The Development of Molecular Markers for Peach Skin Blush and Their Application in Peach Breeding Practice
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Genomic PCR
2.3. Measurement of the Total Anthocyanins
2.4. Quantitative RT-PCR
3. Results
3.1. Comparison of Anthocyanin Accumulation Patterns in Different Peach Skin Color Types
3.2. Expression Analysis of Genes Associated with Peach Skin Blush
3.3. Sequence Variation in PpMYB10.1 Promoter Region
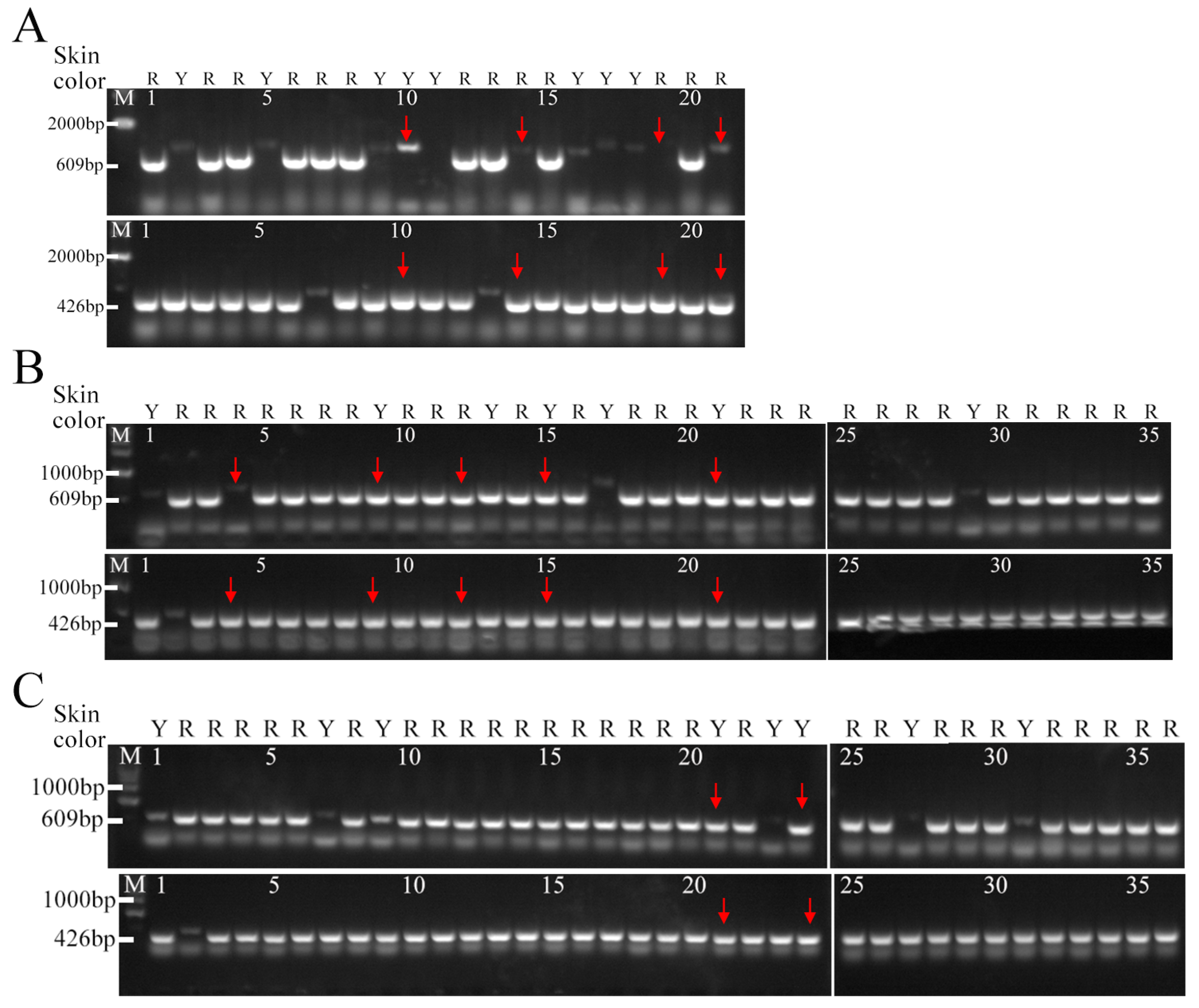
3.4. Development and Validation of Molecular Markers for Peach Skin Blush
3.5. Molecular Identification of the Hybrid Populations in the Promoter of MYB10.1
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kong, J.-M.; Chia, L.-S.; Goh, N.-K.; Chia, T.-F.; Brouillard, R. Analysis and biological activities of anthocyanins. Phytochemistry 2003, 64, 923–933. [Google Scholar] [CrossRef] [PubMed]
- Frett, T.J.; Reighard, G.L.; Okie, W.R.; Gasic, K. Mapping quantitative trait loci associated with blush in peach [Prunus persica (L.) Batsch]. Tree Genet. Genomes 2014, 10, 367–381. [Google Scholar] [CrossRef]
- Eduardo, I.; Pacheco, I.; Chietera, G.; Bassi, D.; Pozzi, C.; Vecchietti, A.; Rossini, L. QTL analysis of fruit quality traits in two peach intraspecific populations and importance of maturity date pleiotropic effect. Tree Genet. Genomes 2011, 7, 323–335. [Google Scholar] [CrossRef]
- Cantín, C.; Crisosto, C.; Ogundiwin, E.; Gradziel, T.; Torrents, J.; Moreno, M.; Gogorcena, Y. Chilling injury susceptibility in an intra-specific peach [Prunus persica (L.) Batsch] progeny. Postharvest Biol. Technol. 2010, 58, 79–87. [Google Scholar] [CrossRef]
- Beckman, T.; Alcazar, J.R.; Sherman, W.; Werner, D. Evidence for qualitative suppression of red skin color in peach. HortScience 2005, 40, 523–524. [Google Scholar] [CrossRef]
- Beckman, T.; Sherman, W. Probable qualitative inheritance of full red skin color in peach. HortScience 2003, 38, 1184–1185. [Google Scholar] [CrossRef]
- Tuan, P.A.; Bai, S.; Yaegaki, H.; Tamura, T.; Hihara, S.; Moriguchi, T.; Oda, K. The crucial role of PpMYB10.1 in anthocyanin accumulation in peach and relationships between its allelic type and skin color phenotype. BMC Plant Biol. 2015, 15, 280. [Google Scholar] [CrossRef]
- Bretó, M.; Cantín, C.M.; Iglesias, I.; Arús, P.; Eduardo, I. Mapping a major gene for red skin color suppression (highlighter) in peach. Euphytica 2017, 213, 14. [Google Scholar] [CrossRef]
- Zhao, L.; Sun, J.; Cai, Y.; Yang, Q.; Zhang, Y.; Ogutu, C.O.; Liu, J.; Zhao, Y.; Wang, F.; He, H.; et al. PpHYH is responsible for light-induced anthocyanin accumulation in fruit peel of Prunus persica. Tree Physiol. 2022, 42, 1662–1677. [Google Scholar] [CrossRef]
- Chen, Z.J. Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu. Rev. Plant Biol. 2007, 58, 377–406. [Google Scholar] [CrossRef]
- Espley, R.V.; Brendolise, C.; Chagne, D.; Kutty-Amma, S.; Green, S.; Volz, R.; Putterill, J.; Schouten, H.J.; Gardiner, S.E.; Hellens, R.P. Multiple repeats of a promoter segment causes transcription factor autoregulation in red apples. Plant Cell 2009, 21, 168–183. [Google Scholar] [CrossRef] [PubMed]
- Chagné, D.; Carlisle, C.M.; Blond, C.; Volz, R.K.; Whitworth, C.J.; Oraguzie, N.C.; Crowhurst, R.N.; Allan, A.C.; Espley, R.V.; Hellens, R.P. Mapping a candidate gene (MdMYB10) for red flesh and foliage colour in apple. BMC Genom. 2007, 8, 212. [Google Scholar] [CrossRef]
- Chagné, D.; Lin-Wang, K.; Espley, R.V.; Volz, R.K.; How, N.M.; Rouse, S.; Brendolise, C.; Carlisle, C.M.; Kumar, S.; De Silva, N. An ancient duplication of apple MYB transcription factors is responsible for novel red fruit-flesh phenotypes. Plant Physiol. 2013, 161, 225–239. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Sha, G.; Wei, T.; Ma, B.; Li, C.; Li, P.; Zou, Y.; Xu, L.; Ma, F. Linkage map and QTL mapping of red flesh locus in apple using a R1R1× R6R6 population. Hortic. Plant J. 2021, 7, 393–400. [Google Scholar] [CrossRef]
- Zhang, L.; Hu, J.; Han, X.; Li, J.; Gao, Y.; Richards, C.M.; Zhang, C.; Tian, Y.; Liu, G.; Gul, H.; et al. A high-quality apple genome assembly reveals the association of a retrotransposon and red fruit colour. Nat. Commun. 2019, 10, 1494. [Google Scholar] [CrossRef]
- Butelli, E.; Licciardello, C.; Zhang, Y.; Liu, J.; Mackay, S.; Bailey, P.; Reforgiato-Recupero, G.; Martin, C. Retrotransposons control fruit-specific, cold-dependent accumulation of anthocyanins in blood oranges. Plant Cell 2012, 24, 1242–1255. [Google Scholar] [CrossRef]
- Ou, C.; Zhang, X.; Wang, F.; Zhang, L.; Zhang, Y.; Fang, M.; Wang, J.; Wang, J.; Jiang, S.; Zhang, Z. A 14 nucleotide deletion mutation in the coding region of the PpBBX24 gene is associated with the red skin of “Zaosu Red” pear (Pyrus pyrifolia White Pear Group): A deletion in the PpBBX24 gene is associated with the red skin of pear. Hortic. Res. 2020, 7, 39. [Google Scholar] [CrossRef]
- Cheng, G.W.; Breen, P.J. Activity of phenylalanine ammonia-lyase (PAL) and concentrations of anthocyanins and phenolics in developing strawberry fruit. J. Am. Soc. Hortic. Sci. 1991, 116, 865–869. [Google Scholar] [CrossRef]
- Verde, I.; Abbott, A.G.; Scalabrin, S.; Jung, S.; Shu, S.; Marroni, F.; Zhebentyayeva, T.; Dettori, M.T.; Grimwood, J.; Cattonaro, F.; et al. The high-quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nat. Genet. 2013, 45, 487–494. [Google Scholar] [CrossRef]
- Fuentes, A.P.; Ruzzante, D.E. Whole-genome sequencing approaches for conservation biology: Advantages, limitations and practical recommendations. Mol. Ecol. 2017, 26, 5369–5406. [Google Scholar] [CrossRef]
- Bentley, D.R. Whole-genome re-sequencing. Curr. Opin. Genet. Dev. 2006, 16, 545–552. [Google Scholar] [CrossRef]
- Lu, Z.H.; Shen, Z.J.; Niu, L.; Pan, L.; Cui, G.C.; Zeng, W.F.; Wang, Z.Q. Molecular Marker-Assisted Identification of Yellow/White Flesh Trait for 122 Peach Cultivars (Lines). Sci. Agric. Sin. 2020, 53, 2929–2940. [Google Scholar]
- Wang, S.; Li, L.-X.; Zhang, Z.; Fang, Y.; Li, D.; Chen, X.-S.; Feng, S.-Q. Ethylene precisely regulates anthocyanin synthesis in apple via a module comprising MdEIL1, MdMYB1, and MdMYB17. Hortic. Res. 2022, 9, uhac034. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Cao, K.; Deng, C.; Li, Y.; Zhu, G.; Fang, W.; Chen, C.; Wang, X.; Wu, J.; Guan, L.; et al. An integrated peach genome structural variation map uncovers genes associated with fruit traits. Genome Biol. 2020, 21, 258. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Qian, J.; Han, Y.; Jia, Y.; Kuang, H.; Chen, J. Alternative splicing triggered by the insertion of a CACTA transposon attenuates LsGLK and leads to the development of pale-green leaves in lettuce. Plant J. 2022, 109, 182–195. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.L.; Diego, M.; John, P.; Richard, V.; Lidia, L.; Richard, E.; Roger, P.H.; David, C.; Daryl, D.R.; Michela, T.; et al. High temperature reduces apple fruit colour via modulation of the anthocyanin regulatory complex. Plant Cell Environ. 2011, 34, 1176–1190. [Google Scholar] [CrossRef]
- Dela, G.; Or, E.; Ovadia, R.; Nissim-Levi, A.; Weiss, D.; Oren-Shamir, M. Changes in anthocyanin concentration and composition in ‘Jaguar’ rose flowers due to transient high-temperature conditions. Plant Sci. 2003, 164, 333–340. [Google Scholar] [CrossRef]
- Wu, X.; Zhou, Y.; Yao, D.; Iqbal, S.; Gao, Z.; Zhang, Z. DNA methylation of LDOX gene contributes to the floral colour variegation in peach. J. Plant Physiol. 2020, 246–247, 153116. [Google Scholar] [CrossRef]
- Qian, M.; Sun, Y.; Allan, A.C.; Teng, Y.; Zhang, D. The red sport of ‘Zaosu’ pear and its red-striped pigmentation pattern are associated with demethylation of the PyMYB10 promoter. Phytochemistry 2014, 107, 16–23. [Google Scholar] [CrossRef]
- Xia, H.; Shen, Y.; Hu, R.; Wang, J.; Deng, H.; Lin, L.; Lv, X.; Deng, Q.; Xu, K.; Liang, D. Methylation of MYBA1 is Associated with the Coloration in “Manicure Finger” Grape Skin. J. Agric. Food Chem. 2021, 69, 15649–15659. [Google Scholar] [CrossRef]
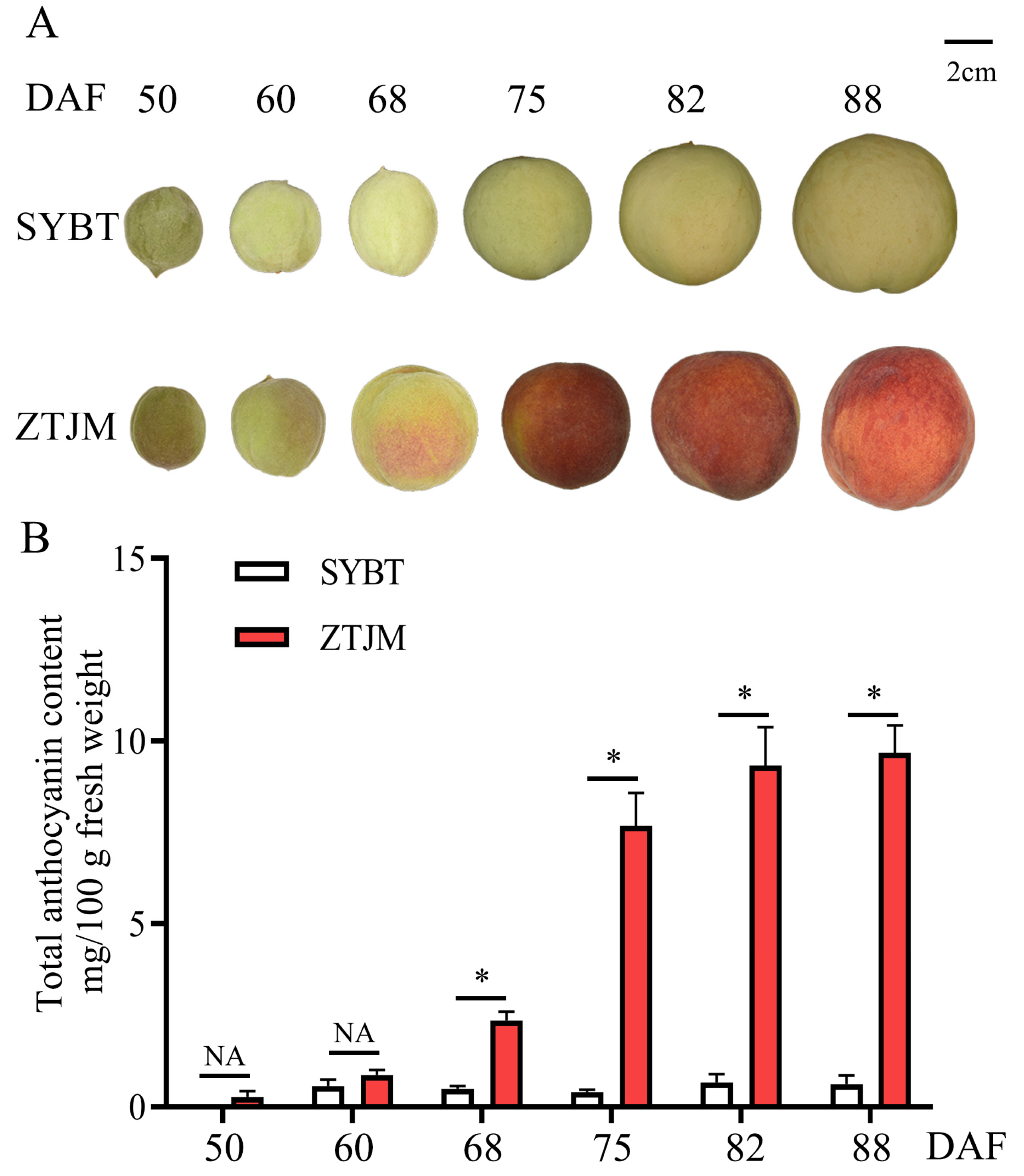
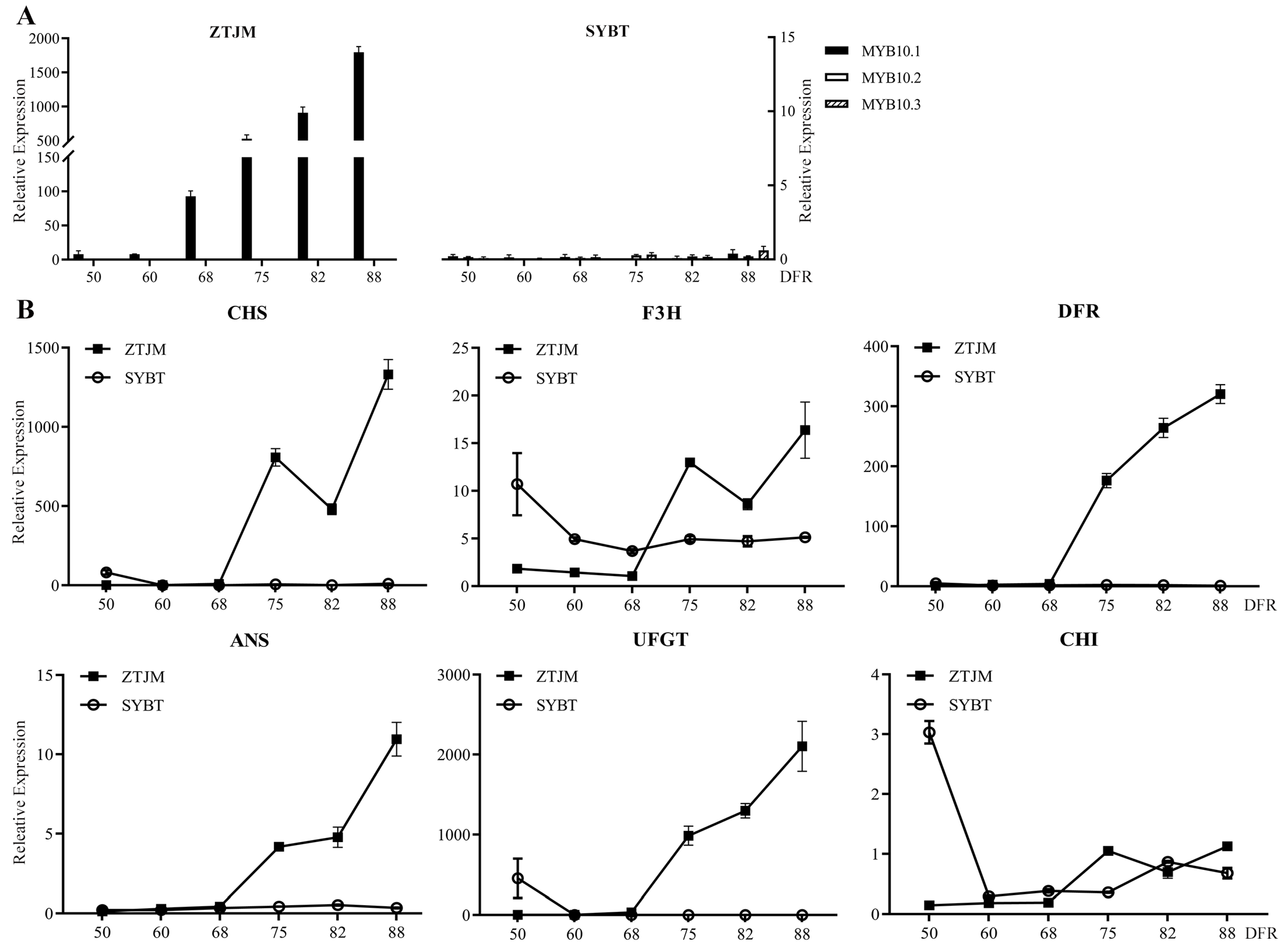
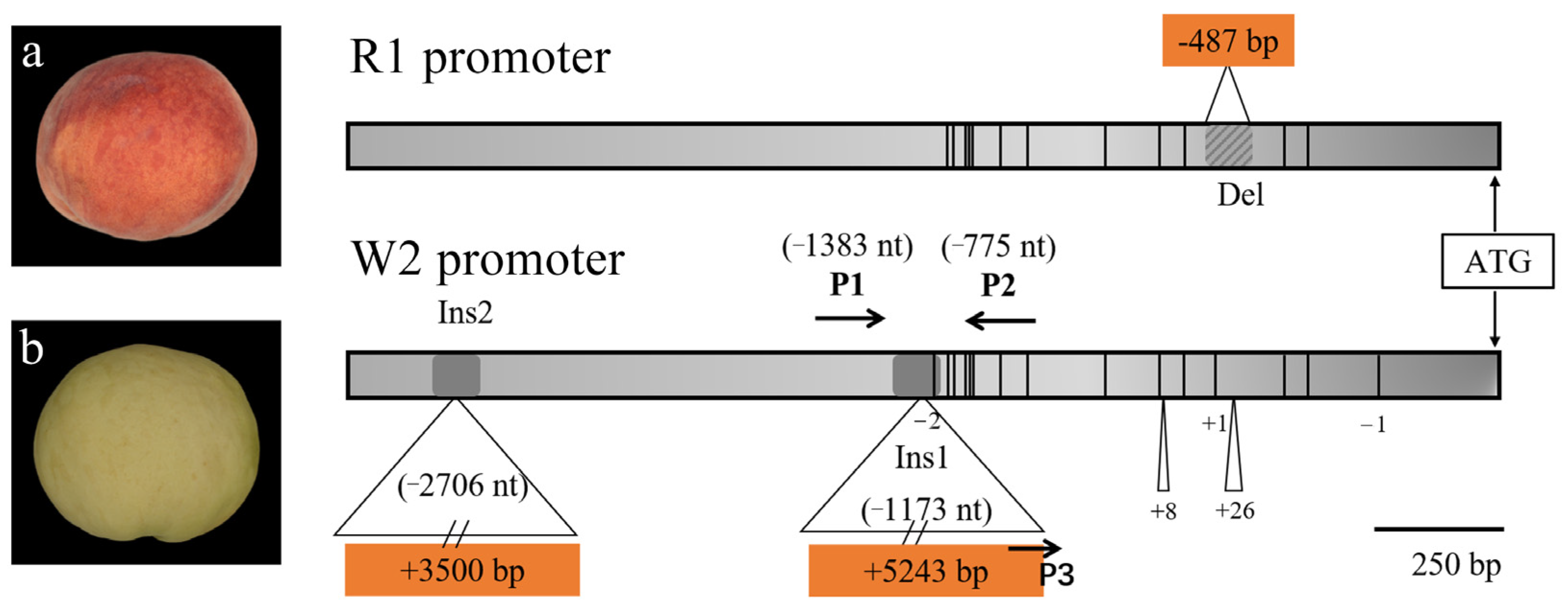
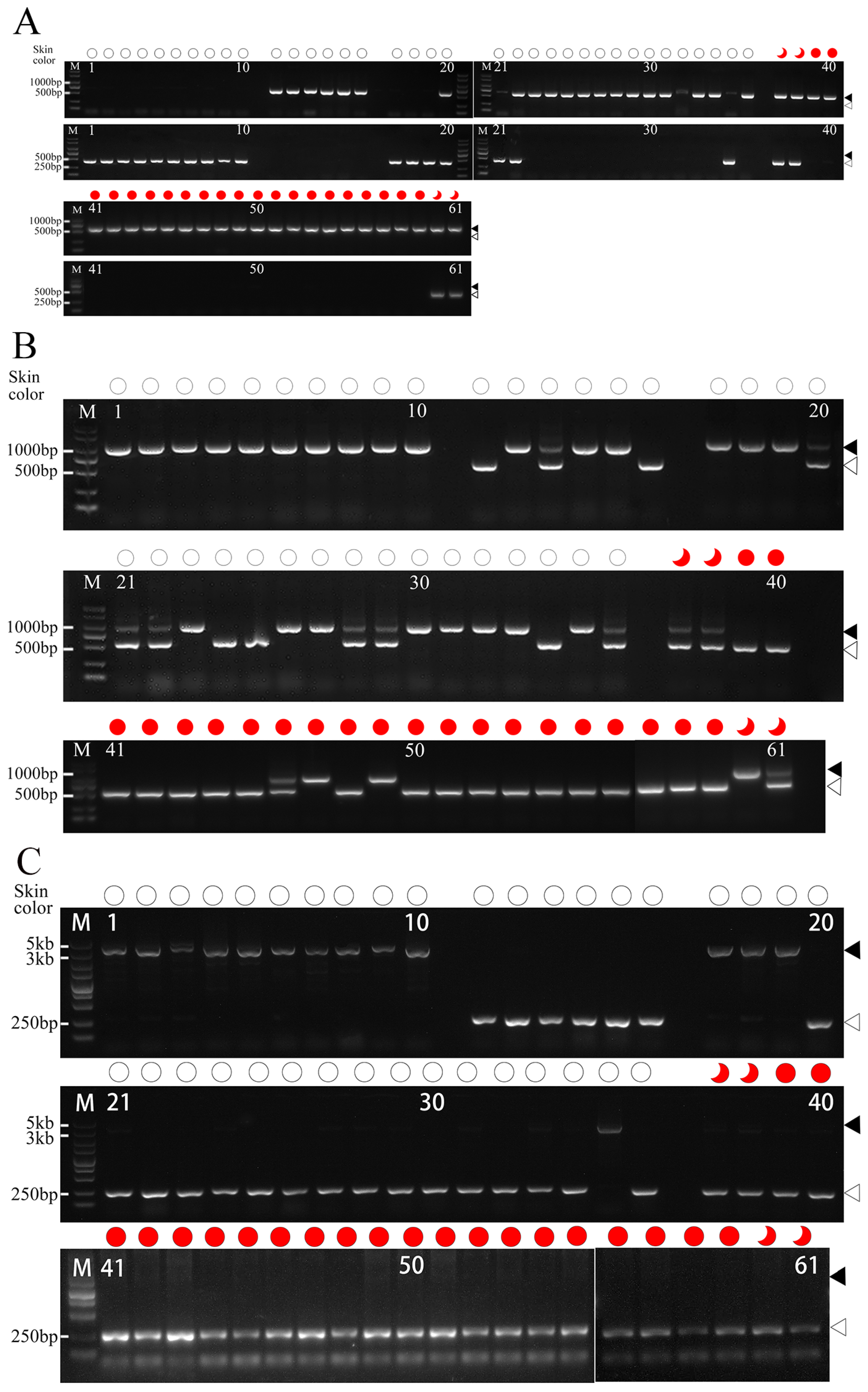
| No. | Accession Name | Resource Type | Sampled Location | Origin | Skin Color | MYB10.1 Allelic Insertion/bp | MYB10.1 Allelic Deletion/bp |
|---|---|---|---|---|---|---|---|
| P1 | Gui Zhou Shui Mi | Landrace variety | Guizhou | China | White | 426/426 | 1053/1053 |
| P2 | Yu Bai | Improved variety | Zhengzhou, Henan | China | White | 426/426 | 1053/1053 |
| P3 | Qing Zhou Bai Pi Mi Tao | Landrace variety | Qingzhou, Shandong | China | White | 426/426 | 1053/1053 |
| P4 | Da Xue Tao | Landrace variety | Mancheng, Hebei | China | White | 426/426 | 1053/1053 |
| P5 | Shui bai Tao | Landrace variety | Najing, Jiangsu | China | White | 426/426 | 1053/1053 |
| P6 | Shi Yu Bai Tao | Improved variety | Shijiazhuang, Hebei | China | White | 426/426 | 1053/1053 |
| P7 | NJC77 | Improved variety | Xinzexizhou | America | Yellow | 426/426 | 1053/1053 |
| P8 | Maria Serena | Improved variety | Florence | Italy | Yellow | 426/426 | 1053/1053 |
| P9 | KXN43-37 | Superior line | Zhengzhou, Henan | China | White | 426/426 | 1053/1053 |
| P10 | KXN43-52 | Superior line | Zhengzhou, Henan | China | White | 426/426 | 1053/1053 |
| P11 | Mi Yang Shan | Landrace variety | Hetian, Xinjiang | China | Green | 609/609 | 487/487 |
| P12 | Bai Li Hu | Landrace variety | Mengzi, Yunnan | China | Green | 609/609 | 1053/1053 |
| P13 | Tu-2 | Landrace variety | Yecheng, Xinjiang | China | Green | 609/609 | 487/1053 |
| P14 | Tie 4-1 | Landrace variety | Yecheng, Xinjiang | China | Green | 609/609 | 1053/1053 |
| P15 | Da Li He Huang Rou | Landrace variety | Hetian, Xinjiang | China | Yellow | 609/609 | 1053/1053 |
| P16 | Gua Tao | Landrace variety | Napo, Guangxi | China | Yellow | 609/609 | 487/487 |
| P17 | Yang Zhou 3 | Improved variety | Yangzhou, Jiangsu | China | White | 426/426 | 1053/1053 |
| P18 | Yun Shu 2 | Improved variety | Hangzhou, Zhejiang | China | Green | 426/426 | 1053/1053 |
| P19 | Pin Ding You Pan Tao | Improved variety | Zhengzhou, Henan | China | White | 426/426 | 1053/1053 |
| P20 | Suan Tao | Landrace variety | Feicheng, Shandong | China | Green | 609/426 | 487/1053 |
| P21 | Chi Yuan Mi | Improved variety | Nanjing, Jiangsu | China | White | 426/426 | 487/1053 |
| P22 | Tai Yuan Shui Mi | Landrace variety | Taiyuan, Shanxi | China | White | 609/426 | 487/1053 |
| P23 | Gao Tai 1 | Landrace variety | Gaotai, Gansu | China | Green | 609/609 | 1053/1053 |
| P24 | Ying Xue | Landrace variety | Beijing | China | Green | 609/609 | 487/487 |
| P25 | Han Lu Mi | Landrace variety | Qingdao, Shandong | China | Green | 609/609 | 487/487 |
| P26 | Dunhuang Dong Tao | Landrace variety | Dunhuang, Gansu | China | White | 609/609 | 1053/1053 |
| P27 | Xiang Tao | Landrace variety | Dalian, Liaoning | China | Green | 609/609 | 1053/1053 |
| P28 | Bai He Tao | Landrace variety | Yunguigaoyuan | China | White | 609/609 | 487/1053 |
| P29 | Bai Nian He | Landrace variety | Mengzi, Yunnan | China | White | 609/609 | 487/1053 |
| P30 | Golden Queen | Improved variety | Unknow | New Zealand | Yellow | 609/609 | 1053/1053 |
| P31 | Everts | Improved variety | Unknow | America | Yellow | 609/609 | 1053/1053 |
| P32 | Huang Yan | Landrace variety | Jingning, Uunnan | China | Yellow | 609/609 | 1053/1053 |
| P33 | Nan Shan 1 | Landrace variety | Ningxian, Gansu | China | Yellow | 609/609 | 1053/1053 |
| P34 | Bei Jing Wan Pan Tao | Landrace variety | Beijing | China | Green | 609/609 | 487/487 |
| P35 | Feicheng Bai Li 10 | Landrace variety | Feicheng, Shandong | China | White | 426/426 | 1053/1053 |
| P36 | Anlong Bai Tao | Landrace variety | Anlong, Guizhou | China | White | 609/609 | 487/1053 |
| P37 | KXN43-04 | Superior line | Zhengzhou, Henan | China | Red | 609/426 | 487/1053 |
| P38 | KXN42-125 | Superior line | Zhengzhou, Henan | China | Red | 609/426 | 487/1053 |
| P39 | KXN43-57 | Superior line | Zhengzhou, Henan | China | Red | 609/609 | 487/487 |
| P40 | KXN43-59 | Superior line | Zhengzhou, Henan | China | Red | 609/609 | 487/487 |
| P41 | Da Hong Tao | Landrace variety | Yuncheng, Yuncheng | China | Red | 609/609 | 487/487 |
| P42 | An Nong Shui Mi | Improved variety | Shouxian, Anhui | China | Red | 609/609 | 487/487 |
| P43 | Xi Nong Shui Mi | Improved variety | Wugong, Shanxi | China | Red | 609/609 | 487/487 |
| P44 | Ba Hua | Landrace variety | Wuxi, Jiangsu | China | Red | 609/609 | 487/487 |
| P45 | Bao Lu | Improved variety | Hangzhou, Zhejiang | China | Red | 609/609 | 487/487 |
| P46 | Hu Jing Mi Lu | Landrace variety | Wuxi, Jiangsu | China | Red | 609/609 | 487/1053 |
| P47 | Yi Xian Hong | Landrace variety | Changli, Hebei | China | Red | 609/609 | 1053/1053 |
| P48 | Qi Tao | Landrace variety | Tianshui, Gansu | China | Red | 609/609 | 487/487 |
| P49 | Ji Zui Bai | Landrace variety | Shangshui, Henan | China | Red | 609/609 | 1053/1053 |
| P50 | Hatsukami | Improved variety | Unknow | Japan | Red | 609/609 | 487/487 |
| P51 | Nagasawa Hakuho | Improved variety | Shanlixian | Japan | Red | 609/609 | 487/487 |
| P52 | Gan Xuan 4 | Landrace variety | Lanzhou, Gansu | China | Red | 609/609 | 487/487 |
| P53 | Hong Gan Lu | Improved variety | Dalian, Liaoning | China | Red | 609/609 | 487/487 |
| P54 | Liquan 54 | Landrace variety | Liquan, Shanxi | China | Red | 609/609 | 487/487 |
| P55 | Datuan Mi Lu | Improved variety | Nanhui, Shanghai | China | Red | 609/609 | 487/487 |
| P56 | Er Zao Tao | Landrace variety | Chenggon, Yunnan | China | Red | 609/609 | 487/487 |
| P57 | Wu Yue Bai | Landrace variety | Xihua, Henan | China | Red | 609/609 | 487/487 |
| P58 | Zhong Tao Jin Mi | Improved variety | Zhengzhou, Henan | China | Red | 609/609 | 487/487 |
| P59 | Fantasia | Improved variety | Jiazhou, Fresno | America | Red | 609/609 | 487/487 |
| P60 | Su Hong | Landrace variety | Jiangsu | China | Red | 609/426 | 1053/1053 |
| P61 | Shi Tou Tao | Landrace variety | Zhengzhou, Henan | China | Red | 609/426 | 487/1053 |
| Prime Name | Primer Sequence | Annealing Temperature (°C) | Size (bp) |
|---|---|---|---|
| Mpro1 F | GGGAAACGATGTAAAGCCAC | 55 °C | 609 |
| Mpro1 R | CGAATATCAATGCAGCATCGTG | ||
| Mpro2 F | CGAATATCAATGCAGCATCGTG | 55 °C | 426 |
| Mpro2 R | CGGTTTTGGTCTTGCGCTAT | ||
| Mpro3 F | GTGGCTACGTACGGTTCTCC | 55 °C | 487/1053 |
| Mpro3 R | TTTTATGCCCTGCCTGCTCA | ||
| CR F | CCTTCTCCTATGGACTTTCTCCC | 58 °C | 269/3500 |
| CR R | AGCGTCGTGGTTTATGAGGG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, T.; Wang, J.; Lu, X.; Wu, J.; Wang, L. The Development of Molecular Markers for Peach Skin Blush and Their Application in Peach Breeding Practice. Horticulturae 2023, 9, 887. https://doi.org/10.3390/horticulturae9080887
Guo T, Wang J, Lu X, Wu J, Wang L. The Development of Molecular Markers for Peach Skin Blush and Their Application in Peach Breeding Practice. Horticulturae. 2023; 9(8):887. https://doi.org/10.3390/horticulturae9080887
Chicago/Turabian StyleGuo, Tianfa, Jiao Wang, Xinxin Lu, Jinlong Wu, and Lirong Wang. 2023. "The Development of Molecular Markers for Peach Skin Blush and Their Application in Peach Breeding Practice" Horticulturae 9, no. 8: 887. https://doi.org/10.3390/horticulturae9080887
APA StyleGuo, T., Wang, J., Lu, X., Wu, J., & Wang, L. (2023). The Development of Molecular Markers for Peach Skin Blush and Their Application in Peach Breeding Practice. Horticulturae, 9(8), 887. https://doi.org/10.3390/horticulturae9080887






