Callus Induction and Adventitious Root Regeneration of Cotyledon Explants in Peach Trees
Abstract
1. Introduction
2. Material and Methods
2.1. Plant Material and Growth Conditions
2.2. Estimation of Callus Induction Rate and Rooting Rate in Peach trees
2.3. Histological and Ultrastructural Analyses
2.4. Measurement of Plant Metabolites
2.5. RNA Extraction, Quantitative Real-Time PCR and Transcriptome Analysis
2.6. Transcriptome Analysis and Identification of Differentially Expressed Genes (DEGs)
3. Results
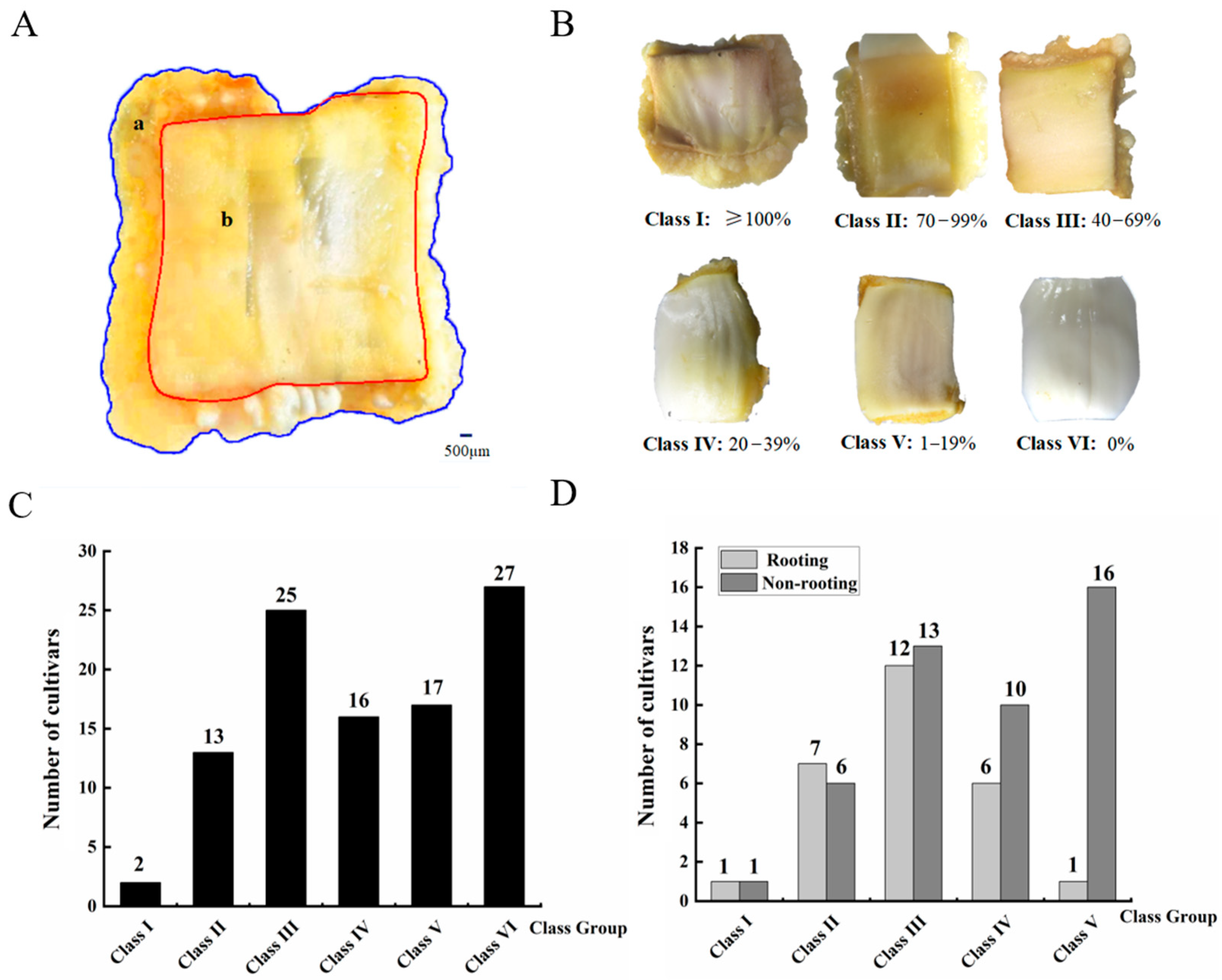
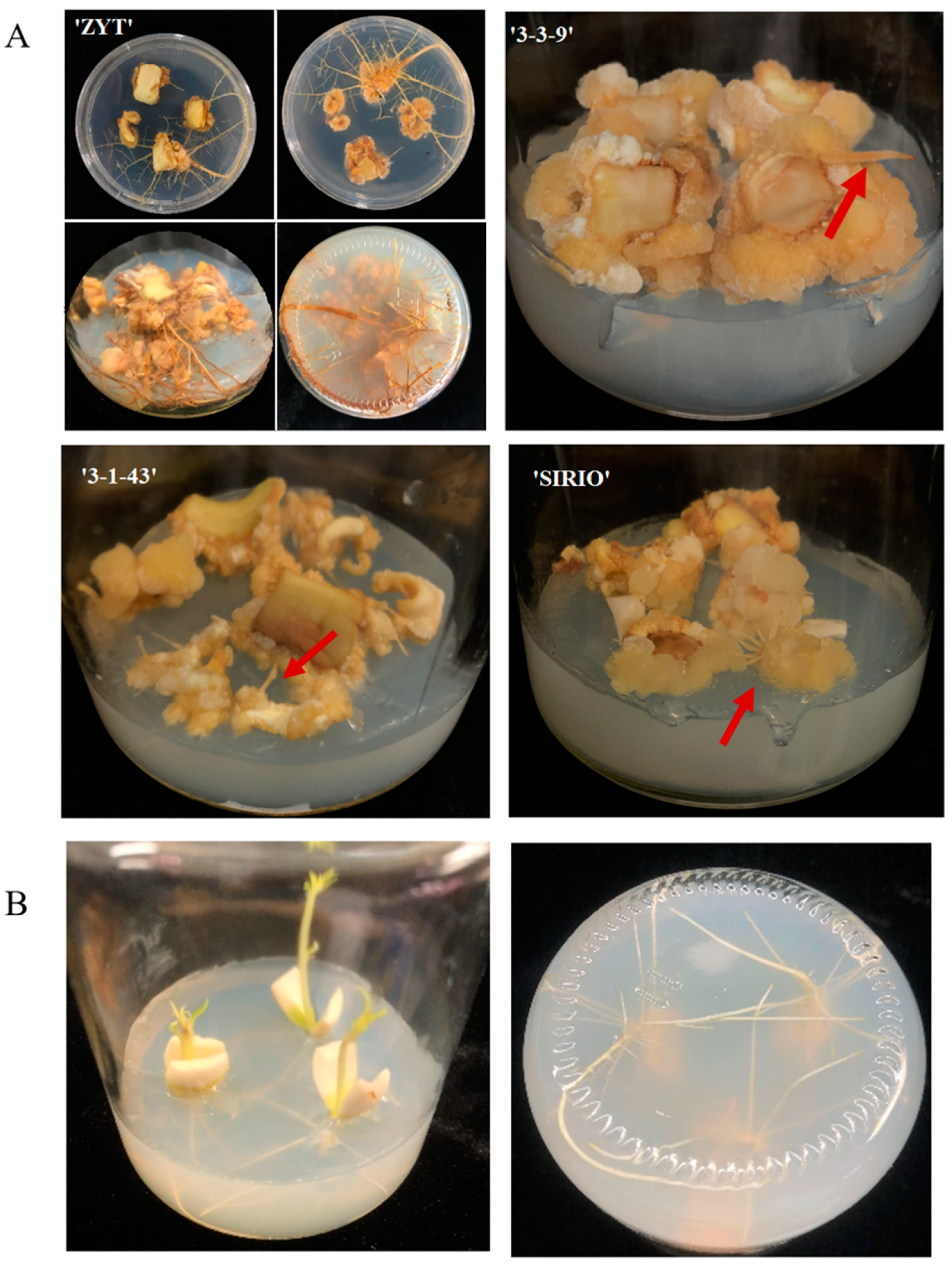
3.1. Difference in Callus Induction Rate and Callus-Based Root Regeneration among Peach Cultivars
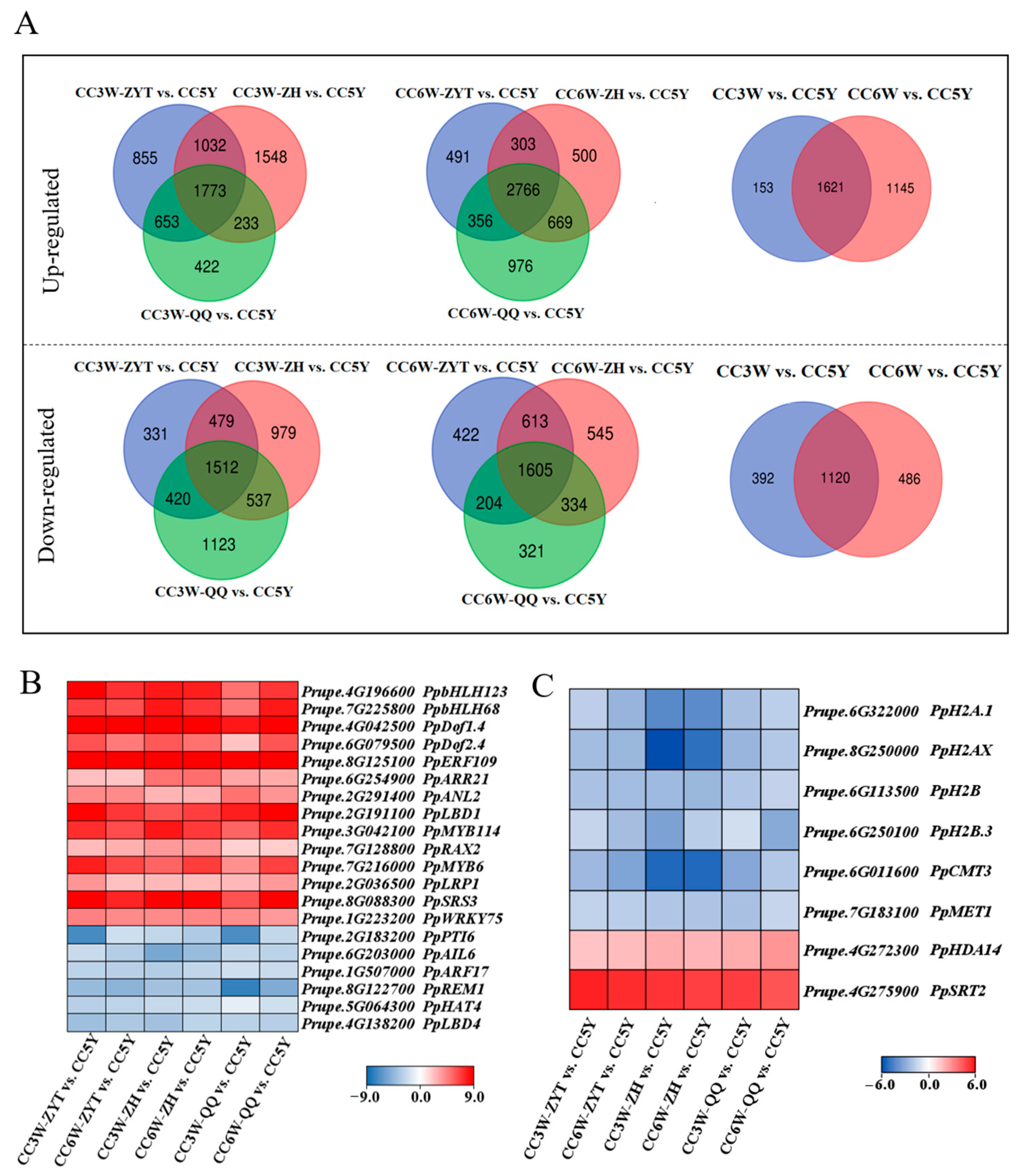
3.2. Identification of Candidate Genes Related to Adventitious Root Formation during Callus Culture
3.3. Expression of Candidate Genes Associated with Callus Adventitious Rooting in the Cotyledon-Derived Callus
3.4. Cellular Morphological Changes in the Cotyledon-Derived Callus during the Process of Root Differentiation
4. Discussion
4.1. Induction of Callus from Cotyledon and Its Root Regeneration in Peach trees
4.2. PpLBD1 Is Likely Involved in Callus Adventitious Rooting in Peach Trees
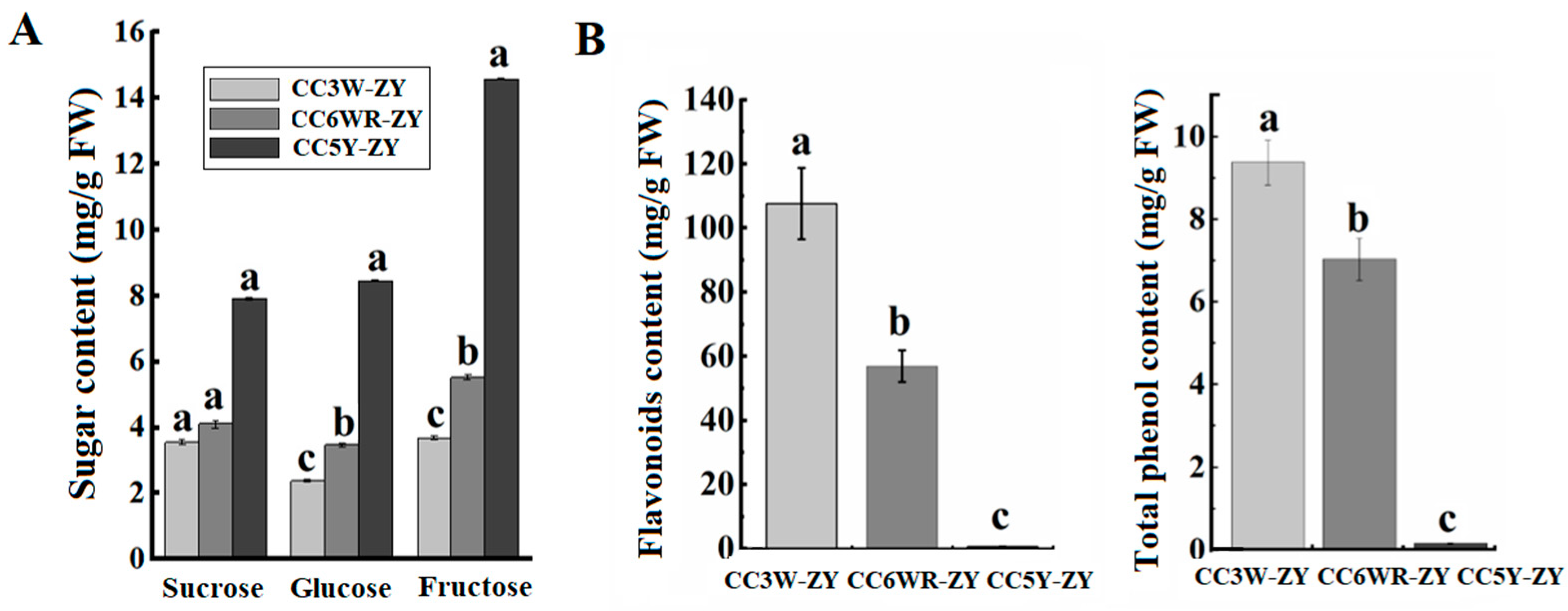
4.3. The Effect of Metabolites on Callus Adventitious Rooting in Peach
4.4. Epigenetic Regulation in Callus Adventitious Rooting in Peach
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Abbott, A.G.; Lecouls, A.C.; Wang, Y.; Georgi, L.; Scorza, R.; Reigherd, G. Peach: The model genome for rosaceae genomics. Acta Hortic. 2002, 592, 199–209. [Google Scholar] [CrossRef]
- Verde, I.; Jenkins, J.; Dondini, L.; Micali, S.; Pagliarani, G.; Vendramin, E.; Paris, R.; Aramini, V.; Gazza, L.; Rossini, L.; et al. The Peach v2. 0 release: High-resolution linkage mapping and deep resequencing improve chromosome-scale assembly and contiguity. BMC Genom. 2017, 18, 225. [Google Scholar] [CrossRef] [PubMed]
- Ricci, A.; Sabbadini, S.; Prieto, H.; Padilla, I.M.; Dardick, C.; Li, Z.J.; Scorza, R.; Limera, C.; Mezzetti, B.; Perez-Jimenez, M.; et al. Genetic transformation in peach (Prunus persica L.): Challenges and ways forward. Plants 2020, 9, 971. [Google Scholar] [CrossRef] [PubMed]
- San, B.; Li, Z.G.; Hu, Q.; Reighard, G.L.; Luo, H. Adventitious shoot regeneration from in vitro cultured leaf explants of peach rootstock Guardian is significantly enhanced by silver thiosulfate. Plant Cell Tissue Organ Cult. 2015, 120, 757–765. [Google Scholar] [CrossRef]
- Ikeuchi, M.; Favero, D.S.; Sakamoto, Y.; Iwase, A.; Coleman, D.; Rymen, B.; Sugimoto, K. Molecular mechanisms of plant regeneration. Annu. Rev. Plant Biol. 2019, 70, 377–406. [Google Scholar] [CrossRef]
- Ikeuchi, M.; Ogawa, Y.; Iwase, A.; Sugimoto, K. Plant regeneration: Cellular origins and molecular mechanisms. Development 2016, 143, 1442–1451. [Google Scholar] [CrossRef]
- Wu, G.Y.; Wei, X.L.; Wang, X.; Wei, Y. Induction of somatic embryogenesis in different explants from Ormosia henryi Prain. Plant Cell Tissue Organ Cult. 2020, 142, 229–240. [Google Scholar] [CrossRef]
- Hammerschlag, F.A.; Bauchan, G.R.; Scorza, R. Regeneration of peach plants from callus derived from immature embryos. Theor. Appl. Genet. 1985, 70, 248–251. [Google Scholar] [CrossRef]
- Yao, J.L.; Cohen, D.; Atkinson, R.; Richardson, K.; Morris, B. Regeneration of transgenic plants from the commercial apple cultivar Royal Gala. Plant Cell Rep. 1995, 14, 407–412. [Google Scholar] [CrossRef]
- Huang, H.; Wei, Y.; Zhai, Y.J.; Ouyang, K.X.; Chen, X.Y.; Bai, L.H. High frequency regeneration of plants via callus-mediated organogenesis from cotyledon and hypocotyl cultures in a multipurpose tropical tree (Neolamarkia Cadamba). Sci. Rep. 2020, 10, 4558. [Google Scholar] [CrossRef]
- Zarinjoei, F.; Rahmani, M.S.; Shabanian, N. In vitro plant regeneration from cotyledon-derived callus cultures of leguminous tree Gleditsia caspica Desf. New For. 2014, 45, 829–841. [Google Scholar] [CrossRef]
- Anandan, R.; Prakash, M.; Deenadhayalan, T.; Nivetha, R.; Kumar, N.S. Efficient invitro plant regeneration from cotyledon-derived callus cultures of sesame (Sesamum indicum L.) and genetic analysis of True-to-Type regenerants using RAPD and SSR markers. S. Afr. J. Bot. 2018, 119, 244–251. [Google Scholar] [CrossRef]
- Parmar, N.; Kanwar, K.; Thakur, A.K. In Vitro Organogenesis from cotyledon derived callus cultures of Punica granatum L. cv. Kandhari Kabuli. Natl. Acad. Sci. Lett. 2012, 35, 215–220. [Google Scholar] [CrossRef]
- Slusarkiewicz-Jarzina, A.; Ponitka, A.; Kaczmarek, Z. Influence of cultivar, explant source and plant growth regulator on callus induction and plant regeneration of Cannabis sativa L. Acta Biol. Crac. Ser. Bot. 2005, 47, 145–151. [Google Scholar]
- Wang, C.; Ma, H.; Zhu, W.; Zhang, J.; Zhao, X.; Li, X. Seedling-derived leaf and root tip as alternative explants for callus induction and plant regeneration in maize. Physiol Plant. 2021, 172, 1570–1581. [Google Scholar] [CrossRef] [PubMed]
- Gentile, A.; Monticelli, S.; Damiano, C. Adventitious shoot regeneration in peach [Prunus persica (L.) Batsch]. Plant Cell Rep. 2002, 20, 1011–1016. [Google Scholar] [CrossRef]
- Smigocki, A.C.; Hammerschlag, F.A. Regeneration of plants from peach embryo cells infected with a shooty mutant strain of Agrobacterium. J. Am. Soc. Hortic. Sci. 1991, 116, 1092–1097. [Google Scholar] [CrossRef]
- Perez-Clemente, R.M.; Perez-Sanjuan, A.; Garcia-Ferriz, L.; Beltran, J.P.; Canas, L.A. Transgenic peach plants (Prunus persica L.) produced by genetic transformation of embryo sections using the green fluorescent protein (GFP) as an in vivo marker. Mol. Breed. 2004, 14, 419–427. [Google Scholar] [CrossRef]
- Xu, S.L.; Lai, E.H.; Zhao, L.; Cai, Y.M.; Ogutu, C.O.; Cherono, S.; Han, Y.P.; Zheng, B.B. Development of a fast and efficient root transgenic system for functional genomics and genetic engineering in peach. Sci. Rep. 2020, 10, 2836. [Google Scholar] [CrossRef]
- Reighard, G.L.; Cain, D.W.; Newall, W.C. Rooting and survival potential of hardwood cuttings of 406 species, cultivars, and hybrids of Prunus. Hortscience 1990, 25, 517–518. [Google Scholar] [CrossRef]
- Justamante, M.S.; Mhimdi, M.; Molina-Perez, M.; Albacete, A.; Moreno, M.A.; Mataix, I.; Perez-Perez, J.M. Effects of auxin (Indole-3-butyric Acid) on adventitious root formation in peach-based Prunus rootstocks. Plants 2022, 11, 913. [Google Scholar] [CrossRef] [PubMed]
- Bellini, C.; Pacurar, D.I.; Perrone, I. Adventitious roots and lateral roots: Similarities and differences. Annu. Rev. Plant Biol. 2014, 65, 639. [Google Scholar] [CrossRef] [PubMed]
- Shuai, B.; Reynaga-Pena, C.G.; Springer, P.S. The lateral organ boundaries gene defines a novel, plant-specific gene family. Plant Physiol. 2002, 129, 747–761. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.W.; Cho, C.; Pandey, S.K.; Park, Y.; Kim, M.J.; Kim, J. LBD16 and LBD18 acting downstream of ARF7 and ARF19 are involved in adventitious root formation in Arabidopsis. BMC Plant Biol. 2019, 19, 46. [Google Scholar] [CrossRef]
- Yamauchi, T.; Tanaka, A.; Inahashi, H.; Nishizawa, N.K.; Tsutsumi, N.; Inukai, Y.; Nakazono, M.N. Fine control of aerenchyma and lateral root development through AUX/IAA-and ARF-dependent auxin signaling. Proc. Natl. Acad. Sci. USA 2019, 116, 20770–20775. [Google Scholar] [CrossRef]
- Inukai, Y.; Sakamoto, T.; Ueguchi-Tanaka, M.; Shibata, Y.; Gomi, K.; Umemura, I.; Hasegawa, Y.; Ashikari, M.; Kitano, H.; Matsuoka, M. Crown rootless1, which is essential for crown root formation in rice, is a target of an AUXIN RESPONSE FACTOR in auxin signaling. Plant Cell. 2005, 17, 1387–1396. [Google Scholar] [CrossRef]
- Liu, H.J.; Wang, S.F.; Yu, X.B.; Yu, J.; He, X.W.; Zhang, S.L.; Shou, H.X.; Wu, P. ARL1, a LOB-domain protein required for adventitious root formation in rice. Plant J. 2005, 43, 47–56. [Google Scholar] [CrossRef]
- Bernhardt, C.; Lee, M.M.; Gonzalez, A.; Zhang, F.; Lloyd, A.; Schiefelbein, J. The bHLH genes GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Development 2003, 130, 6431–6439. [Google Scholar] [CrossRef]
- Ramsay, N.A.; Glover, B.J. MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 2005, 10, 63–70. [Google Scholar] [CrossRef]
- Zheng, B.B.; Liu, J.J.; Gao, A.Q.; Chen, X.M.; Gao, L.L.; Liao, L.; Luo, B.W.; Ogutu, C.O.; Han, Y.P. Epigenetic reprogramming of H3K27me3 and DNA methylation during leaf-to-callus transition in peach. Hortic. Res. 2022, 9, uhac132. [Google Scholar] [CrossRef]
- Zhou, H.C.; Li, M.; Zhao, X.; Fan, X.; Guo, A.G. Plant regeneration from in vitro leaves of the peach rootstock ‘Nemaguard’ (Prunus persica × P. davidiana). Plant Cell Tissue Organ Cult. 2010, 101, 79–87. [Google Scholar] [CrossRef]
- Perez-Jimenez, M.; Lopez-Soto, M.B.; Cos-Terrer, J. In vitro callus induction from adult tissues of peach (Prunus persica L. Batsch). Vitr. Cell. Dev. Biol. -Plant 2013, 49, 79–84. [Google Scholar] [CrossRef]
- Zhen, Q.L.; Fang, T.; Peng, Q.; Liao, L.; Zhao, L.; Owiti, A.; Han, Y.P. Developing gene-tagged molecular markers for evaluation of genetic association of apple SWEET genes with fruit sugar accumulation. Hortic. Res. 2018, 5, 14. [Google Scholar] [CrossRef] [PubMed]
- Tong, Z.; Gao, Z.; Wang, F.; Zhou, J.; Zhang, Z. Selection of reliable reference genes for gene expression studies in peach using real-time PCR. BMC Mol. Biol. 2009, 10, 71. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 1–21. [Google Scholar] [CrossRef]
- Varet, H.; Brillet-Guéguen, L.; Coppée, J.Y.; Dillies, M.A. SARTools: A DESeq2-and EdgeR-based R pipeline for comprehensive differential analysis of RNA-Seq data. PLoS ONE 2016, 11, e0157022. [Google Scholar] [CrossRef]
- Paul, P. Using ANOVA for gene selection from microarray studies of the nervous system. Methods 2003, 31, 282–289. [Google Scholar] [CrossRef]
- Liu, L.; Fan, X.; Zhang, J.W.; Yan, M.L.; Bao, M.Z. Long-term cultured callus and the effect factor of high-frequency plantlet regeneration and somatic embryogenesis maintenance in Zoysia japonica. In Vitro Cell. Dev. Biol.-Plant 2009, 45, 673–680. [Google Scholar] [CrossRef]
- Feng, C.P.; Andreasson, E.; Maslak, A.; Mock, H.P.; Mattsson, O.; Mundy, J. Arabidopsis MYB68 in development and responses to environmental cues. Plant Sci. 2004, 167, 1099–1107. [Google Scholar] [CrossRef]
- Liberman, L.M.; Sparks, E.E.; Moreno-Risueno, M.A.; Petricka, J.J.; Benfey, P.N. MYB36 regulates the transition from proliferation to differentiation in the Arabidopsis root. Proc. Natl. Acad. Sci. USA 2015, 112, 12099–12104. [Google Scholar] [CrossRef]
- Lee, M.M.; Schiefelbein, J. WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell 1999, 99, 473–483. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Mitsuda, N.; Yoshizumi, T.; Horii, Y.; Oshima, Y.; Ohme-Takagi, M.; Matsui, M.; Kakimoto, T. Two types of bHLH transcription factor determine the competence of the pericycle for lateral root initiation. Nat. Plants 2021, 7, 633–643. [Google Scholar] [CrossRef] [PubMed]
- Miyashima, S.; Roszak, P.; Sevilem, I.; Toyokura, K.; Blob, B.; Heo, J.O.; Mellor, N.; Help-Rinta-Rahko, H.; Otero, S.; Smet, W.; et al. Mobile PEAR transcription factors integrate positional cues to prime cambial growth. Nature 2019, 565, 490–494. [Google Scholar] [CrossRef]
- Smith, D.L.; Fedoroff, N.V. LRP1, a gene expressed in lateral and adventitious root primordia of Arabidopsis. Plant Cell. 1995, 7, 735–745. [Google Scholar] [CrossRef]
- Kubo, H.; Peeters, A.J.M.; Aarts, M.G.M.; Pereira, A.; Koornneef, M. ANTHOCYANINLESS2, a homeobox gene affecting anthocyanin distribution and root development in Arabidopsis. Plant Cell 1999, 11, 1217–1226. [Google Scholar] [CrossRef]
- Takahashi, N.; Kajihara, T.; Okamura, C.; Kim, Y.; Katagiri, Y.; Okushima, Y.; Matsunaga, S.; Hwang, I.; Umeda, M. Cytokinins control endocycle onset by promoting the expression of an APC/C activator in Arabidopsis roots. Curr. Biol. 2013, 23, 1812–1817. [Google Scholar] [CrossRef]
- Cai, X.T.; Xu, P.; Zhao, P.X.; Liu, R.; Yu, L.H.; Xiang, C.B. Arabidopsis ERF109 mediates cross-talk between jasmonic acid and auxin biosynthesis during lateral root formation. Nat. Commun. 2014, 5, 5833. [Google Scholar] [CrossRef]
- Ye, L.L.; Wang, X.; Lyu, M.; Siligato, R.; Eswaran, G.; Vainio, L.; Blomster, T.; Zhang, J.; Mahonen, A.P. Cytokinins initiate secondary growth in the Arabidopsis root through a set of LBD genes. Curr. Biol. 2021, 31, 3365–3373.e7. [Google Scholar] [CrossRef] [PubMed]
- Devaiah, B.N.; Karthikeyan, A.S.; Raghothama, K.G. WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol. 2007, 143, 1789–1801. [Google Scholar] [CrossRef]
- Raines, T.; Shanks, C.; Cheng, C.Y.; McPherson, D.; Argueso, C.T.; Kim, H.J.; Franco-Zorrilla, J.M.; Lopez-Vidriero, I.; Solano, R.; Vankova, R.; et al. The cytokinin response factors modulate root and shoot growth and promote leaf senescence in Arabidopsis. Plant J. 2016, 85, 134–147. [Google Scholar] [CrossRef]
- Du, Y.; Scheres, B. PLETHORA transcription factors orchestrate de novo organ patterning during Arabidopsis lateral root outgrowth. Proc. Natl. Acad. Sci. USA 2017, 114, 11709–11714. [Google Scholar] [CrossRef] [PubMed]
- Sorin, C.; Bussell, J.D.; Camus, I.; Ljung, K.; Kowalczyk, M.; Geiss, G.; McKhann, H.; Garcion, C.; Vaucheret, H.; Sandberg, G.; et al. Auxin and light control of adventitious rooting in Arabidopsis require ARGONAUTE1. Plant Cell 2005, 17, 1343–1359. [Google Scholar] [CrossRef] [PubMed]
- Ke, M.Y.; Ma, Z.M.; Wang, D.Y.; Sun, Y.B.; Wen, C.J.; Huang, D.Q.; Chen, Z.C.; Yang, L.; Tan, S.T.; Li, R.X.; et al. Salicylic acid regulates PIN2 auxin transporter hyperclustering and root gravitropic growth via Remorin-depende-nt lipid nanodomain organisation in Arabidopsis thaliana. New Phytol. 2021, 229, 963–978. [Google Scholar] [CrossRef]
- Kollmer, I.; Werner, T.; Schmulling, T. Ectopic expression of different cytokinin-regulated transcription factor genes of Arabidopsis thaliana alters plant growth and development. J. Plant Physiol. 2011, 168, 1320–1327. [Google Scholar] [CrossRef]
- Smit, M.E.; McGregor, S.R.; Sun, H.; Gough, C.; Bagman, A.M.; Soyars, C.L.; Kroon, J.H.; Gaudinier, A.; Williams, C.J.; Yang, X.Y.; et al. A PXY-mediated transcriptional network integrates signaling mechanisms to control vascular development in Arabidopsis. Plant Cell 2020, 32, 319–335. [Google Scholar] [CrossRef]
- Wu, R.; Citovsky, V. Adaptor proteins GIR1 and GIR2. II. Interaction with the co-repressor TOPLESS and promotion of histone deacetylation of target chromatin. Biochem. Biophys. Res. Commun. 2017, 488, 609–613. [Google Scholar] [CrossRef] [PubMed]
- Johnson, L.M.; Bostick, M.; Zhang, X.; Kraft, E.; Henderson, I.; Callis, J.; Jacobsen, S.E. The SRA methyl-cytosine-binding domain links DNA and histone methylation. Curr. Biol. 2007, 17, 379–384. [Google Scholar] [CrossRef]
- Panda, K.; McCue, A.D.; Slotkin, R.K. Arabidopsis RNA polymerase IV generates 21–22 nucleotide small RNAs that can participate in RNA-directed DNA methylation and may regulate genes. Philos. Trans. R. Soc. B 2020, 375, 20190417. [Google Scholar] [CrossRef]
- Laparra, H.; Bronner, R.; Hahne, G. Amyloplasts as a possible indicator of morphogenic potential in sunflower protoplasts. Plant Sci. 1997, 122, 183–192. [Google Scholar] [CrossRef]
- Efferth, T. Biotechnology applications of plant callus cultures. Engineering 2019, 5, 50–59. [Google Scholar] [CrossRef]
- Al Ghasheem, N.; Stanica, F.; Peticila, A.G. Overview of studies on in vitro propagation of peach. Acta Hortic. 2021, 1304, 147–154. [Google Scholar] [CrossRef]
- Michel, Z.; Hilaire, K.T.; Mongomake, K.; Georges, A.N.; Justin, K.Y. Effect of genotype, explants, growth regulators and sugars on callus induction in cotton (Gossypium hirsutum L.). Aust. J. Crop Sci. 2008, 2, 1–9. [Google Scholar]
- Gandonou, C.H.; Errabii, T.; Abrini, J.; Idaomar, M.; Chibi, F.; Senhaji, S. Effect of genotype on callus induction and plant regeneration from leaf explants of sugarcane (Saccharum sp.). Afr. J. Biotechnol. 2005, 4, 1250–1255. [Google Scholar]
- Ozyigit, I.I.; Gozukirmizi, N.; Semiz, B.D. Genotype dependent callus induction and shoot regeneration in sunflower (Helianthus annuus L.). Afr. J. Biotechnol. 2007, 6, 1498–1502. [Google Scholar]
- Nguyen, T.H.N.; Tanzer, S.; Rudeck, J.; Winkelmann, T.; Debener, T. Genetic analysis of adventitious root formation in vivo and in vitro in a diversity panel of roses. Sci. Hortic. 2020, 266, 109277. [Google Scholar] [CrossRef]
- Mohebodini, M.; Mokhtar, J.J.; Mahboudi, F.; Alizadeh, H. Effects of genotype, explant age and growth regulators on callus induction and direct shoot regeneration of Let-tuce (Lactuca sativa L.). Aust. J. Crop Sci. 2011, 5, 92–95. [Google Scholar]
- Sugimoto, K.; Jiao, Y.; Meyerowitz, E.M. Arabidopsis regeneration from multiple tissues occurs via a root development pathway. Dev. Cell 2010, 18, 463–471. [Google Scholar] [CrossRef]
- Ikeuchi, M.; Sugimoto, K.; Iwase, A. Plant callus: Mechanisms of induction and repression. Plant Cell 2013, 25, 3159–3173. [Google Scholar] [CrossRef]
- Fan, M.Z.; Xu, C.Y.; Xu, K.; Hu, Y.X. LATERAL ORGAN BOUNDARIES DOMAIN transcription factors direct callus formation in Arabidopsis regeneration. Cell Res. 2012, 22, 1169–1180. [Google Scholar] [CrossRef]
- Ariel, F.D.; Diet, A.; Crespi, M.; Chan, R.L. The LOB-like transcription factor MtLBD1 controls Medicago truncatula root architecture under salt stress. Plant Signal. Behav. 2010, 5, 1666–1668. [Google Scholar] [CrossRef]
- Yordanov, Y.S.; Regan, S.; Busov, V. Members of the LATERAL ORGAN BOUNDARIES DOMAIN transcription factor family are involved in the regulation of secondary growth in Populus. Plant Cell 2010, 22, 3662–3677. [Google Scholar] [CrossRef]
- Xu, C.Y.; Cao, H.F.; Zhang, Q.Q.; Wang, H.Z.; Xin, W.; Xu, E.J.; Zahng, S.Q.; Yu, D.X.; Hu, Y.X. Control of auxin-induced callus formation by bZIP59-LBD complex in Arabidopsis regeneration. Nat. Plants 2018, 4, 108–115. [Google Scholar] [CrossRef]
- Carciofi, M.; Blennow, A.; Nielsen, M.M.; Holm, P.B.; Hebelstrup, K.H. Barley callus: A model system for bioengineering of starch in cereals. Plant Methods 2012, 8, 36. [Google Scholar] [CrossRef] [PubMed]
- Czernicka, M.; Chlosta, I.; Keska, K.; Kozieradzka-Kiszkurno, M.; Abdullah, M.; Popielarska-Konieczna, M. Protuberances are organized distinct regions of long-term callus: Histological and transcriptomic analyses in kiwifruit. Plant Cell Rep. 2021, 40, 637–665. [Google Scholar] [CrossRef] [PubMed]
- Feng, M.Q.; Lu, M.D.; Long, J.M.; Yin, Z.P.; Jiang, N.; Wang, P.B.; Liu, Y.; Guo, W.W.; Wu, X.M. miR156 regulates somatic embryogenesis by modulating starch accumulation in citrus. J. Exp. Bot. 2022, 73, 6170–6185. [Google Scholar] [CrossRef] [PubMed]
- Osterc, G.; Trobec, M.; Usenik, V.; Solar, A.; Stampar, F. Changes in polyphenols in leafy cuttings during the root initiation phase regarding various cutting types at Castanea. Phyton-Ann. Rei Bot. 2004, 44, 109–119. [Google Scholar]
- Trobec, M.; Stampar, F.; Veberic, R.; Osterc, G. Fluctuations of different endogenous phenolic compounds and cinnamic acid in the first days of the rooting process of cherry rootstock ‘GiSelA 5′leafy cuttings. J. Plant Physiol. 2005, 162, 589–597. [Google Scholar] [CrossRef] [PubMed]
- Denaxa, N.K.; Roussos, P.A.; Vemmos, S.N. Assigning a role to the endogenous phenolic compounds on adventitious root formation of olive stem cuttings. J. Plant Growth Regul. 2020, 39, 411–421. [Google Scholar] [CrossRef]
- Curir, P.; Sulis, S.; Mariani, F.; Vansumere, C.F.; Marchesini, A.; Dolci, M. Influence of endogenous phenols on rootability of chamaelaucium-uncinatum schauer stem cuttings. Sci. Hortic. 1993, 55, 303–314. [Google Scholar] [CrossRef]
- Pfeiffer, P.; Hegedus, A. Review of the molecular genetics of flavonoid biosynthesis in fruits. Acta Aliment. 2011, 40, 150–163. [Google Scholar] [CrossRef]
- Bradai, F.; Pliego-Alfaro, F.; Sanchez-Romero, C. Long-term somatic embryogenesis in olive (Olea europaea L.): Influence on regeneration capability and quality of regenerated plants. Sci. Hortic. 2016, 199, 23–31. [Google Scholar] [CrossRef]
- Pila Quinga, L.A.; Pacheco de Freitas Fraga, H.; do Nascimento Vieira, L.; Guerra, M.P. Epigenetics of long-term somatic embryogenesis in Theobroma cacao L.: DNA methylation and recovery of embryogenic potential. Plant Cell Tissue Organ Cult. 2017, 131, 295–305. [Google Scholar] [CrossRef]
- Peng, X.; Zhang, T.T.; Zhang, J. Effect of subculture times on genetic fidelity, endogenous hormone level and pharmaceutical potential of Tetrastigma hemsleyanum callus. Plant Cell Tissue Organ Cult. 2015, 122, 67–77. [Google Scholar] [CrossRef]
- Konar, S.; Adhikari, S.; Karmakar, J.; Ray, A.; Bandyopadhyay, T.K. Evaluation of subculture ages on organogenic response from root callus and SPAR based genetic fidelit-y assessment in the regenerants of Hibiscus sabdariffa L. Ind. Crops Prod. 2019, 135, 321–329. [Google Scholar] [CrossRef]
- Pi, L.; Aichinger, E.; Graaff, E.; Llavata-Peris, C.I.; Weijers, D.; Hennig, L.; Groot, E.; Laux, T. Organizer-derived WOX5 signal maintains root columella stem cells through chromatin-mediated repression of CDF4 expression. Dev. Cell 2015, 33, 576–588. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, L.; Liu, J.; Liao, L.; Gao, A.; Njuguna, B.N.; Zhao, C.; Zheng, B.; Han, Y. Callus Induction and Adventitious Root Regeneration of Cotyledon Explants in Peach Trees. Horticulturae 2023, 9, 850. https://doi.org/10.3390/horticulturae9080850
Gao L, Liu J, Liao L, Gao A, Njuguna BN, Zhao C, Zheng B, Han Y. Callus Induction and Adventitious Root Regeneration of Cotyledon Explants in Peach Trees. Horticulturae. 2023; 9(8):850. https://doi.org/10.3390/horticulturae9080850
Chicago/Turabian StyleGao, Lingling, Jingjing Liu, Liao Liao, Anqi Gao, Beatrice Nyambura Njuguna, Caiping Zhao, Beibei Zheng, and Yuepeng Han. 2023. "Callus Induction and Adventitious Root Regeneration of Cotyledon Explants in Peach Trees" Horticulturae 9, no. 8: 850. https://doi.org/10.3390/horticulturae9080850
APA StyleGao, L., Liu, J., Liao, L., Gao, A., Njuguna, B. N., Zhao, C., Zheng, B., & Han, Y. (2023). Callus Induction and Adventitious Root Regeneration of Cotyledon Explants in Peach Trees. Horticulturae, 9(8), 850. https://doi.org/10.3390/horticulturae9080850








