglani, P.; Diao, E.; Xiang, X.; Liang, X.; Mandoulakani, B.A.; Qian, S. Investigation of pectolytic and PR genes expression, quality and phytochemical contents in organic and non-organic table grapes at harvest and during storage. Food Res. Int. 2023, 167, 112717. [Google Scholar] [CrossRef]
- Suriyamoorthy, P.; Madhuri, A.; Tangirala, S.; Michael, K.R.; Sivanandham, V.; Rawson, A.; Anandharaj, A. Comprehensive review of banana fruit allergy: Pathogenesis, diagnosis, management, and potential modification of allergens through food processing. Plant Foods Hum. Nutr. 2022, 77, 159–171. [Google Scholar] [CrossRef]
- Fernández-Rivas, M.; Bolhaar, S.; González-Mancebo, E.; Asero, R.; van Leeuwen, A.; Bohle, B.; Ma, Y.; Ebner, C.; Rigby, N.; Sancho, A.I.; et al. Apple allergy across Europe: How allergen sensitization profiles determine the clinical expression of allergies to plant foods. J. Allergy Clin. Immunol. 2006, 118, 481–488. [Google Scholar] [CrossRef] [PubMed]
- Gamboa, P.M.; Cáceres, O.; Antepara, I.; Sánchez-Monge, R.; Ahrazem, O.; Salcedo, G.; Barber, D.; Lombardero, M.; Sanz, M.L. Two different profiles of peach allergy in the north of Spain. Allergy 2007, 62, 408–414. [Google Scholar] [CrossRef] [PubMed]
- Reuter, A.; Lidholm, J.; Andersson, K.; Ostling, J.; Lundberg, M.; Scheurer, S.; Eneique, E.; Cistero-Bahima, A.; Miguel-Moncin, M.S.; Ballmer-Weber, B.K.; et al. A critical assessment of allergen component-based in vitro diagnosis in cherry allergy across Europe. Clin. Exp. Allergy 2006, 36, 815–823. [Google Scholar] [CrossRef] [PubMed]
- Dramburg, S.; Hilger, C.; Santos, A.F.; Vecillas, L.d.L.; Aalberse, R.C.; Acevedo, N.; Aglas, L.; Altmann, F.; Arruda, K.L.; Asero, R.; et al. EAACI Molecular Allergology User’s Guide 2.0. Pediatr. Allergy Immunol. 2023, 34, e13854. [Google Scholar]
- Asero, R.; Mistrello, G.; Roncarolo, D.; de Vries, S.C.; Gautier, M.F.; Ciurana, C.L.; Verbeek, E.; Mohammadi, T.; Knul-Brettlova, V.; Akkerdaas, J.H.; et al. Lipid transfer protein: A pan-allergen in plant-derived foods that is highly resistant to pepsin digestion. Int. Arch. Allergy Immunol. 2000, 122, 20–32. [Google Scholar] [CrossRef]
- Tordesillas, L.; Cubells-Baeza, N.; Gómez-Casado, C.; Berin, C.; Esteban, V.; Barcik, W.; O’Mahony, L.; Ramirez, C.; Pacios, L.F.; Garrodo-Arandia, M.; et al. Mechanisms underlying induction of allergic sensitization by Pru p 3. Clin. Exp. Allergy 2017, 47, 1398–1408. [Google Scholar] [CrossRef]
- Asero, R.; Mistrello, G.; Amato, S.; Roncarolo, D.; Martinelli, A.; Zaccarini, M. Peach fuzz contains large amounts of lipid transfer protein: Is this the cause of the high prevalence of sensitization to LTP in Mediterranean countries? Eur. Ann. Allergy Clin. Immunol. 2006, 38, 118–121. [Google Scholar]
- Zuidmeer, L.; Goldhahn, K.; Rona, R.J.; Gislason, D.; Madsen, C.; Summers, C.; Sodergren, E.; Dahlstrom, J.; Lindner, T.; Sigurdardottir, S.T.; et al. The prevalence of plant food allergies: A systematics review. J. Allergy Clin. Immunol. 2008, 121, 1210–1218. [Google Scholar] [CrossRef]
- Burney, P.G.; Summers, C.; Chinn, S.; Hooper, R.; van Ree, R.; Lidholm, J. Prevalence and distribution of sensitization to foods in the European Community Respiratory Health Suvey: A EuroPrevall Analysis. Allergy 2010, 65, 1182–1188. [Google Scholar]
- Burney, P.G.; Potts, J.; Kummeling, I.; Mills, E.N.; Clausen, M.; Dubakiene, R.; Barreales, L.; Fernández-Pérez, C.; Fernández-Rivas, M.; Le, T.M.; et al. The prevalence and distribution of food sensitization in European adults. Allergy 2014, 69, 365–371. [Google Scholar] [CrossRef] [Green Version]
- Fernández-Rivas, M.; González-Mancebo, E.; Rodríguez-Pérez, R.; Benito, C.; Sánchez-Monge, R.; Salcedo, G.; Alonso, M.D.; Rosadeo, A.; Tejedor, M.A.; Vila, C.; et al. Clinically relevant peach allergy is related to peach lipid transfer protein, Pru p 3, in the Spanish population. J. Allergy Clin. Immunol. 2003, 112, 789–795. [Google Scholar] [CrossRef] [PubMed]
- Boyano-Martinez, T.; Pedrosa, M.; Belver, T.; Quirce, S.; García-Ara, C. Peach allergy in Spanish children: Tolerance to the pulp and molecular sensitization profile. Pediatr. Allergy Immunol. 2013, 24, 168–172. [Google Scholar] [CrossRef] [PubMed]
- Pastorello, E.A.; Farioli, L.; Stafylaraki, C.; Mascheri, A.; Scibilia, J.; Pravettoni, L.; Piantanida, M.; Nichelatti, M.; Aseo, R. Anti-Pru p 3 IgE levels are inversely related to the age at oncet of peach-induced severe symptoms reported by peach-allergic adults. Int. Arch. Allergy Immunol. 2013, 162, 45–49. [Google Scholar] [CrossRef] [PubMed]
- Le, T.M.; Bublin, M.; Breiteneder, H.; Fernández-Rivas, M.; Asero, R.; Ballmer-Weber, B.; Barreales, L.; Bures, P.; Belohlavkova, S.; de Blay, F.; et al. Kiwifruit allergy across Europe: Clinical manifestation and IgE recognition patterns to kiwifruit allergens. J. Allergy Clin. Immunol. 2013, 131, 164–171. [Google Scholar] [CrossRef] [PubMed]
- González-Mancebo, E.; González-de-Olano, D.; Trujillo, M.J.; Santos, S.; Gandolfo-Cano, M.; Meléndez, A.; Juárez, R.; Morales, P.; Calso, A.; Mazuela, O.; et al. Prevalence of sensitization to lipid transfer proteins and profilins in a population of 430 patients in the south of Madrid. J. Investig. Allergol. Clin. Immunol. 2011, 21, 278–282. [Google Scholar]
- Klingebiel, C.; Chantran, Y.; Arif-Lusson, R.; Ehrenberg, A.; Östling, J.; Poisson, A.; Liabeuf, V.; Agabriel, C.; Birnbaum, J.; Porri, F.; et al. Pru p 7 sensitization is predominant cause of severe, cypress pollen-associated peach allergy. Clin. Exp. Allergy 2019, 49, 526–536. [Google Scholar] [CrossRef]
- Feng, H.; Zhou, J.; Lu, Y.; Zhao, Q.; Yang, Z.; Xiong, X.; Luo, N.; Chen, Y.; Wang, L.; Wu, Y. Prevalence of self-reported food allergy among adults in Jiangxi, China. World Allergy Organ. J. 2023, 16, 100773. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL-X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tool. Nucleic Acids Res. 1997, 15, 4876–4882. [Google Scholar] [CrossRef] [Green Version]
- Page, R.D.M. TREEVIEW: An application to display phylogenetic trees on personal computers. Computers Appl. BioSci. 1996, 12, 357–358. [Google Scholar]
- Kofler, S.; Asam, C.; Eckhard, U.; Wallner, M.; Ferreira, F.; Brandstetter, H. Crystallographically mapped ligand binding differs in high and low IgE binding isoforms of birch pollen allergen Bet v 1. J. Mol. Biol. 2012, 422, 109–123. [Google Scholar] [CrossRef] [Green Version]
- Rose, P.W.; Prlic, A.; Bi, C.; Bluhm, W.F.; Christie, C.H.; Dutta, S.; Green, R.K.; Goodsell, D.S.; Westbrook, J.D.; Woo, J.; et al. The RCSB Protein Data Bank: Views of structural biology for basic and applied research and education. Nucleic Acids res. 2015, 43, D345–D356. [Google Scholar] [CrossRef]
- Krieger, E.; Koraimann, G.; Vriend, G. Increasing the precision of comparative models with YASARA NOVA—A self-parameterizing force field. Proteins 2002, 47, 393–402. [Google Scholar] [CrossRef]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemistry of protein structures. J. Appl. Cryst. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Melo, F.; Feytmans, E. Assessing protein structures with a non-local atomic interaction energy. J. Mol. Biol. 1998, 277, 1141–1152. [Google Scholar] [CrossRef] [PubMed]
- Benkert, P.; Biasini, M.; Schwede, T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 2011, 27, 343–350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arnold, K.; Bordoli, L.; Kopp, J.; Schwede, T. The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics 2006, 22, 195–201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021, 30, 70–82. [Google Scholar] [CrossRef]



















Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barre, A.; Benoist, H.; Rougé, P. An Overview of Fruit Allergens: Structural, Functional, Phylogenetical, and Clinical Aspects. Allergies 2023, 3, 134-176. https://doi.org/10.3390/allergies3030010
Barre A, Benoist H, Rougé P. An Overview of Fruit Allergens: Structural, Functional, Phylogenetical, and Clinical Aspects. Allergies. 2023; 3(3):134-176. https://doi.org/10.3390/allergies3030010
Chicago/Turabian StyleBarre, Annick, Hervé Benoist, and Pierre Rougé. 2023. "An Overview of Fruit Allergens: Structural, Functional, Phylogenetical, and Clinical Aspects" Allergies 3, no. 3: 134-176. https://doi.org/10.3390/allergies3030010
APA StyleBarre, A., Benoist, H., & Rougé, P. (2023). An Overview of Fruit Allergens: Structural, Functional, Phylogenetical, and Clinical Aspects. Allergies, 3(3), 134-176. https://doi.org/10.3390/allergies3030010






